Abstract
Real time fluorescent quantitative PCR (universal QPCR) methods are used routinely in both academic and clinical research to measure HIV cDNA. Fast QPCR allows for faster ramping times between cycles and smaller reaction volumes, but may lose sensitivity and accuracy. We demonstrate that primer sets for HIV late reverse transcripts and 2-LTR circles have similar sensitivity and accuracy with either universal or fast QPCR methods. However, both cost and time are reduced with fast QPCR.
Keywords: HIV, Quantitative PCR, Fast PCR
Real time fluorescent quantitative PCR (QPCR) is a key method for both research and clinical laboratories to detect and measure the presence of HIV cDNA (Butler et al., 2001; Desire et al., 2001). Recent advances in QPCR instruments have led to an increase of thermal ramping capability from less than 2°C per second to 5°C per second, yielding faster times to results. New "fast" QPCR protocols also allow for reduced reaction volumes and reduced cost.
Not all universal QPCR assays for viruses translate to a fast QPCR platform. Previous studies have shown that some Epstein-Barr virus (EBV) primer sets had less sensitivity and greater variability with fast QPCR conditions (Hilscher et al., 2005). These same EBV primer sets did retain their specificity under fast conditions, but the utility of these assays is questionable unless a universal QPCR protocol is employed. Here we show that two primer sets that detect HIV cDNA retain their sensitivity and accuracy under fast QPCR conditions.
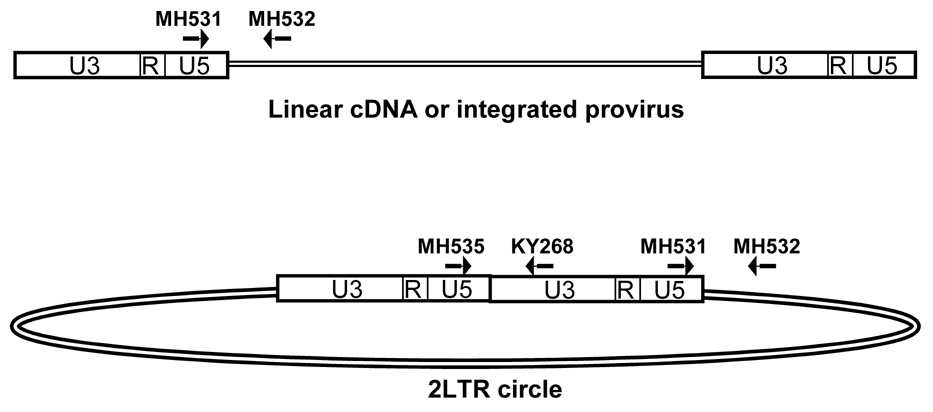
HIV late reverse transcripts (LRTs) may be quantified by primers that span the reverse transcriptase primer binding site (Figure 1; Butler et al., 2001). This region is completed after the second strand transfer of reverse transcription and is present in all full length HIV cDNA molecules. During infection, HIV cDNA may be linear double-stranded DNA, provirus integrated into the host chromosome, or extrachromosomal dead-end circular products with 1LTR or 2LTRs (Coffin et al., 1997). The HIV LRT primer set measures all HIV cDNA forms (Butler et al., 2001).
Fig. 1.
QPCR primer sets and linear or circular HIV cDNA. HIV late reverse transcripts are measured by the primers MH531 and MH532. This amplicon spans the HIV primer binding site and is present in all forms of HIV cDNA, including linear cDNA, integrated provirus, 2-LTR circles, and 1-LTR circles. All complete reverse transcripts are measured by the primer set MH531 and MH532. 2-LTR circles are specifically amplified by the primers MH535 and KY268. The junction of U5 and U3 is unique to 2-LTR circles. The 2-LTR circle primer pair spans this unique junction in 2-LTR circles and will not amplify alternative HIV cDNA forms.
Although somewhat controversial, quantitation of HIV 2-LTR circles has been used to measure continuing infection in patient samples (Bushman, 2003; Sharkey et al., 2000; Shaunak et al., 2003). When host enzymes ligate the two ends of the linear HIV cDNA before integration, the result is a circular DNA with two complete LTRs. The junction of the two LTRs provides a unique target for PCR (Figure 1). Primers that span the 2-LTR circle ligation junction will only amplify HIV 2-LTR circles.
