Abstract
Lipases play key roles in nearly all cells and organisms. Potent and selective inhibitors help to elucidate their physiological functions and associated metabolic pathways. Organophosphorus (OP) compounds are best known for their anticholinesterase properties but selectivity for lipases and other targets can also be achieved through structural optimization. This review considers several lipid systems in brain modulated by highly OP-sensitive lipases. Neuropathy target esterase (NTE) hydrolyzes lysophosphatidylcholine as a preferred substrate. Gene deletion of NTE in mice is embryo lethal and the heterozygotes are hyperactive. NTE is very sensitive in vitro and in vivo to direct-acting OP delayed neurotoxicants and the related NTE-R is also inhibited in vivo. KIAA1363 hydrolyzes acetyl monoalkylglycerol ether of the platelet-activating factor de novo biosynthetic pathway and is a marker of cancer cell invasiveness. It is also a detoxifying enzyme that hydrolyzes chlorpyrifos oxon (CPO) and some other potent insecticide metabolites. Monoacylglycerol lipase and fatty acid amide hydrolase regulate endocannabinoid levels with roles in motility, pain and memory. Inhibition of these enzymes in mice by OPs, such as isopropyl dodecylfluorophosphonate (IDFP), leads to dramatic elevation of brain endocannabinoids and distinct cannabinoid-dependent behavior. Hormone-sensitive lipase that hydrolyzes cholesteryl esters and diacylglycerols is a newly-recognized in vivo CPO- and IDFP-target in brain. The OP chemotype can therefore be used in proteomic and metabolomic studies to further elucidate the biological function and toxicological significance of lipases in lipid metabolism. Only the first steps have been taken to achieve appropriate selective action for OP therapeutic agents.
Keywords: acetylcholinesterase, brain, chemical warfare agent, fatty acid amide hydrolase, hormone-sensitive lipase, insecticide, KIAA1363, monoacylglycerol lipase, neuropathy target esterase, organophosphorus, serine hydrolase
1. Introduction
1.1 OP-Sensitive Lipases
The serine hydrolase superfamily is one of the largest and most diverse enzyme classes in mammalian proteomes with key roles in all cells and organisms. The lipase subfamily hydrolyzes a myriad of lipid substrates important in cellular structure and as signaling molecules. Disruption of lipid homeostasis is implicated in neurodegeneration [1], cancer [2], obesity [3], and atherosclerosis [4]. Despite their physiological, toxicological and clinical significance, there are gaps in our knowledge of lipases and the lipids they regulate. The human genome encodes more than 1000 serine hydrolases, including lipases, most of which are unannotated or poorly characterized in terms of their physiological functions [5].
1.2 OP Chemotype
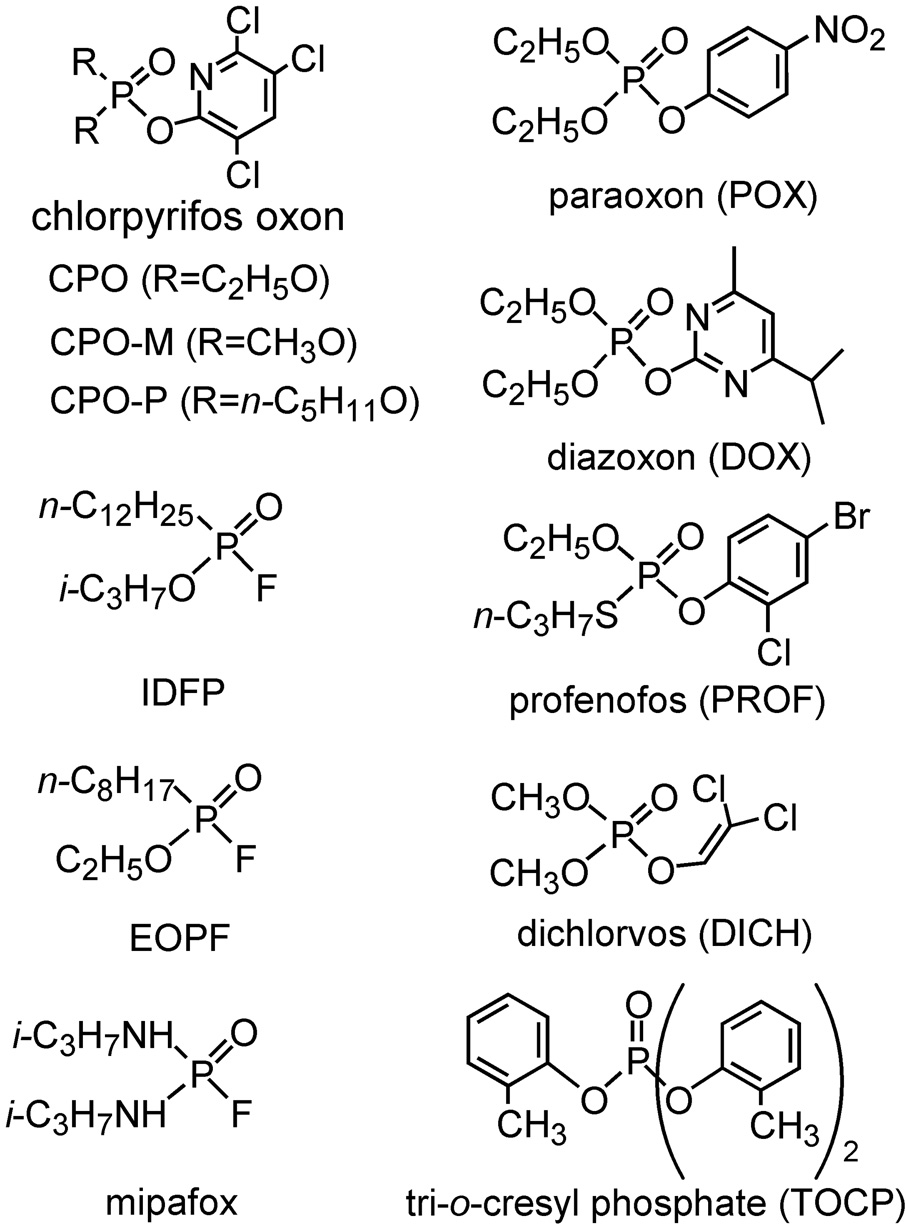
The organophosphorus (OP) chemotype (Fig. 1) contains important insecticides and chemical warfare agents all of which act as acetylcholinesterase (AChE) inhibitors [6]. OP research for the past seven decades emphasized AChE inhibition and disruptions in the cholinergic system on acute or chronic exposure. During this time, several billion pounds of OP pesticides were used to protect crops and livestock from pest insects. Hundreds of thousands of OPs were screened for potency and selective toxicity to develop the 100 or so currently-used insecticides. OPs account for about 25% of the insecticide world market value and there are also important OP herbicides and fungicides. The OP chemical scaffold allows the study of all serine hydrolases including lipases due to their unique reactivity with the active site serine [7]. Structurally-diverse OP inhibitors optimized for potency and selectivity can be used to study the physiological, toxicological and therapeutic relevance of lipases and their associated systems.
Fig. 1.
Structures of OP insecticides and analogs. CPO, POX and DOX are metabolites of the important insecticides chlorpyrifos (CPF), parathion (PT) and diazinon, respectively. Profenofos and dichlorvos are insecticides while most of the others are compounds designed to achieve potency or selectivity. TOCP, mipafox, EOPF and IDFP are delayed neurotoxicants.
