Genetic research is increasingly turning to studies of sequence variation in genes encoding proteins of known structure and function. The principal question in these studies is whether sequence variation affects protein structure or function, and, for certain genes, whether sequence variation affects human health. The proliferation of published sequence data and the growth in the number of publications is a boon to this research, but also makes it difficult to keep track of what is known about a gene. The primary sequence databases of the International Nucleic Acid Sequence Data Library (for example, GenBank) provide powerful sequence-similarity search tools that help researchers deduce the functions of newly identified proteins1, 2, 3. However, they do not contain the annotation required to map sequence variation within a single gene, or to correlate such variation with data about the gene's product or the phenotype arising from variation in the gene4, 5.
Sequence variation is particularly relevant to infectious pathogens that mutate in response to antimicrobial therapy. Sequence variations in human immunodeficiency virus (HIV)-1 reverse transcriptase (RT) and protease, the molecular targets of anti-retroviral drug therapy, are prime examples of genes in which sequence variation has both biological and medical implications. Although HIV-1-infected individuals with drug-susceptible HIV-1 isolates experience substantial reductions in morbidity and mortality with appropriate anti-retroviral drug therapy, individuals infected with drug-resistant isolates generally do not respond to drug therapy6.
There are more than 10,000 published HIV-1 RT and protease sequences in GenBank and more than 2,000 additional amino-acid sequences published in biology and medical journals. Genetic analysis of HIV-1 isolates has demonstrated at least 9 different group M (main) subtypes, differing from one another by 10-30%, and several highly divergent group O and group N outlier sequences7. Sequences of HIV-1 isolates collected from around the world provide evidence of naturally occurring genetic polymorphisms; sequences of isolates from patients receiving anti-HIV drug therapy indicate genetic changes selected under anti-retroviral drug pressure that may be associated with drug resistance. The variety and variability of these changes mean that no single researcher can keep track of the consequences of even the more common changes. The combination of this complexity and the lack of annotation in public databases means that most of these published data on repetitively sequenced genes are essentially lost to the research community.
Secondary databases with curated sequences are required to explicate the experimental results stored in the large public primary sequence databases. For example, a publicly available database at the Los Alamos National Laboratory provides HIV sequence annotation and analysis, and catalogs HIV sequence variation throughout the viral genome, particularly in untreated HIV-1-infected individuals (http://hiv-web.lanl.gov)7. To allow researchers to analyze new HIV-1 RT and protease sequences arising in patients receiving anti-retroviral drug therapy, we have developed an online search program coupled to an extensive database of HIV-1 RT and protease mutations. The program, called HIV-SEQ (for ‘HIV mutation search engine for queries’) compares an HIV-1 sequence submitted by the researcher to a consensus reference sequence. Differences between the sequences are used as query parameters for ‘interrogating’ the database8, which links HIV-1 RT and protease sequence variation to the anti-retroviral drug treatment histories of patients from whom sequenced virus isolates were obtained. The program is available at http://hivdb.stanford.edu/hiv/programs.htm.
Discussion
HIV-SEQ demonstrates that the body of published sequence data on a gene can be made available in real time to researchers sequencing new isolates of that gene. The ability to examine new sequences in the context of published sequence data has two main advantages. Unusual sequence results can be detected, allowing the person sequencing the gene to recheck the primary sequence output (for example, electropherogram) while it is on his/her desktop16. Moreover, unexpected associations between sequences or isolates can be discovered, because the person analyzing the sequence can immediately retrieve data on isolates sharing one or more mutations with the new sequence.
Although HIV-SEQ provides a brief description of experimental data on more than 60 protease and RT mutations, the program's strength lies in its use of annotated sequence data on isolates from individuals receiving known anti-retroviral drug treatments. This is an advantage because several of the mutations that have been characterized in vitro rarely arise in vivo, and many of the mutations that occur commonly in vivo have not yet been characterized in vitro. Sequences of HIV-1 isolates from patients receiving anti-retroviral drug therapy are observations of HIV-1 evolution that show the genetic variants selected under anti-retroviral drug pressure in vivo. These sequence data are a necessary complement to in vitro experimental data.
Assays for sequencing HIV-1 RT and protease are commercially available and are widely used in clinical settings. However, the optimal means of interpreting these sequences is not known. HIV-SEQ identifies mutations and helps in their interpretation, but does not have the logical or heuristic power to provide a complete sequence interpretation. Retrieving drug susceptibility data on HIV-1 isolates with mutation patterns similar to the submitted sequence is a logical next step for future versions of the program. Such a program has been described, but both the program and the underlying database are proprietary17.
