Abstract
Though β-rich self-assemblies comprise a major structural class of polypeptides, a detailed understanding of the determinants of their structure and stability is lacking. In particular, the roles of repetitive stretches of side-chains running the long axis of these β-sheets, termed “cross-strand ladders,” remain poorly characterized due to the inherently insoluble and heterogeneous nature of self-assemblies. To overcome these experimental challenges, we have established a complementary experimental system termed “peptide self-assembly mimics” (PSAMs). The PSAMs capture a defined number of self-assembly-like peptide repeats within a soluble β-rich protein, making structural and energetic studies possible. In this work, we investigated the role of cross-strand ladders containing aromatic residues, which are prominent in self-assembling peptides. A combination of solution data and high-resolution crystal structures revealed that a single cross-strand ladder consisting solely of Tyr significantly stabilized, rigidified and flattened the PSAM β-sheet. These characteristics would stabilize each β-sheet layer of a self-assembly and direct sheet conformations compatible with lamination. Our results therefore provide a rationale for the abundance of aromatic amino acids in fibril-forming peptides and establish important roles of cross-strand Tyr ladders in the structure and stability of β-rich peptide self-assemblies.
Keywords: β-sheet, fibril, tyrosine, protein engineering, protein nanomaterial
It has become clear that polypeptides have inherent ability to self-associate into β-rich structures. β-rich peptide self-assemblies are associated with a wide range of human protein-misfolding diseases1; 2 and have been explored as a potential source for nanomaterials.3; 4 Yet despite considerable interest in self-assemblies, there are few systems that enable quantitative characterization of either their structure or stability. Many self-assembled peptides are by their nature heterogeneous in terms of size and conformation, insoluble and non-crystalline. These properties make it difficult to apply high-resolution structural tools that have been developed for soluble proteins, such as X-ray crystallography and solution NMR spectroscopy. Equally importantly, these properties make energetic characterization of the self-assembly processes extremely difficult.
β-rich self-assemblies commonly adopt the “cross-β” structure, characterized by an extendeG β-conformation with the strands running perpendicular to the long axis of the assembly and lamination of multiple β-sheets.5 Solid-state NMR spectroscopy and X-ray crystallography studies have shown that each β-sheet layer can be either parallel or anti-parallel.6-9 Either orientation of β-strands creates a specific arrangement of side-chains in self-assemblies, termed “cross-strand ladders”.10 These cross-strand ladders consist of repeating side-chain interactions running across β-strands within a β-sheet layer (i.e. parallel to the long axis of the assembly) and arise from the inherently repetitive nature of self-assembly. These side-chain patterns occur regardless of peptide sequence, as confirmed by recent crystal structures of small fibril-forming peptides,7-9 and may play a fundamental structural role in β-rich self-assemblies (Fig. 1A). However, there has been no quantitative characterization of the structural and energetic contributions of these motifs. Mutations in self-assembling peptides often result in drastic perturbations that either completely disrupt, destabilize or alter the mode of assembly, making it extremely difficult to deduce direct effects of mutations in the context of the self-assembly. Cross-strand ladders (also termed “stacks”) found in the hydrophobic core of β-helical proteins could potentially be used as a model system to understand these interactions, but because of extensive interactions of cross-strand ladder residues with the rest of the protein, most mutations cause severe defects in folding and render detailed characterization and interpretation impossible.11
Fig. 1.