1.3 Functional Proteomics and Metabolomics
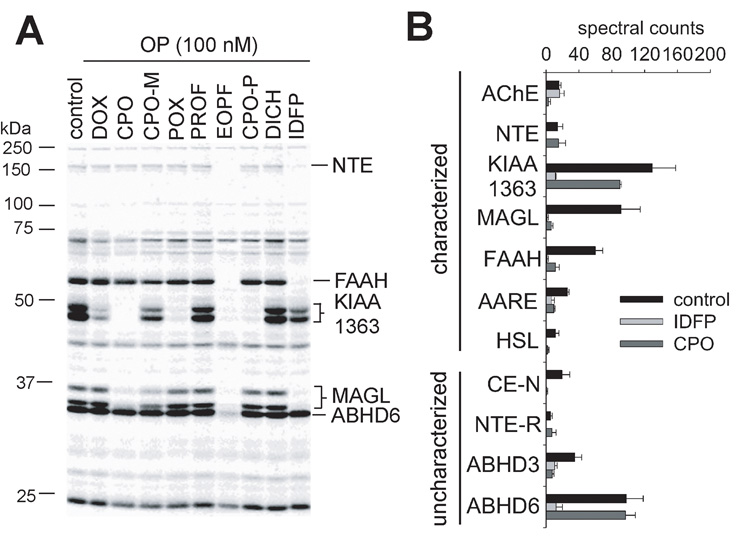
OPs have been extensively used to profile serine hydrolases through functional proteomic and metabolomic platforms. Activity-based protein profiling (ABPP) [7–9] examines their functional state using a fluorophosphonate (FP) reactive group conjugated to a spacer arm and analytical handle (e.g. rhodamine or biotin). ABPP in a competitive mode helps discover potent and selective probes and elucidate toxicological secondary targets. Gel-based ABPP with FP-rhodamine is illustrated in Fig. 2A for several OP insecticides (i.e. DOX, CPO, CPO-M, POX, PROF and DICH) and designer compounds [ethyl octylphosphonofluoridate (EOPF) and isopropyl dodecylfluorophosphonate (IDFP)] as in vitro inhibitors of the mouse brain proteome. Each serine hydrolase has a different OP inhibition profile. Neuropathy target esterase (NTE) and fatty acid amide hydrolase (FAAH) are most sensitive to EOPF and IDFP; KIAA1363 to DOX, CPO, POX, EOPF and CPO-P; monoacylglycerol lipase (MAGL) to CPO, CPO-M, EOPF and IDFP; and ABHD6 to EOPF. ABPP coupled to mass spectrometry (MS) [(Multidimensional Protein Identification Technology (MudPIT)] (ABPP/MudPIT) [9,10] with FP-biotin allows increased resolution and sensitivity and thereby more comprehensive selectivity profiles. For example, assays of mouse brain membranes 4 h after CPO and IDFP are administered ip to mice reveals major in vivo disruptions in the serine hydrolase activity profiles (Fig. 2B). CPO inhibits not only AChE, but also MAGL, FAAH, KIAA1363, and other serine hydrolases considered later. IDFP inhibits NTE, NTE-related esterase (NTE-R), MAGL, FAAH, KIAA1363, and several additional targets. The physiological functions of the new OP-sensitive target enzymes discovered by ABPP can be elucidated with metabolomic liquid chromatography (LC)-MS and gas chromatography (GC)-MS platforms.
Fig. 2.
OP sensitivity of mouse brain serine hydrolases determined by ABPP analysis. A. In vitro inhibition assessed by in-gel ABPP using FP-rhodamine. B. In vivo inhibition 4 h after ip treatment with IDFP (10 mg/kg) and CPO (4 mg/kg) compared with control determined by ABPP-MudPIT using FP-biotin. Spectral counts refer to the total amount of uninhibited enzyme.
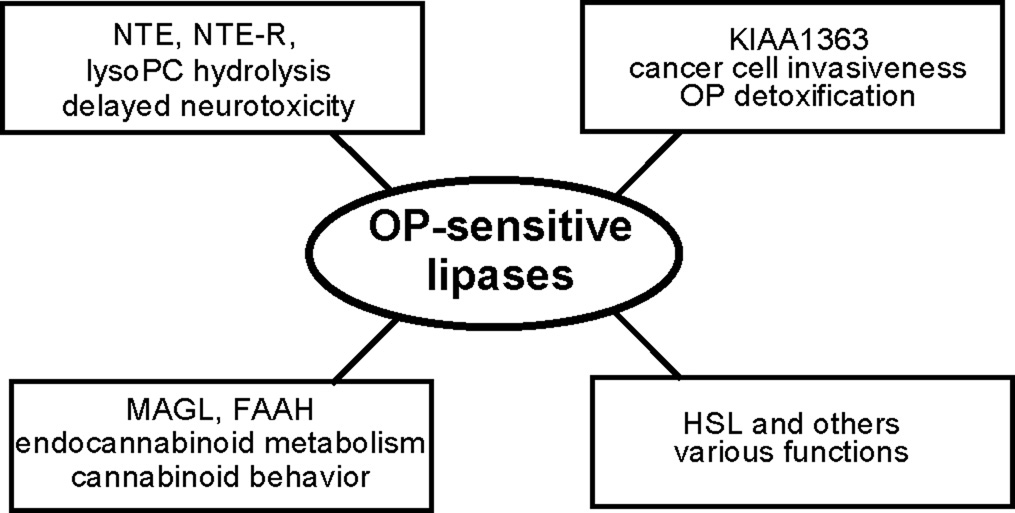
This review considers six OP-sensitive lipases, i.e. NTE, NTE-R, KIAA1363, MAGL, FAAH and hormone-sensitive lipase (HSL) (Fig. 3) (Table 1). The endogenous substrates are known in most cases. Structures of these enzymes are defined by crystallography (FAAH) or deduced from models (except NTE-R). They vary in molecular weight from 37 to 150 kDa (without glycosylation) and in number of amino acids from 339 to 1352. The GXSXG motif is evident (except for FAAH) with variations in the catalytic dyad or triad. Knockout mouse models are developed other than for NTE-R and MAGL. All six lipases are highly OP sensitive in vitro or in vivo prompting detailed analysis of their recognition, structure, function and toxicology or pharmacology.
Fig. 3.
Selected OP-sensitive lipase targets and their functions.
Table 1.
Properties of six OP-sensitive lipasesa
| Property | NTE | NTE-R | KIAA1363 | MAGL | FAAH | HSL |
|---|---|---|---|---|---|---|
| IPI | IPI00128034 | IPI00331610 | IPI00403586 | IPI00132874 | IPI00117176 | IPI00228826 |
| substrate | ||||||
| endogenous | lysoPC | unknown | AcMAGE | monoacylglycerols | acylethanolamides | diacylglycerols, cholesteryl esters |
| assay | phenyl valerate b | phenyl valerate | AcMAGE CPO | 2-AG | AEA | diacylglycerols, cholesteryl esters |
| structure c | model | none | model | Model | crystal | model |
| kDa (mouse) | 147 | 150 | 46 | 37 | 63 | 88 |
| amino acids | 1329 | 1352 | 408 | 339 | 579 | 802 |
| GXSXG motif | GTSIG | GTSIG | GDSAG | GHSMG | GGSIR | GDSAG |
| catalytic | mouse | mouse | mouse | rat | rat | human |
| residues | S966 | S983 | S191 | S122 | S241 | S424 |
| D1086 | D1103 | D348 | D239 | S217 | D693 | |
| H378 | H269 | K142 | H723 | |||
| knockout moused | yes | no | yes | no | yes | yes |
| OP sensitivity | ||||||
| in vitro inhibition (IC50, nM) | ||||||
| CPO | 180 | 8 | 34 | 40 | ||
| EOPF | 0.05 | 52 | 3 | 0.6 | ||
| IDFP | ca.0.2 | 271 | 0.8 | 2 | ||
| POX | >105 | 14 | 2300 | 540 | ||
| in vivo inhibition (%) | ||||||
| CPO | 0 | 0 | 30 | 93 | 81 | 85 |
| 4 mg/kg | ||||||
| IDFP | 100 | 100 | 91 | 99 | 98 | 100 |
| 10 mg/kg | ||||||
Three other OP-sensitive proteins recognized by ABPP are as follows [designation, IPI, kDa (mouse)]: CE-N, IPI00138342, 61; ABHD3, IPI00131173, 46; ABHD6, IPI00321386, 38. They vary in OP sensitivity in vivo (% inhibition, CPO 4 mg/kg and IDFP 10 mg/kg, respectively): CE-N 97, 100; ABHD3, 79, 70; ABHD6, 1, 88.