In conclusion, we foresee the development of mutation search programs similar to HIV-SEQ for other frequently sequenced polymorphic genes of biomedical importance. The prerequisites for creating such programs include a reference sequence for the gene of interest, a system for classifying the source of a sequence, an accepted ontology for correlating sequences with extra-sequence features and an annotated database. The development of public databases and their associated applications will encourage researchers to adopt standard approaches for reporting biological data and, in doing so, new findings will be more effectively subjected to peer review and to subsequent analyses and use by others.
Methods
User interface
Users submit single sequences or sets of multiple sequences encompassing HIV-1 protease and/or RT by typing nucleotides into a text box or by uploading a nucleotide text file. The program output includes the amino-acid translation; a list of amino-acid differences (called mutations here) between the submitted sequence and the HIV-1 subtype B consensus sequence7; a list of problematic positions indicative of poor sequence quality; a comparison of the submitted sequence to a set of reference sequences of known subtype9; data on the frequency with which each mutation occurs in individuals according to HIV-1 subtype and type of drug therapy; background data on potential drug-resistance mutations; and hyperlinks to the Medline references and GenBank entries associated with published sequences. Submitted sequences are not added to the database or stored on the server.
Sequence translation and alignment
The program identifies the submitted sequence's correct reading frame by determining which reading frame contains conserved amino-acid motifs (Table 1). The presence of motifs in more than one reading frame indicates a reading frame shift. When this occurs, the program locates, records and removes such shifts by applying an optimal sequence alignment algorithm10 to the region between the closest motifs in different reading frames. Although reading frame shifts in HIV-1 RT and protease have not been reported, they may conceivably result from defective virions or sequencing errors.
Table 1.
Amino-acid motifs used to align HIV-1 protease and reverse transcriptase sequences
| Motifa | Positions | Functionb | % Conserved in HIV-1c | % Conserved in HIV-2 |
|---|---|---|---|---|
| Protease | ||||
| PQ(I|V)T | 1-4 | Dimer interface | >99 | 0 |
| DTG | 25-27 | Active site | 100 | 100 |
| GCTLN | 94-98 | Dimer interface | >99 | 0 |
| RT | ||||
| PISP | 1-4 | Part of protease/RT cleavage site | >99 | 0 |
| QWPL(T|S) | 23-27 | β1-αA turn | >99 | 100 |
| D(V|I)GDA | 110-114 | Active site | >99 | 100 |
| QY(.)DD(L|I) | 182-187 | Active site | >99 | 100 |
| WMG(Y|F)) | 229-232 | β12-α13 turn | >99 | 100 |
I|V, “I” means that either I or V is acceptable at that position; (.), any amino acid is acceptable at the position.
DTG constitutes the active site of the protease. The three catalytic aspartates (D) at codons 110, 185 and 186 constitute the active site of the RT.
Each motif not only is highly conserved among HIV-1 RT and protease, but also is not found at any other position within these two genes.
The program then uses the position of conserved amino-acid motifs within the submitted sequence to infer the starting positions of the protease and RT. Amino-acid insertions or deletions will cause the distances between conserved motifs to be different from the inter-motif distances in the consensus reference sequence. When an amino-acid insertion or deletion is detected, the program locates, records and removes it using an optimal sequence alignment algorithm. RT insertions and deletions have been reported in 1-2% of isolates from individuals who have received prolonged treatment with multiple nucleoside RT inhibitors. The occurrence of reading frame shifts and amino-acid insertions or deletions is reported at the beginning of the program's output.
Representation of amino acid mixtures
Because of its high mutation rate, HIV-1 exists within an individual as a mixture of genetically distinguishable variants. Sequencing is often done using uncloned, PCR-amplified cDNA (population-based sequencing), and therefore nucleotide mixtures are often detected in clinical plasma virus samples. Such mixtures are represented using the nucleotide ambiguity code (for example, M = A/C, R = A/G and W = A/T). HIV-SEQ translates nucleotide triplets containing ambiguities into each of the possible amino acids they encode. For example, the nucleotide triplet WMC is translated to NTYS (N for AAC, T for ACC, Y for TAC and S for TCC).