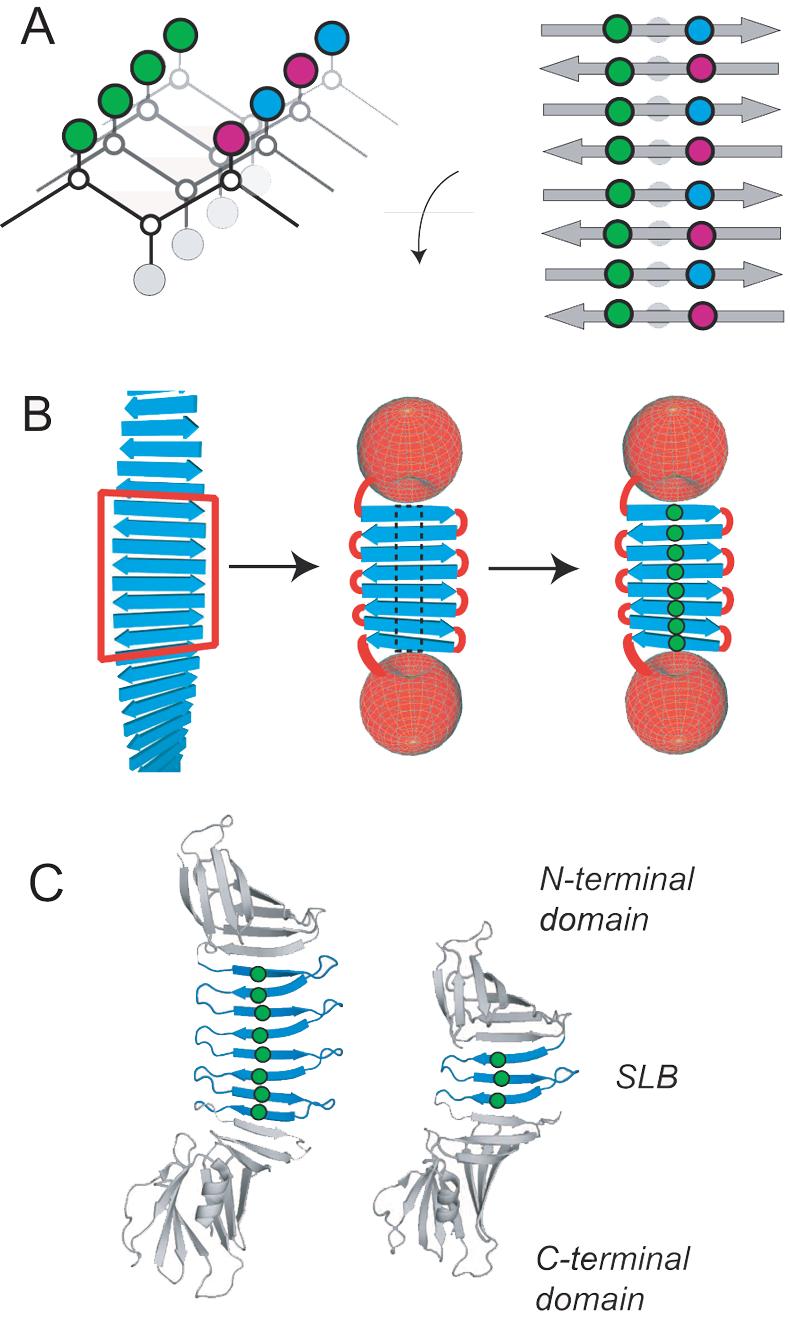
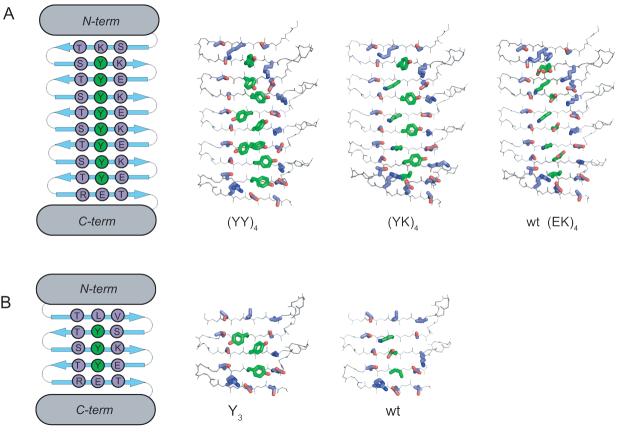
Cross-stand ladders in self-assembled β-sheets and the PSAM system. (A) Schematic drawings showing how the sequence and register of a self-assembling peptide determines ladder composition. A simplified β-sheet is shown as a connection of α-carbons, with side-chain spheres colored to indicate chemical identity. Only an anti-parallel assembly is shown. (B) Scheme illustrating the PSAM concept and ladder mutations: a segment of peptide self-assembly is excised, linked, and capped. Mutations, shown as green spheres, can then be introduced into ladder positions within the PSAM scaffold. (C) Cartoon representations of the PSAM scaffolds used (large scaffold i.e. OspA+3bh shown at left; small scaffold i.e. natural OspA shown at right). The SLB in each scaffold is colored blue with the α-carbons of mutated ladder positions shown as spheres. The side-chains of these ladders are all located on the same face of the β-sheet.