POX (40 µM) resistant and mipafox-sensitive (50 µM) portion
2. NTE and Lysophospholipids
2.1 Recognition
TOCP added to drinks as a substitute for ginger extract in “ginger jake” in the prohibition-era 1930s induced a delayed neurotoxicity in thousands of people. Recurring incidents with TOCP, used as a cooking oil, high pressure lubricant, and in the manufacture of plastics or hydraulic oils, and a few with insecticides during manufacturing (mipafox) or application led to a total of at least 75,000 victims in the past 75 years. Research in the Medical Research Council Toxicology Unit in England defined the target protein in brain associated with this toxicological problem and designated it NTE; phosphorylation of NTE correlated with delayed neurotoxicity [23]. NTE is usually assayed with brain membranes and a model substrate, phenyl valerate (PV), with specificity achieved by considering only the POX-resistant and mipafox-sensitive portion of the total hydrolysis. NTE is expressed not only in brain, but also in heart, lung, liver, kidney, muscle and lymphocytes, as well as other tissues [24]. NTE-R is highly homologous in sequence to NTE around the GXSXG active site motif [25]. It is highly expressed in adipose, pancreas and prostate and less so in other tissues [24–26] and the C-terminal domain exhibits POX-resistant mipafox-sensitive PV hydrolysis activity [26].
2.2 Structure
NTE is a 147 kDa membrane-bound protein of the patatin-like phospholipase (PNPLA) family, named for their common patatin-like domain [24]. Patatin is a glycoprotein in potatoes with a ser-asp catalytic dyad and defined crystal structure [27]. Other family members include NTE-R, calcium-independent phospholipase A2γ (iPLA2γ), calcium-dependent phospholipase A2 group VI (PLA2G6), and adiponutrin (ADPN) and its family members [24]. The modeled active site of NTE consists of a S966, D1086 catalytic dyad, although D960 is critical for catalysis [11,28]. In addition to the C-terminal patatin-like esterase domain, NTE has three cyclic nucleotide binding domains and is anchored to the cytoplasmic face of the endoplasmic reticulum by an amino-terminal transmembrane segment [16,23]. The C-terminal esterase domain is conserved in bacteria, yeast and nematodes [23]. Orthologs of NTE are found in chicken, mouse, rat, dog and chimpanzee. NTE-R retains the cyclic nucleotide binding domains found in NTE and is membrane-associated [16,25]. NTE-R is 65 and 73 % homologous to NTE at the amino acid level in human and mice [16].
2.3 Function
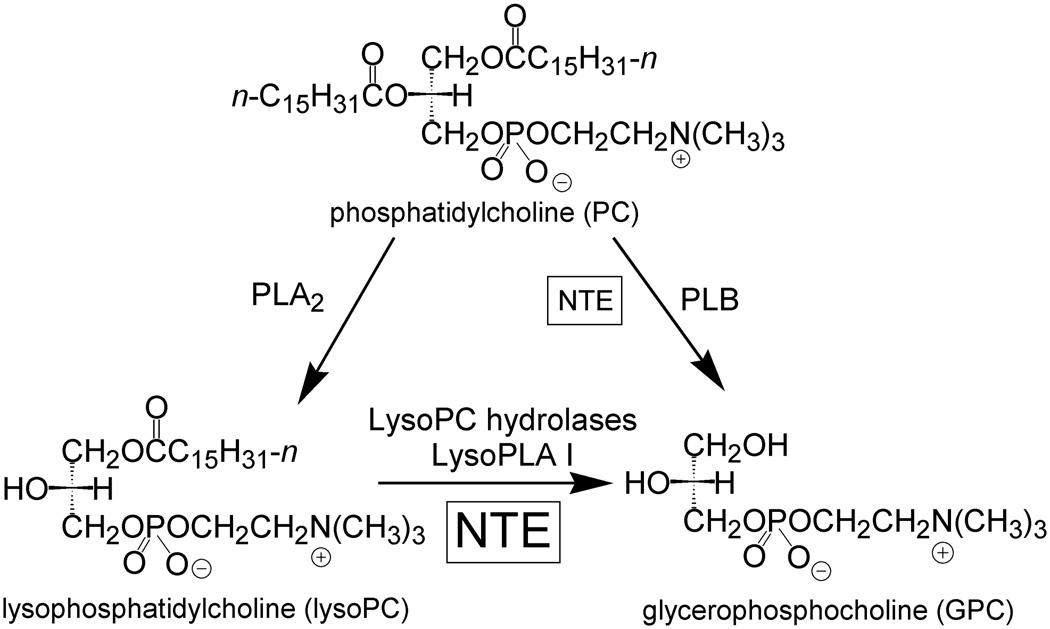
NTE regulates phospholipid metabolism (Fig 4). It accounts for a significant portion of total brain lysophosphatidylcholine (lysoPC) hydrolysis [29] and may also act on phosphatidylcholine (PC) with lower affinity [28]. NTE gene knockout in mice is embryo lethal due to impaired vasculogenesis and the heterozygote is hyperactive [16,17]. NTE is upregulated in lymphocytes from patients with chronic fatigue syndrome [30]. In kidney cells exposed to high sodium chloride levels, NTE catalyzes the production of GPC to maintain osmolyte levels [31]. NTE may be controlled by protein kinase C (PKC) since it is downregulated by PKC activator phorbol 12-myristate 13-acetate in a manner blocked by PKC inhibitor staurosporine [32]. There is a direct interaction between NTE and G protein beta-2 which may regulate NTE activity [33]. Addition of cGMP to brain homogenates does not increase NTE activity, and no direct binding of cAMP to NTE is observed [28, Vose unpublished results]. The function of NTE-R is not defined but is likely to be similar to that of NTE.
Fig. 4.
NTE regulates phospholipid metabolism as a lysoPC hydrolase.
The PNPLA family hydrolyzes a wide variety of lipid substrates. Patatin hydrolyzes acyl lipids [27] and ADPN hydrolyzes triacylglycerols in adipocytes [34]. iPLA2γ selectively hydrolyzes 1-palmitoyl-2-arachidonoyl-sn-glycerophosphocholine to produce 2-arachidonoyl LysoPC, and PLA2G6 hydrolyzes various phosphatidylcholine species as well as platelet activating factor (PAF) [35,36]. Mutations in PLA2G6 are associated with neurodegenerative diseases involving elevated brain iron, such as infantile neuroaxonal dystrophy and Karak syndrome [37].