Problematic positions
The program output includes a list of problematic positions in each sequence. These positions include those coding for stop codons, those containing mutations at highly conserved sites and those containing highly ambiguous nucleotides: N (A/C/G/T), B (C/G/T), D (A/G/T), H (A/C/T) and V (A/C/G). Sequence quality is likely to be inversely correlated with the number of these problematic positions.
Mutation frequency data
Each sample mutation is used as a query parameter to ‘interrogate’ the HIV RT and Protease Sequence Database (Fig. 1). Within the database, mutation frequency tables contain data on the frequency with which each mutation occurs in different categories of HIV-1 isolates. For protease isolates, these categories include subtype B protease isolates from ‘untreated’ individuals (those who have not received a protease inhibitor, but may have received an RT inhibitor); non-subtype B protease isolates from untreated individuals; isolates from individuals who have received at least one protease inhibitor; and isolates from individuals who have received at least three protease inhibitors.
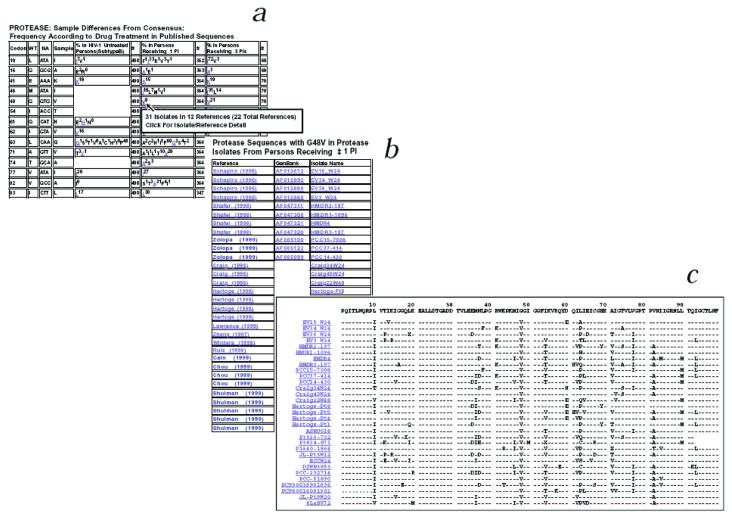
Fig. 1.
One analysis returned by HIV-SEQ. a, List of HIV-1 protease amino acids in a submitted sequence that differ from the subtype B consensus reference sequence (columns 1-4). Right (columns 5-10), tables containing the percent occurrence (the superscript number) of each mutation (difference from consensus) in protease isolates from untreated individuals, protease isolates from individuals who have received one protease inhibitor or more, and protease isolates from individuals who have received three protease inhibitors or more. (Additional sections of the output contain similar data for the RT gene, as well as frequency data for each mutation in non-subtype B isolates). By moving the cursor onto a mutation, users are shown a ‘tool-tip’ with the number of individuals and the number of references in which the selected mutation has been reported. b and c, By clicking on a mutation, users are shown a second page that includes links to the appropriate entries in Medline, GenBank and the HIV RT and Protease Sequence Database for each of the reported isolates. At the top of this second page, users are given the option of retrieving the sequences in an alignment format (c) or in plain text format.
For RT isolates, the categories include subtype B RT isolates from ‘untreated’ individuals (those who have not received an RT inhibitor); non-subtype B isolates from untreated individuals; isolates from individuals who have received at least one nucleoside RT inhibitor, but have not received a non-nucleoside RT inhibitor; isolates from individuals who have received at least four nucleoside RT inhibitors, but have not received a non-nucleoside RT inhibitor; and isolates from individuals who have received a non-nucleoside RT inhibitor. The numbers of individuals and literature references associated with each category as of 1 March, 2000 are shown in Table 2.
Table 2.
Mutation frequency data sets used by HIV-SEQ
| Gene | Subtype | Treatment | Number of isolatesa | Number of referencesa |
|---|---|---|---|---|
| Protease | B | Nonea | 436 | 55 |
| Protease | Non-B | Nonea | 163 | 39 |
| Protease | B | >=1 protease inhibitor | 471 | 32 |
| Protease | B | >=3 protease inhibitors | 80 | 11 |
| RT | B | Noneb | 136 | 42 |
| RT | Non-B | Noneb | 117 | 31 |
| RT | B | >=1 nucleoside RT inhibitorsc | 381 | 42 |
| RT | B | >=4 nucleoside RT inhibitorsc | 90 | 14 |
| RT | B | Non-nucleoside RT inhibitorsd | 121 | 11 |
These numbers are based on data through 3/1/2000. The most recent figures can be found in the ‘Release Notes’ section (http://hivdb.stanford.edu/hiv/programs.htm#ReleaseNotes).