In order to elucidate the significance of cross-strand ladder interactions, it is essential to employ an experimental system in which both constructive and destructive perturbations to self-assembly structure and stability are accessible to quantitative characterization, while maintaining a common mode of assembly. To this end, we have developed “peptide self-assembly mimics” (PSAMs) that capture a self-assembly-like β-sheet structure within a water-soluble protein (Fig. 1B).12 The PSAMs eliminate the heterogeneity of self-assembly by covalently linking a fixed number of peptide units into a flat, single-layer β-sheet (SLB) of the natural protein OspA. The two edges of the linked peptide units are capped by N- and C-terminal globular domains that prevent lateral assembly and lamination. Using PSAMs as “host” scaffolds, we can introduce a variety of “guest” mutations into the ladders and study their effects on sheet structure and stability using well-established biophysical methods. Unlike other experimental studies of self-assembly, the SLB allows us to isolate intra-sheet interactions from the inter-sheet interactions, making it possible to characterize the effects of cross-strand ladders on the basic unit of β-rich self-assemblies. Our hypothesis is that the side-chains of these ladders influence β-sheet backbone conformation and dynamics, which in turn affect the structure and stability of a multi-layer assembly. Although the current generation of our PSAM system only addresses the role of side-chains in the structural context of anti-parallel self-assemblies due to the strand topology of the OspA host, it can still provide fundamental information that would be otherwise unattainable.
In this work, we investigated the contribution of cross-strand ladders consisting of Tyr residues. Because of their prominence in self-assembling peptides, aromatic residues have garnered considerable attention in models of the nucleation and propagation. For example, it is hypothesized that π-stacking contributes to inter-strand stabilization as well as fibril lamination13-17 and that aromatic residues high β-sheet propensity may also be a factor.18 Though aromatic cross-strand ladders are abundantly observed in β-rich assemblies,7-9 the inability to experimentally probe their roles in a well-defined environment has made it difficult to test these assumptions. We therefore investigated a series of Tyr-rich ladders grafted into the PSAM scaffold. Atomic structures and thermodynamic characterization of these molecules provide a rationale for the importance of aromatics in peptide self-assembly.
Results
Design of aromatic ladders in a PSAM scaffold
We selected a PSAM containing four homologous β-hairpins comprising a total of eight β-strands as the host scaffold. It is referred to as “OspA+3bh” (i.e. OspA plus a three β-hairpin insertion) in our previous publication.12 This large β-sheet scaffold minimizes the context effects from the N- and C-terminal globular domains (Fig. 1C), and its sequence can be manipulated efficiently. Furthermore, we have successfully crystallized this PSAM by employing surface engineering of the globular domains,12 suggesting a good likelihood of obtaining atomic structures of mutants.
We selected eight residues that constitute a cross-strand ladder in the center of the β-sheet as the host positions. These positions exhibit particularly regular β-sheet geometry because they are centrally located within the β-strands (Fig. 1; Table 1). The wild-type host ladder consists of three repeats of Glu and Lys followed by Glu and Ile (i.e. “EKEKEKEI”), which for brevity is denoted as (EK)4. Note that this nomenclature does not denote the amino acid sequence of a contiguous peptide segment, but rather it represents a motif formed across β-strands in the folded PSAM (Fig. 1C). We produced a total of three mutants. (YY)4 contains eight Tyr substitutions that form a contiguous ladder. (YK)4 and (EY)4 each contain four Tyr substitutions and form an alternating pattern of two amino acids. These are all representative motifs that can exist in self-assemblies of a short peptide containing a Tyr residue (Fig. 1A).
Table 1.