The first QPCR method described for HIV cDNA (referred to here as "universal QPCR") included 50µl reactions incubated at 95°C for 10 min. followed by 40 cycles of 95°C for 15 sec. and 60°C for 1 min. Similar universal QPCR protocols utilize reaction volumes ranging from 20µl to 50µl. The total cycling time is approximately 2 hours. In contrast, fast QPCR reactions can be only 10µl incubated at 95°C for 20 sec. followed by 40 cycles of 95°C for 1 sec. and 60°C for 20 sec. This modified cycling time allows completion of the PCR reactions in less than 35 min.
Absolute quantitation requires a standard curve of known amounts of the amplicon per reaction. The standard curve substrates in this study were generated by subcloning the amplicon in a plasmid (pGEM-Teasy, Promega). A standard curve of the HIV LRT plasmid was quantified with the universal QPCR method in a 50µl reaction volume including 1X Taqman Universal PCR Master Mix (Applied Biosystems) and previously described primers 300nM MH531 (5' TGTGTGCCCGTCTGTTGTGT 3'), 300nM MH532 (5' GAGTCCTGCGTCGAGAGAGC 3'), and fluorescent probe 100nM LRT-P (5' FAM-CAGTGGCGCCCGAACAGGGA-TAMRA 3') (Integrated DNA Technologies) (Butler et al., 2001). Serial ten-fold dilutions of HIV LRT plasmid covered a range of 100 molecules to 108 molecules per reaction. Triplicate reactions of each dilution were pipetted into a 96 well plate and amplified in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems) at 95°C for 10 min. followed by 40 cycles of 95° for 10 sec. and 60° for 1min. The data was analyzed with Sequence Detection Systems (SDS) Software (Applied Biosystems) (Figure 2A and Table 1). These reactions yielded a standard curve with a slope of −3.08 and R2 value 0.9986 (Figure 3A).
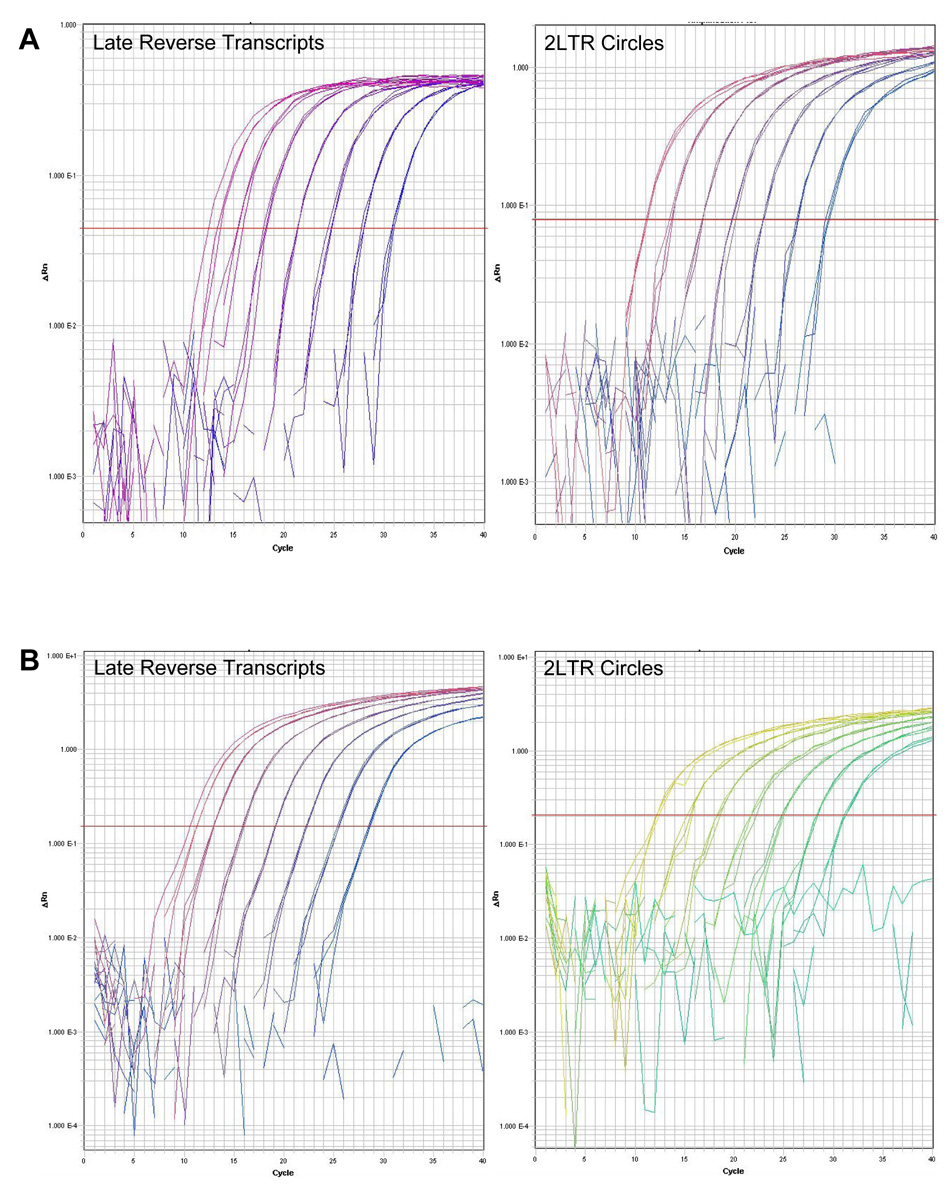
Fig. 2.