2.4 OP Toxicology
Neuropathic OPs result in axonal degeneration with a dying-back type appearance [28]. Peripheral paralysis and pathological lesions in humans become apparent between one and three weeks after exposure [28] and are essentially irreversible. The hen is the preferred model for the human OP-induced delayed neuropathy syndrome, with mice displaying different neurotoxic responses to the same compounds [1]. The molecular mechanism leading from NTE inhibition to delayed neuropathy or neurotoxicity is not resolved, although hypotheses include defective axonal transport leading to impaired nutrition of distal axons [23], or localized alterations in lysoPC metabolism or signal transduction pathways [1,29].
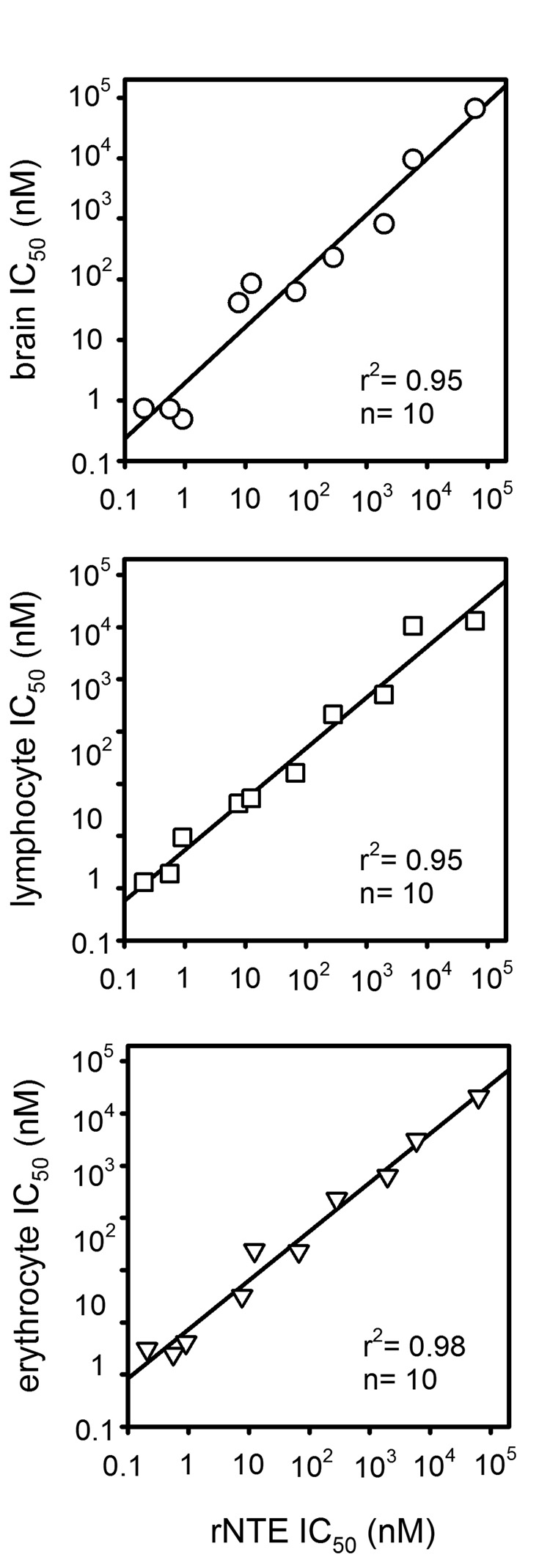
The relationship between NTE and lysophospholipases is of particular interest since they both hydrolyze LysoPC and similar OP inhibitor profiles are obtained with mouse brain NTE and LysoPC hydrolase assays [29] and recombinant NTE (rNTE) (Vose, unpublished results); surprisingly this extends to human erythrocyte and lymphocyte LysoPC hydrolases [38] (Fig. 5). Known lysoPC hydrolases include group IV cytosolic phospholipase A2 [39] and the dedicated lysophospholipase I (LysoPLA I). NTE and LysoPLA I may have a similar function but they are very different in structure. LysoPLA I has the canonical alpha/beta hydrolase fold and a catalytic Ser-Asp-His triad [38]. Because blood LysoPC hydrolase activity is similar to rNTE in OP inhibitor sensitivity and specificity, the blood enzymes were evaluated for possible biomonitoring of exposure to OP pesticides and neurotoxicants using mice as the model. Mice treated with the highest tolerated doses of widely-used OP insecticides showed only moderate reductions in blood LysoPC hydrolase activity, indicating that this is not an appropriate marker for OP pesticide exposure [38]. However, EOPF gives dose-dependent reduction in brain NTE and blood LysoPC hydrolase activities [38]. For this compound, lysoPC hydrolase inhibition in blood is as good as that in brain to predict delayed neurotoxicity [38].
Fig. 5.
Correlation of inhibitor sensitivity and specificity profiles of LysoPC hydrolase activities of rNTE with those of human brain, erythrocytes and lymphocytes. The inhibitors in each case are nine OPs and one carbamate.
3. KIAA1363 and Ether Lipids
3.1 Recognition
KIAA1363 first appeared on a list of unannotated proteins based on the human genome [40]. ABPP revealed high levels of KIAA1363 in human invasive breast, ovarian and melanoma cancer cell lines suggesting involvement in tumor invasiveness [41]. Independently, it was established as the primary brain detoxifying enzyme for CPO [19]. KIAA1363 is expressed in brain, heart, kidney, lung, spinal cord and testes [12].
3.2 Structure
KIAA1363 is an about 50 kDa membrane-bound protein with two glycosylation states. The primary sequence places KIAA1363 in the arylacetamide deacetylase (AADA) class of which there are five members in humans, i.e. AADA, AADA-like 1 (KIAA1363), AADA-like 2, AADA-like 3 and AADA-like 4. Structural analysis of a KIAA1363 model [12] reveals an alpha/beta hydrolase fold, GDSAG motif and S191-D348-H378 catalytic triad. Docking of the two KIAA1363 substrates acetyl monoalkylglycerol ether (AcMAGE) (see below) and CPO into the active site model places the alkyl chain of AcMAGE and the diethyl moieties of CPO in the entry gorge, the acetate carbonyl carbon of AcMAGE and the phosphorus of CPO 2–4 Å from the S191 hydroxyl oxygen, and the acetyl moiety of AcMAGE and the trichloropyridinyl leaving group of CPO deep in the enzyme pocket [42].