PSAM aromatic ladder mutations used in this work
| Mutant | Mutations |
|---|---|
| Large scaffold | |
| (EY)4 | K136Y, K159Y, K182Y, I205Y |
| (YK)4 | E123Y, E146Y, E169Y, E192Y |
| (YY)4 | E123Y, K136Y, E146Y, K159Y, E169Y, K182Y, E192Y, I205Y |
| Small scaffold | |
| Y3 | K113Y, E123Y, I136Y |
Tyr ladder stabilizes and rigidifies the PSAM
The three PSAM mutants expressed in E. coli were soluble and monomeric, as determined with size-exclusion chromatography. Urea denaturation experiments, analyzed using a three-state model containing native, intermediate and unfolded species,19 demonstrated the (YY)4 and (YK)4 mutants were stabilized by 3.3 and 2.2 kcal/mol, respectively, whereas (EY)4 was destabilized by 1.7 kcal/mol (Table 2 and Supplementary Fig. 1). These results indicate that a Tyr ladder can greatly influence the stability of a self-assembly layer.
Table 2.
Stability of PSAMs measured by urea-induced unfolding.
| Mutant | ΔGUI3M (kcal/mol) |
ΔGNU3M (kcal/mol) |
mNI | ΔΔGNU3M (kcal/mol) |
|---|---|---|---|---|
| Large scaffold | ||||
| wt | 0.40 (±0.06) | 1.54 (±0.10) | −1.60 (±0.06) | 0 |
| (EY)4 | 0.79 (±0.05) | 3.26 (±0.10) | −2.29 (±0.06) | 1.72 (±0.14) |
| (YK)4 | 0.88 (±0.06) | −0.70 (±0.06) | −1.77 (±0.06) | −2.24 (±0.12) |
| (YY)4 | 1.87 (±0.05) | −1.75 (±0.06) | −2.29 (±0.05) | −3.29 (±0.12) |
| Small scaffold | ||||
| wt | 1.74 (±0.04) | 2.71 (±0.01) | −2.41 (±0.02) | 0 |
| Y3 | 1.48 (±0.05) | 2.29 (±0.05) | −1.91 (±0.04) | 0.42 (±0.05) |
ΔGUI3M is the free energy difference between the unfolded state and the intermediate state at 3M urea. ΔGNU3M is the free energy difference between the unfolded state and the native state at 3M urea. mNI is the m-value for the transition between the native and intermediate states. The m-values for the N-I and I-U transitions (mNI and mIU) were allowed to vary, but the total m-value (mNU = mNI + mIU) was fixed at −4.4 and −5.3 for the small and large scaffolds, respectively, following our previously established method.19 ΔΔGNU is the difference in GNU3M between the wild-type and a mutant (ΔΔGNU = ΔGNU3M(wild-type) − ΔGNU3M(mutant)). A positive ΔΔGNU value indicates destabilization by a mutation. The values for the wild-type small scaffold were taken from Yan et al.19
We performed NMR measurements on the wild-type (EK)4 and (YY)4 PSAMs to characterize the effects of the Tyr ladder on the solution conformation of the β-sheet. Both proteins exhibited highly dispersed HSQC spectra characteristic of a well-folded β-rich protein (Supplementary Fig. 2). Resonances for a total of 48 residues from the invariant globular domains were well isolated and could be readily assigned from comparisons with previous assignments of smaller PSAMs.12 The presence of four nearly identical β-hairpin segments in these proteins made it impossible to assign many resonances. Nevertheless, the available assignments allowed us to investigate the relative orientations and conformational dynamics between the two globular domains using residual dipolar couplings (RDCs). Previous studies have established that the two globular domains are rigid and RDC measurements of residues in the globular domains can probe the properties of the SLB region (i.e. the region that contains the cross-strand ladder) that connects the two globular domains.12; 20
We found that while the overall patterns of RDCs for the two proteins are generally similar, RDCs of the N-terminal globular domain of (YY)4 had larger absolute values than corresponding RDCs of the wild-type (EK)4 protein (Fig. 2), particularly for residues that are farther away from the SLB region. We observed a similar trend in comparing the wild-type OspA (i.e. no β-hairpin insertion) to OspA+2bh (i.e. two β-hairpin insertions), and have attributed the RDC decreases of OspA+2bh to conformational averaging resulting from an increased degree of flexibility in the SLB of OspA+2bh.12 The RDC results here likewise strongly suggest that the globular domains were held in more rigid conformations relative to each other in the (YY)4 mutant. Therefore, the Tyr ladder rigidified the PSAM.