Individuals may have receivod an RT inhibitor.
Individuals in these categories did not receive nonnucleoside RT inhibitors.
Individuals in this category usually also received nucleoside RT inhibitors.
To minimize reporting bias, the mutation frequency tables contain one sequence per individual. For individuals with multiple isolates, the tables include the earliest isolate from untreated persons and the latest isolate from persons receiving anti-retroviral therapy. To exclude technical sequencing errors and cases of circulating virus containing unusual variants, the tables include only mutations reported in at least two isolates and mutations present as the main form whenever multiple clones from the same isolate were sequenced.
Background mutation data
Identified mutations are further annotated with background data that include brief summaries of associations between mutations and drug resistance based on in vitro susceptibility test results obtained from recent literature reviews13, 14 and scientific meetings. Background data are available for mutations at 27 of the 99 protease positions and 34 of the 560 RT positions.
Application deployment
HIV-SEQ is coded using Perl and CGI programming and runs on a Windows NT operating system15. The Win32::ODBC Perl module links the program to tables in the HIV RT and Protease Sequence Database.
Acknowledgments
We thank W. Wells for critical review of the manuscript.
REFERENCES
- 1.Benson DA, et al. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoesser G, Tuli MA, Lopez R, Sterk P. The EMBL nucleotide sequence database. Nucleic Acids Res. 1999;27:18–24. doi: 10.1093/nar/27.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara H, Miyazaki S, Gojobori T, Tateno Y. DNA Data Bank of Japan dealing with large scale data submission. Nucleic Acids Res. 1999;27:25–28. doi: 10.1093/nar/27.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner M, Goodman N. Informatics—genome and genetic databases. Curr. Opin. Gen. Dev. 1997;7:750–756. doi: 10.1016/s0959-437x(97)80036-6. [DOI] [PubMed] [Google Scholar]
- 5.Frishman D, Heumann K, Lesk A, Mewes H-W. Comprehensive, comprehensible, distributed and intelligent databases: Current status. BioInformatics. 1998;14:551–561. doi: 10.1093/bioinformatics/14.7.551. [DOI] [PubMed] [Google Scholar]
- 6.Panel on Clinical Practices for Treatment of HIV Infection . Guidelines for the use of antiretroviral agents in HIV-infected adults adolescents. Department of Health and Human Services; 2000. ( http://www.hivatis.org/trtgdlns.html) [Google Scholar]
- 7.Korber B, et al. Human retroviruses and AIDS: A compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory; Los Alamos, New Mexico: 1998. ( http;//hiv-web.lanl.gov) [Google Scholar]
- 8.Shafer RW, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Center for Biotechnology Information HIV-1 Subtyping Tool. 1998 www.ncbi.nlm.nih.gov/retroviruses/HIV1.
- 10.Setubal J, Meidanis J. Introduction to Computational Molecular Biology. PWS; Boston Massachusetts: 1997. pp. 61–78. [Google Scholar]
- 11.Winters MA, et al. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Invest. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamichi T, et al. High-level resistance to 3-azido-3-deoxythimidine due to a deletion in the reverse transcriptase gene human immunodeficiency virus type 1. J. Virol. 2000;74:1023–1028. doi: 10.1128/jvi.74.2.1023-1028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond J, Calef C, Larder B, Schinazi R, Mellors JW. In: Human Retroviruses and AIDS: A Compilation and Analysis of Nucleic and Amino Acid Sequences. Korber BT, et al., editors. Theoretical Biology and Biophysics, Los Alamos National Laboratory; Los Alamos, New Mexico: 1998. pp. III-36–III-79. [Google Scholar]
- 14.Hirsch MS, et al. Antiretroviral drug resistance testing in adult HIV-1 infection; recommendations of an International AIDS Society-USA Panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 15.Hardt D, Olson E, Furtado D, Jepson B. Perl Resource Kit - Win32 Edn. O'Reilly & Associates; Sebastopol, California: 1998. [Google Scholar]
- 16.Learn GH, et al. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 1997;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B, et al. Predicting HIV-1 phenotypic resistance from genotype using a large phenotype-genotype relational database. Antiviral Ther. 1999;4:41. [Google Scholar]