3.3 Function
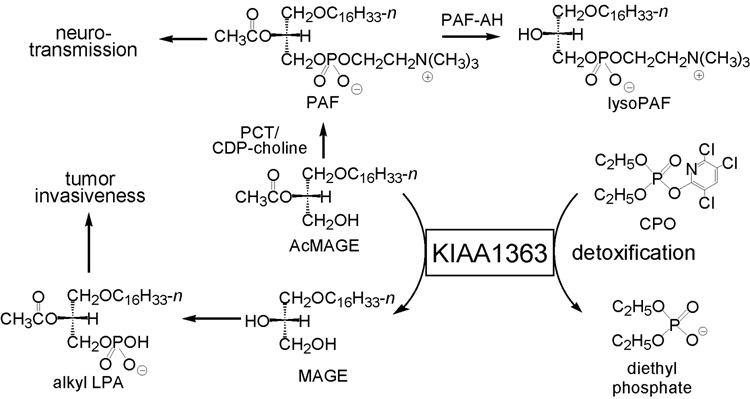
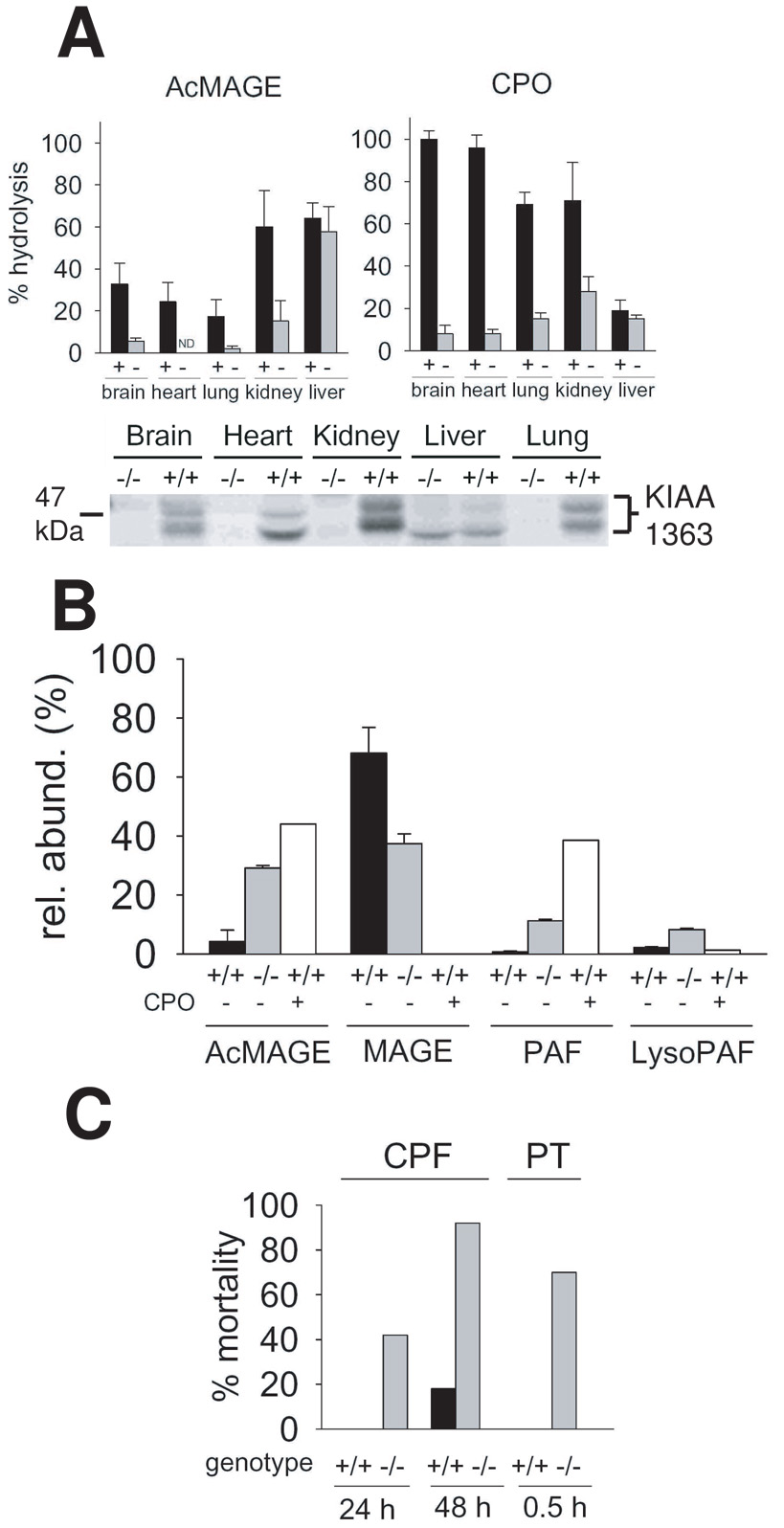
KIAA1363 plays a regulatory role in ether lipid metabolism. In mice and cancer cells, it hydrolyzes AcMAGE to acetate and monoalkylglycerol ether (MAGE) (Fig. 6) [2,42]. A KIAA1363 knockout mouse is completely normal in all outward respects [19]. In comparing knockout and wildtype mice (Fig. 7A) or inhibited versus control cancer cells, KIAA1363 is the primary AcMAGE hydrolase in mouse brain, heart, lung and kidney and in invasive breast and ovarian cancer cell lines. AcMAGE is the penultimate precursor in the de novo biosynthesis pathway for PAF and the product MAGE can be further metabolized to alkyl lysophosphatidic acid (alkyl LPA). Therefore, this enzyme regulates the levels of two highly bioactive ether lipids. In the brain, PAF levels may be restricted by AcMAGE degradation with KIAA1363 in addition to PAF-acetylhydrolase (PAF-AH). In vitro studies with brain membranes show a dramatic increase in PAF and lysoPAF levels and decrease in MAGE levels in −/− compared to +/+ mice upon coincubation with AcMAGE and the cytidine 5’- diphosphocholine (CDP-choline) cofactor for phosphocholine transferase (PCT) with or without CPO. PAF stimulates neurotransmission at moderate levels but is highly neurotoxic at high levels. Consistent with its potential role in regulating brain PAF levels and neurotranmission, KIAA1363 is highly clustered in the hippocampal regions of mouse brain along with the PAF receptor and PAF-AH [42]. CPO inhibits KIAA1363 activity leading to an increase in AcMAGE, PAF and lyso-PAF levels in vitro (Fig. 7B). In tumor cells, RNA knockdown of KIAA1363 leads to a decrease in MAGE and alkyl LPA and an associated decrease in cancer cell migration and growth in a tumor xenograft model [2]. The presumed mechanism of KIAA1363 effect on cancer cells is through alkyl LPA receptors. KIAA1363 is therefore a potential therapeutic target for modulation of PAF action in brain and for invasive tumors.
Fig. 6.
KIAA1363 modulates ether lipid metabolism and OP detoxification.
Fig. 7.
KIAA1363 tissue distribution and role in ether lipid metabolism and OP detoxification determined with +/+ and −/− mice.
A. Tissue distribution for AcMAGE and CPO hydrolysis and FP-rhodamine labeling of membrane proteins
B. Involvement in PAF biosynthetic pathway shown by products from in vitro incubation of mouse brain membranes with 0 or 100 µM CPO (preincubation), 10 µM AcMAGE and 100 µM CDP-choline.
C. CPF and PT are less toxic to +/+ than −/− mice
3.4 OP Toxicology
The principal CPO-hydrolyzing enzyme in mouse brain, heart, lung and kidney is KIAA1363. It is unusual in its ability to rapidly and spontaneously reactivate from diethylphosphorylation of the catalytic Ser compared to most serine hydrolases such as AChE and butyrylcholinesterase that require hours to days. Both AChE and KIAA1363 have similar OP sensitivity profiles for O,O-dimethyl, O,O-diethyl and O,O-dipropyl OP insecticides and analogs. Extensive OP structure-activity studies reveal that KIAA1363 prefers longer alkyl chain substituents compared to AChE. KIAA1363 −/− mice are more sensitive than wildtype to CPF and PT as evidenced by increased mortality (Fig. 7C), consistent with its role in hydrolyzing diethylphosphates. KIAA1363 is known to detoxify CPO and DOX [12,19] and most likely POX [42]. This enzyme serves as a potential structural scaffold for modifications to protect against poisoning by improved OP detoxification [12,19,42].