Fig 2.
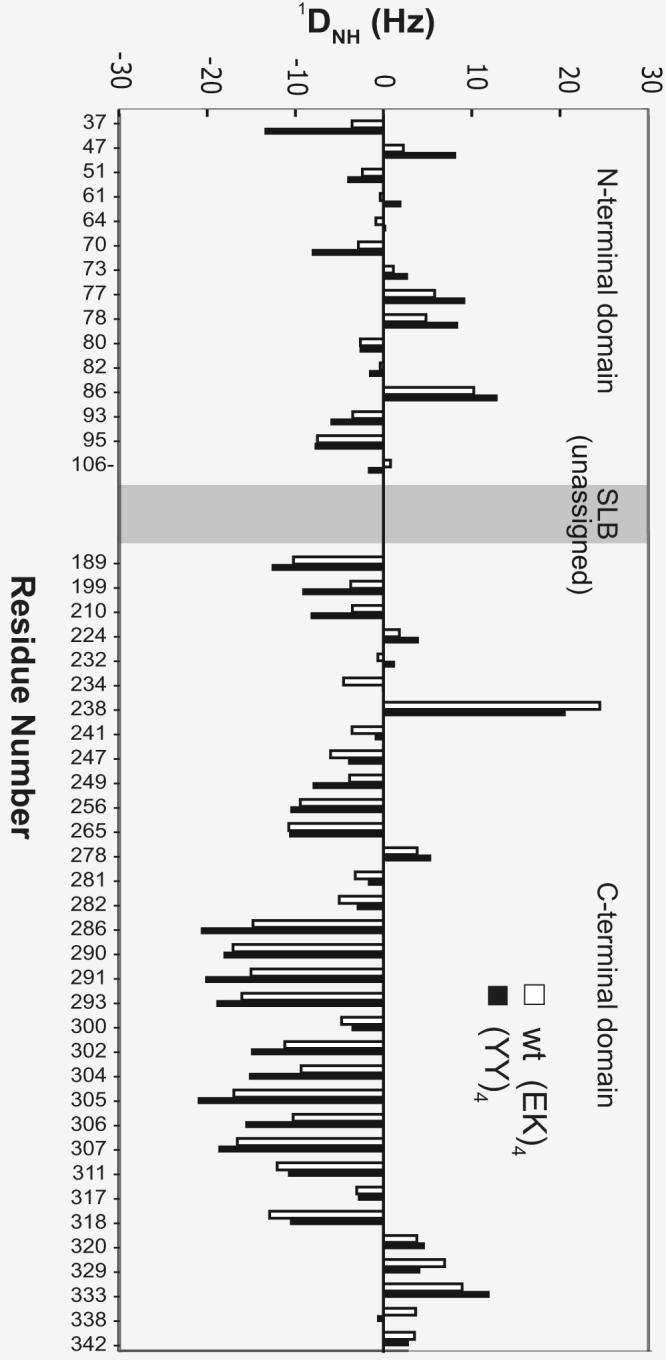
Residual dipolar couplings (RDCs) of the backbone amide 1H and 15N pairs of the PSAMs plotted as a function of residue number. Data for the N- and C-terminal caps of wild-type (EK)4 and (YY)4 PSAMs are colored red and blue, respectively. The break between the two corresponds to the SLB, for which amino acid sequence degeneracy prevented resonance assignments and accordingly no RDC data were obtained.
Aromatic ladders control β-sheet twistedness
To characterize structural effects of the ladder mutations in detail, we determined x-ray crystal structures of the (YY)4 and (YK)4 PSAMs at 1.55Å and 2.00Å resolutions, respectively. The wild-type (EK)4 structure has previously been determined at 1.60Å (Fig. 3A).12 We have not been successful in crystallizing (EY)4. All structures crystallized in the P212121 space group and share a similar mode of crystal packing (Table 3), providing an opportunity to characterize structural consequences of replacing a cross-strand ladder in a well-defined environment. When the entire molecules were superimposed, (YY)4 showed a considerable difference (Cα RMSD = 2.74Å) relative to the wild-type structure, while (YK)4 was virtually identical to the wild-type structure (Cα RMSD = 0.28Å). As expected, the conformations of the N- and C-terminal globular domains were unaffected by the ladder mutations, as manifested in small RMSD values (Table 4). Thus, it appears crystal packing does not dictate the global conformation of these PSAMs, and so the differences between mutants are primarily due to structural perturbations introduced by the Tyr ladder. Indeed, there are clear differences in the SLB conformation between (YY)4 and other PSAMs (Fig. 4A). While wild-type (EK)4 and probably (YK)4 have higher levels of conformational dynamics as described above, the fact that both crystallized in a similar conformation that is distinct from (YY)4 suggest that the observed crystal structures of these three proteins highly likely reflect their respective low-energy structures.