Universal QPCR and fast QPCR amplification plots. (A) Universal QPCR amplification plots of HIV late reverse transcripts (left panel) and HIV 2-LTR circles (right panel). (B) Fast QPCR amplification plot of HIV late reverse transcripts (left panel) and HIV 2-LTR circles (right panel). Triplicate samples of 102, 103, 104, 105, 106, 107, and 108 target molecules were amplified for 40 cycles. The horizontal red line indicates the threshold determined by the SDS software.
Table 1.
| Universal Conditions | Fast Conditions | |||||||
|---|---|---|---|---|---|---|---|---|
| R2 | Slope | Reaction volume | Time | R2 | Slope | Reaction volume | Time | |
| LRT | 0.9986 | −3.08 | 50 µl | 2 hours | 0.9958 | −3.00 | 10 µl | 35 min. |
| 2LTR circles | 0.9954 | −3.02 | 50 µl | 2 hours | 0.9990 | −3.19 | 10 µl | 35 min. |
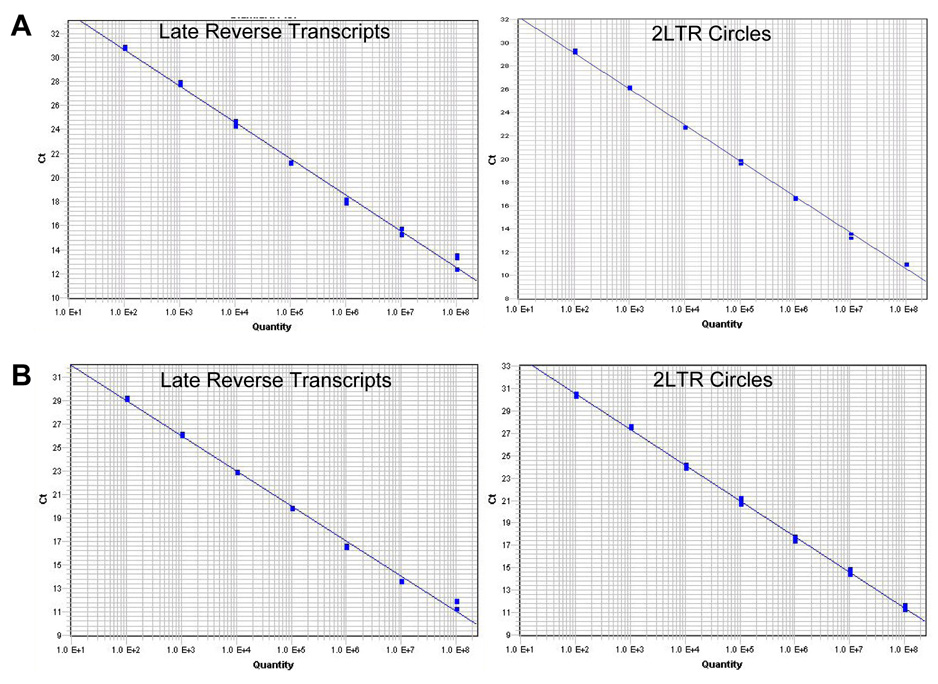
Fig. 3.
Standard curves from universal QPCR and fast QPCR. (A) Universal QPCR derived standard curves for HIV late reverse transcripts (left panel) and HIV 2-LTR circles (right panel). (B) Fast QPCR derived standard curves for HIV late reverse transcripts (left panel) and HIV 2-LTR circles (right panel). Triplicate samples of 102, 103, 104, 105, 106, 107, and 108 target molecules were amplified for 40 cycles. Best fit lines were generated by the SDS software. For all standard curves R2 > 0.995.