4. MAGL and FAAH and Endocannabinoids
4.1 Recognition
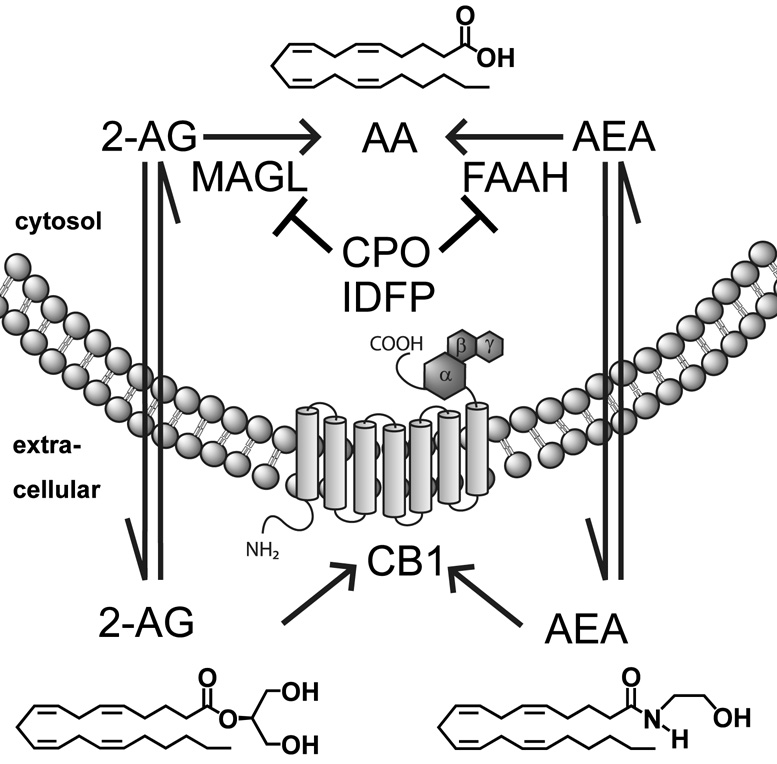
The first steps in defining the cannabinoid system (Fig. 8) involved recognition that tetrahydrocannabinol (THC) was the active component in marijuana and the cloning of the cannabinoid receptor 1 (CB1) responsible for a majority of the psychoactive effects of THC [43]. Two endogenous CB1 ligands, 2-arachidonoyl-sn-glycerol (2-AG) and anandamide (AEA), were then isolated [44, 45] followed soon thereafter by identification of FAAH as the predominant enzyme for hydrolysis of fatty acid amides and AEA [46]. MAGL was first characterized in adipose tissue important for fat mobilization [47] and subsequently in brain involved in 2-AG hydrolysis for endocannabinoid inactivation [48]. Additional endocannabinoids (e.g. arachidonoyldopamine), receptors (e.g. CB2 and GPR55) and metabolizing enzymes discovered since then have allowed a more complete understanding of the systems biology [43].
Fig. 8.
MAGL and FAAH regulate endocannabinoid levels.
4.2 Structure
MAGL is a membrane bound and cytosolic protein with a suggested alpha/beta hydrolase fold, a GXSXG motif, a typical catalytic triad (S122-D239-H269) and essential sulfhydryl groups [13]. A crystal structure of MAGL would greatly aid in discovery of potent and selective inhibitors. FAAH is the only mammalian member of the amidase signature class of the serine hydrolase superfamily [14]. It is a membrane-bound dimeric protein with an unusual S241-S217-K142 catalytic triad. The protein core is a twisted sheet consisting of 11 mixed strands surrounded by 24 α-helices. The putative substrate entry gorge is amphipathic in nature, accommodating the admission and movement of polar fatty acid amide head groups toward the active site. The crystal structure of FAAH phosphorylated with methyl arachidonylfluorophosphonate (MAFP) defines the substrate binding pocket surrounding the arachidonyl chain and S241 covalently bonded to the phosphorus atom.
4.3 Function
2-AG and AEA in brain modulate neurotransmitter release and are synthesized “on demand” and released upon post-synaptic depolarization and calcium mobilization to bind presynaptic CB1, subsequently decreasing the release of either glutamate or γ-aminobutyric acid [43]. This endocannabinoid-mediated retrograde neurotransmission is involved in synaptic plasticity, motility, pain, excitotoxicity, food intake and energy processing [43, 49]. 2-AG and AEA levels are changed under multiple pathological disease states (e.g. pain, multiple sclerosis, Alzheimer’s disease, Parkinson’s disease, epilepsy, diabetes and obesity) as a protective defense mechanism to restore the body to homeostasis [43,49]. MAGL and FAAH are key players in controlling endocannabinoid levels and their basal signaling tone. The phenotypic and metabolic consequences of selective MAGL inhibition are poorly understood due to the lack of a gene-deficient mouse model and selective in vivo inhibitors. On the other hand, the physiological phenotypes associated with FAAH inhibition are well defined through the use of FAAH knockout (−/−) mice and selective inhibitors (e.g. URB597). FAAH −/− mice have a normal appearance but are hypoalgesic, anxiolytic and supersensitive to exogenously administered AEA [50]. FAAH is the sole hydrolase for the N-acylethanolamides [20]. Manipulating endocannabinoid levels through MAGL and FAAH inhibition offers promise for treating neurological disorders [51].
4.4 OP Pharmacology
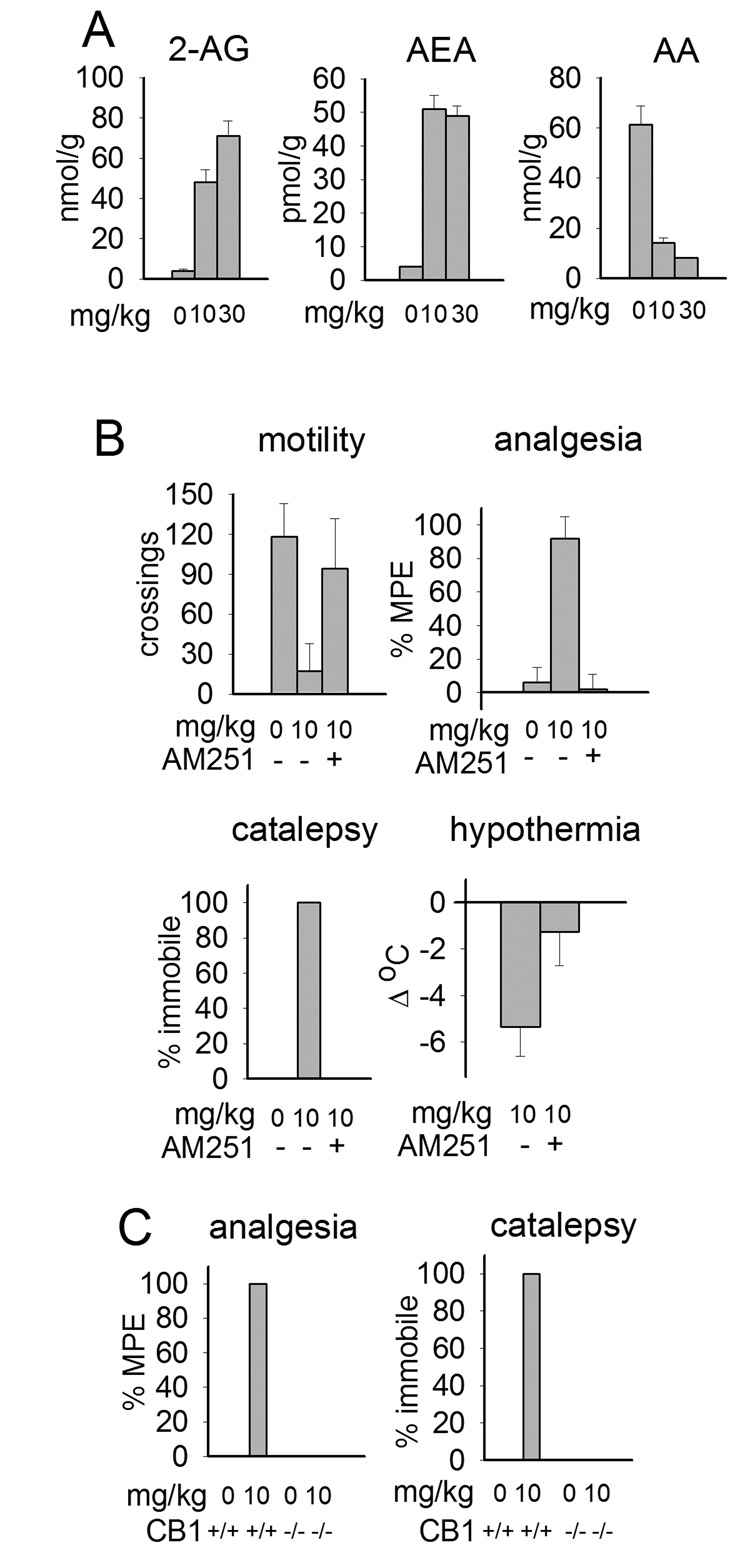
MAGL and FAAH are highly sensitive to many OP compounds as first recognized with MAFP [52,53]. ABPP and substrate hydrolysis assays reveal some OP MAGL and FAAH inhibitors acting at 0.1– 1 nM in vitro and 1-10 mg/kg in vivo, with moderate selectivity versus other serine hydrolase targets [54]. IDFP (10 mg/kg ip) in mice completely blocks brain MAGL and FAAH (Fig. 2), dramatically elevates 2-AG and AEA levels (with an associated reduction in AA) and produces full-blown cannabinoid pharmacology (i.e. hypomotility, analgesia, catalepsy and hypothermia) (Fig. 9) similar to the effects observed with THC. This behavior is averted by CB1 antagonist-pretreatment and is not apparent in CB1 −/− mice. Since FAAH −/− mice are outwardly normal, the overt cannabinoid behavior associated with dual MAGL and FAAH inhibition may be mostly due to the MAGL/2-AG component. Although not shown, the major insecticide CPF and its bioactivated metabolite CPO also give marked endocannabinoid elevation and distinct cannabinoid behavior [54].
Fig. 9.
IDFP-induced activation of the endocannabinoid system 4 h posttreatment.
A. Elevation of 2-AG and AEA levels with corresponding lowering of AA.
B. CB1 antagonist (AM251) blocks IDFP-induced changes in behavior.
C. CB1 knockout mice are protected from IDFP cannabinoid effects.
5. Concluding Remarks
There are many secondary targets for OP insecticides [5,6] in addition to those considered above. ABPP analysis of the brain proteome from CPO- and IDFP-treated mice shows that CPO inhibits AARE, ABHD3, CE-N and HSL while IDFP inhibits all of these targets plus ABHD6 (Fig. 2). AARE is a known OP target and hydrolyzes N-acetyl peptides [6]. CE-N plays a role in lung surfactant transformation but the substrate is not defined [55]. The metabolic roles of ABHD3 and ABHD6 are also unknown. HSL has a modeled S424-D693-H723 catalytic triad and alpha/beta hydrolase fold. The endogenous substrates are diacylglycerols and cholesteryl esters [22]. The brain enzyme is very sensitive in vivo to both IDFP and CPO [54]. A knockout mouse shows diglyceride and cholesteryl ester accumulation in adipocytes, muscle and testes with associated male sterility [21,22]. OP-induced inhibition of HSL may therefore lead to lipid accumulation with adverse effects. Therefore, the OP chemotype can be used to elucidate the biological function of lipases and other serine hydrolases by proteomic or metabolomic studies.
The serine hydrolases are sensitive to a large number and great variety of inhibitors including OPs, carbamates, sulfonyl fluorides and trifluoromethylketones with the OPs generally most potent. OP lipase inhibitors are potential therapeutic agents for cancer invasiveness, obesity, atherosclerosis, pain relief and neuromotor diseases. OP potency, selectivity and suitable pharmacokinetics can potentially be achieved for most lipase targets.
Acknowledgements
Research in this review was supported in part by National Institutes of Health grants ES08762 (J.E.C., D.K.N. and K.F.) and ES09605 (S.C.V.), University of California School of Public Health Center for Children’s Environmental Health Research (S.C.V.) and University of California Toxic Substances Research and Teaching fellowship (D.K.N.). We thank Andreas Zimmer and Carl Lupica of the National Institutes of Health for providing CB1 +/− breeding pairs. We acknowledge major contributions from our University of California at Berkeley colleagues Gary Quistad, Yoffi Segall, Nina Holland and Brenda Eskenazi and Scripps Research Institute at La Jolla, California collaborator Benjamin Cravatt.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Quistad GB, Barlow C, Winrow CJ, Sparks SE, Casida JE. Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7983–7987. doi: 10.1073/pnas.1232473100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Burn P. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat. Rev. Drug Discov. 2004;3:695–710. doi: 10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- 4.Macphee CH, Nelson J, Zalewski A. Role of lipoprotein-associated phospholipase A2 in atherosclerosis and its potential as a therapeutic target. Curr. Opin. Pharmacol. 2006;7:154–161. doi: 10.1016/j.coph.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Casida JE, Quistad GB. Serine hydrolase targets of organophosphorus toxicants. Chem. Biol. Interact. 2005;157–158:277–283. doi: 10.1016/j.cbi.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem. Res. Tox. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 7.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 8.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 9.Barglow KT, Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat. Meth. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J. Am. Chem. Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 11.Wijeyesakere SJ, Richardson RJ, Stuckey JA. Modeling the tertiary structure of the patatin domain of neuropathy target esterase. The Protein J. 2007;26:165–172. doi: 10.1007/s10930-006-9058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura DK, Durkin KA, Chiang KP, Quistad GB, Cravatt BF, Casida JE. Serine hydrolase KIAA1363: toxicological and structural features with emphasis on organophosphate interactions. Chem. Res. Tox. 2006;19:1142–1150. doi: 10.1021/tx060117m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saario SM, Salo OMH, Nevalainen T, Poso A, Laitinen JT, Järvinen T, Niemi R. Characterization of the sulfhydryl-sensitive site in the enzyme responsible for hydrolysis of 2-arachidonoyl-glycerol in rat cerebellar membranes. Chem. Biol. 2005;12:649–656. doi: 10.1016/j.chembiol.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 15.Contreras JA, Karlsson M, Østerlund T, Laurell H, Svensson A, Holm C. Hormone-sensitive lipase is structurally related to acetylcholinesterase, bile salt-stimulated lipase, and several fungal lipases, Building of a three-dimensional model for the catalytic domain of hormone-sensitive lipase. J. Biol. Chem. 1996;271:31426–31430. doi: 10.1074/jbc.271.49.31426. [DOI] [PubMed] [Google Scholar]
- 16.Winrow CJ, Hemming ML, Allen DM, Quistad GB, Casida JE, Barlow C. Loss of neuropathy target esterase in mice links organophosphate exposure to hyperactivity. Nat. Genet. 2003;33:477–485. doi: 10.1038/ng1131. [DOI] [PubMed] [Google Scholar]
- 17.Moser M, Li Y, Vaupel K, Kretzschmar D, Kluge R, Glynn P, Buettner R. Placental failure and impaired vasculogenesis result in embryonic lethality for neuropathy target esterase-deficient mice. Mol. Cell. Biol. 2004;24:1667–1679. doi: 10.1128/MCB.24.4.1667-1679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5075–5080. doi: 10.1073/pnas.0401030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomura DK, Leung D, Chiang KP, Quistad GB, Cravatt BF, Casida JE. A brain detoxifying enzyme for organophosphorus nerve poisons. Proc. Natl. Acad Sci, U.S.A. 2005;102:6195–6200. doi: 10.1073/pnas.0501915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc. Natl. Acad. Sci., U.S.A. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osuga J, Ishibashi S, Oka T, Yagyu H, Tozawa R, Fujiomoto A, Shionoiri F, Yahagi N, Kraemer FB, Tsutsumi O, Yamada N. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci., U.S.A. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haemmerle G, Zimmerman R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J. Biol. Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 23.Glynn P. A mechanism for organophosphate-induced delayed neuropathy. Toxicol. Lett. 2006;162:94–97. doi: 10.1016/j.toxlet.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. Characterization of the human patatin-like phospholipase family. J. Lipid Res. 2006;47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Xie M, Yang D, Matoney L, Yan B. Rat NTE-related esterase is a membrane-associated protein, hydrolyzes phenyl valerate, and interacts with diisopropylfluorophosphate through a serine catalytic machinery. Arch. Biochem. Biophys. 2003;416:137–146. doi: 10.1016/s0003-9861(03)00264-9. [DOI] [PubMed] [Google Scholar]