HIV 2-LTR circles were also quantified by universal QPCR. 2-LTR circle primers are MH535 (5' AACTAGGGAACCCACTGCTTAAG 3'), KY268 (5' CCAGAGAGACCCAGTACAAGCA 3'), and MH603 (5' FAM-ACACTACTTGAAGCACTCAAGGCAAGCTTT-TAMRA 3') (Integrated DNA Technologies). The serial dilution standard for these reactions is the 2LTR circle amplicon subcloned to pGEMT-easy (Promega) and evaluated as 10 fold serial dilutions from 100 to 108 molecules per reaction. The primer concentrations, reaction conditions, and data analysis were the same as with the HIV LRT universal QPCR reactions (Figure 2A). The HIV 2-LTR circle reactions yielded a standard curve with a slope of −3.02 and R2 value 0.9954 (Figure 3A).
The same primers and standard plasmids were tested with a fast QPCR method. This modified QPCR technique included 1X Taqman Fast Universal PCR Master Mix (Applied Biosystems) and the same molar concentrations of primers in 10µl total volume reactions. Triplicate reactions of each serial dilution of standard plasmids were pipetted into a 96 well plate and amplified in a 7900HT Fast Real-Time PCR System (Applied Biosystems). Samples were at 95°C for 20 sec followed by 40 cycles of 95°C for 1 sec. and 60°for 20sec. Data was analyzed by SDS Software (Figure 2B). The slope for HIV LRT reactions was −3.00 and the R2 value was 0.9958 (Figure 3B and Table 1). The slope for the HIV 2-LTR circle reactions was −3.19 and the R2 value was 0.9990 (Figure 3B and Table 1).
The slope of each QPCR standard curve is an indication of the efficiency of the PCR reaction. If the reactions are 100% efficient, then the slope will equal −3.3. The HIV LRT standard reactions had a slope of −3.08 under universal QPCR conditions and −3.00 with the fast QPCR conditions. The slopes of the HIV 2-LTR circle standard reactions were −3.02 and −3.19 under universal and fast QPCR conditions, respectively. The slopes of the standard curves obtained by universal QPCR and fast QPCR suggest that they are comparably efficient. The R2 values for all of the standard dilution series were greater than 0.995 indicating that the fast QPCR method retains the accuracy of the universal QPCR technique (Table 1). Thus these HIV primer sets in fast QPCR have comparable efficiency and accuracy to the universal QPCR reaction conditions. However, the time of amplification is reduced by approximately 4 fold (Table 1). While the Taqman Fast Universal PCR Master Mix is somewhat more expensive than the Taqman Universal PCR Master Mix, the actual cost per reaction is less due to the reduction in reaction volume from 20–50µl with universal QPCR to 10µl with fast QPCR. Due to the small reaction volumes of the fast QPCR method, it is possible to use 384 well plates instead of 96 well plates. This would allow even more samples to be analyzed in a shorter time.
Quantitative PCR methods for detection of HIV cDNA have proved highly valuable for both research and clinical laboratories. These methods are readily suited for decreased reaction volumes and faster cycling times. RT-PCR methods may also be adapted to a faster DNA amplification protocol although the primary reverse transcription step should remain the same. The fast QPCR method allows for both faster and more cost effective detection of HIV cDNA through reduction of cycling times and reaction volumes.
Acknowledgments
We thank Hansjuerg Alder, Dharshna Bhatt, Sarah Miller, and the OSU Comprehensive Cancer Center Nucleic Acid Shared Resource for support and helpful discussions.
This work was supported by NIH grant CA56542 to R.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bushman F. Measuring covert HIV replication during HAART: the abundance of 2-LTR circles is not a reliable marker. Aids. 2003;17:749–750. doi: 10.1097/01.aids.0000050868.71999.09. [DOI] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Desire N, Dehee A, Schneider V, Jacomet C, Goujon C, Girard PM, Rozenbaum W, Nicolas JC. Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J Clin Microbiol. 2001;39:1303–1310. doi: 10.1128/JCM.39.4.1303-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, Bucy RP, Kostrikis LG, Haase A, Veryard C, Davaro RE, Cheeseman SH, Daly JS, Bova C, Ellison RT, 3rd, Mady B, Lai KK, Moyle G, Nelson M, Gazzard B, Shaunak S, Stevenson M. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaunak S, Teo I, Choi JW, Gazzard B. HIV DNA two long terminal repeat circles: observations and interpretations. Aids. 2003;17:2273–2274. doi: 10.1097/00002030-200310170-00021. [DOI] [PubMed] [Google Scholar]