- 26.Chang P-A, Long D-X, Wu Y-J. Molecular cloning and expression of the C-terminal domain of mouse NTE-related esterase. Mol. Cell Biochem. 2007 doi: 10.1007/s11010-007-9550-2. [DOI] [PubMed] [Google Scholar]
- 27.Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a ser-asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 28.Glynn P. Neuropathy target esterase and phospholipid deacylation. Biochim. Biophys. Acta. 2005;1736:87–93. doi: 10.1016/j.bbalip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Quistad GB, Casida JE. Lysophospholipase inhibition by organophosphorus toxicants. Toxicol. Appl. Pharmacol. 2004;196:319–326. doi: 10.1016/j.taap.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Kaushik N, Fear D, Richards SCM, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DAJ, Holgate ST, Kerr JR. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J. Clin. Pathol. 2005;58:826–832. doi: 10.1136/jcp.2005.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallazzini M, Ferraris JD, Kunin M, Morris RG, Burg MB. Neuropathy target esterase catalyzes osmoprotective renal synthesis of glycerophosphocholine in response to high NaCl. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15260–15265. doi: 10.1073/pnas.0607133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Chang P-A, Long D-X, Yang L, Wu Y-J. Down-regulation of neuropathy target esterase by protein kinase C activation with PMA stimulation. Mol. Cell Biochem. 2007;302:179–185. doi: 10.1007/s11010-007-9439-0. [DOI] [PubMed] [Google Scholar]
- 33.Chen R, Chang P-A, Long D-X, Liu C-Y, Yang L, Wu Y-J. G protein β2 subunit interacts directly with neuropathy target esterase and regulates its activity. Int. J. Biochem. Cell Biol. 2007;39:124–132. doi: 10.1016/j.biocel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 35.Yan W, Jenkins CM, Han X, Mancuso DJ, Sims HF, Yang K, Gross RW. The highly selective production of 2-arachidonoyl lysophosphatidylcholine catalyzed by purified calcium-independent phospholipase A2γ. J. Biol. Chem. 2005;280:26669–26679. doi: 10.1074/jbc.M502358200. [DOI] [PubMed] [Google Scholar]
- 36.Tang J, Kriz RW, Wolfman N, Shaffer M, Seehra J, Jones SS. A novel cytosolic calcium-independent Phospholipase A2 contains eight ankyrin motifs. J. Biol. Chem. 1997;272:8567–8575. doi: 10.1074/jbc.272.13.8567. [DOI] [PubMed] [Google Scholar]
- 37.Morgan NV, Westaway SK, Morton JEV, Gregory A, Gissen P, Sonek S, Cangul H, Coryell J, Canham N, Nardocci N, Zorzi G, Pasha S, Rodriguez D, Desguerre I, Mubaidin A, Bertini E, Trembath RC, Simonati A, Schanen C, Johnson CA, Levinson B, Woods CG, Wilmot B, Kramer P, Gitschier J, Maher ER, Hayflick SJ. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat. Genet. 2006;38:752–754. doi: 10.1038/ng1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vose SC, Holland NT, Eskenazi B, Casida JE. Lysophosphatidylcholine hydrolases of human erythrocytes, lymphocytes and brain: Sensitive targets of conserved specificity for organophosphorus delayed neurotoxicants. Toxicol. Appl. Pharmacol. 2007;224:98–104. doi: 10.1016/j.taap.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macdonald DJ, Boyle RM, Glen ACA, Horrobin DF. Cytosolic phospholipase A2 type IVA is present in human red cells. Blood. 2004;103:3562–3564. doi: 10.1182/blood-2002-09-2698. [DOI] [PubMed] [Google Scholar]
- 40.Nagase T, Kikuno R, Ishikawa K, Hirosawa M, Ohara O. Prediction of the coding sequences of unidentified human genes. XVI. The complete sequences of 150 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:65–73. doi: 10.1093/dnares/7.1.65. [DOI] [PubMed] [Google Scholar]
- 41.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc. Natl. Acad. Sci, U.S.A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomura DK, Fuijioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Dual roles of brain serine hydrolase KIAA1363 in ether lipid metabolism and organophosphate detoxification. Toxicol. Appl. Pharmacol. 2008;228:42–48. doi: 10.1016/j.taap.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Marzo V, Bisogno T, De Petrocellis L. Endocannabinoids and related compounds: walking back and forth between plant natural products and animal physiology. Chem. Biol. 2007;14:741–756. doi: 10.1016/j.chembiol.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Devane WA, Hanuš L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 45.Mechoulam R, Ben-Shabat S, Hanuš L, Ligumsky M, Kaminski NE, Schatz AR, Gopher A, Almog S, Martin BR, Compton DR, Pertwee RG, Griffin G, Bayewitch M, Barg J, Vogel Z. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- 46.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoacylglyceride lipase. J. Biol. Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 48.Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc. Natl. Acad. Sci., U.S.A. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Marzo V, Petrosino S. Endocannabinoids and the regulation of their levels in health and disease. Curr. Opin. Lipid. 2007;18:129–140. doi: 10.1097/MOL.0b013e32803dbdec. [DOI] [PubMed] [Google Scholar]
- 50.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 51.Saario SM, Laitinen JT. Therapeutic potential of endocannabinoid-hydrolysing enzyme inhibitors. Basic Clin. Pharmacol. Toxicol. 2007;101:287–293. doi: 10.1111/j.1742-7843.2007.00130.x. [DOI] [PubMed] [Google Scholar]
- 52.Deutsch DG, Omeir R, Arreaza G, Salchani D, Prestwich GD, Huang Z, Howlett A. Methyl arachidonyl fluorophosphonate: a potent irreversible inhibitor of anandamide amidase. Biochem. Pharmacol. 1997;53:255–260. doi: 10.1016/s0006-2952(96)00830-1. [DOI] [PubMed] [Google Scholar]
- 53.Quistad GB, Klintenberg R, Caboni P, Liang SN, Casida JE. Monoacylglycerol lipase inhibition by organophosphorus compounds leads to elevation of brain 2-arachidonoylglycerol and the associated hypomotility in mice. Toxicol. Appl. Pharmacol. 2006;211:78–83. doi: 10.1016/j.taap.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat. Chem. Biol. 2008 doi: 10.1038/nchembio.86. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross NJ. Extracellular metabolism of pulmonary surfactant: the role of a new serine protease. Annu. Rev. Physiol. 1995;57:135–150. doi: 10.1146/annurev.ph.57.030195.001031. [DOI] [PubMed] [Google Scholar]