Fig. 3.
Crystal structures of aromatic ladder PSAMs. (A) Large scaffold PSAMs. The N- and C-temini are omitted for clarity. The SLB is in the plane of the page, with the N-terminus above and the C-teminus below. The side-chain carbon atoms of the central ladder to which Tyr was introduced are colored in green and those of all neighboring residues are colored in lavender. (B) Small scaffold PSAMs, illustrated in the same manner. The sequences of the (YY)4 and Y3 proteins are also shown schematically.
Table 3.
Statistics for crystal structures of PSAM mutants
| Protein | (YK)4 | (YY)4 | OspA Y3 |
|---|---|---|---|
| PDB code | 2OY8 | 2OY7 | 2OYB |
| Data collection statistics | |||
| Space group | P212121 | P212121 | P21 |
| Cell parameters | a=37.234 | a=37.055 | a=33.125 |
| b=76.645 | b=93.103 | b=54.686 | |
| c=120.348 | c=106.101 | c=66.466 | |
| β=99.96 | |||
| Beamline | 22-ID | 22-ID | 23-ID |
| Wavelength | 1.0000 | 0.97105 | 0.9793 |
| Resolution (Å) a) | 50-2.0 (2.07-2.00) | 50-1.5 (1.59-1.55) | 50-1.3 (1.35-1.30) |
| Completeness(%) | 98.6 (98.8) | 97.9 (95.5) | 99.9 (100) |
| I/σ(I) | 20.35 (7.02) | 19.82 (2.98) | 14.01 (5.36) |
| Rmerge b) | 0.096 (0.304) | 0.066 (0.497) | 0.111 (0.286) |
| Average redundancy | 7.4 (7.3) | 4.1 (2.9) | 3.4 (3.2) |
| Refinement statistics | |||
| Resolution range (Å) | 20.00-2.00 | 20.00-1.55 | 20.00-1.30 |
| Reflections used (free) | 22533 (1209) | 50395 (2690) | 54499 (2900) |
| R factor c) | 0.21440 | 0.19061 | 0.15535 |
| Rfree d) | 0.25456 | 0.21986 | 0.18830 |
| RMS deviations | |||
| Bonds (Å) | 0.017 | 0.012 | 0.011 |
| Angles (°) | 1.773 | 1.450 | 1.393 |
| No. protein residues | 315 | 315 | 246 |
| No. waters | 109 | 404 | 450 |
| Average B factor (Å2) | 40.024 | 25.758 | 11.737 |
| Ramachandran plot statistics | |||
| Most favored (%) | 91.7 | 91.3 | 91.9 |
| Additionally allowed (%) | 8.0 | 8.7 | 7.6 |
| Generally allowed (%) | 0.3 | 0.0 | 0.4 |
Highest resolution shell is shown in parenthesis.
R-merge = ΣhklΣi| I(hkl)i - <I(hkl)> | /ΣhklΣi<(hkl)i> over i observations of a reflection hkl.
R-factor = Σ | |F(obs)| - |F(calc)| |/Σ|F(obs)|.
Rfree is R with 5% of reflections sequestered before refinement. Crystallization conditions are: Y3, 10.0mg/ml protein, 37% PEG400, 0.1M Tris-HCl pH7; (YY)4, 41.4mg/ml protein, 22% PEG1000, 0.1M HEPES pH7.5; (YK)4, 37.9mg/ml, 31% PEG400, 0.1M Tris-HCl pH9, 2% MPD. Both Y3 and (YK)4 used the crystallization buffer itself as a cryo-protectant, whereas for (YY)4 used a mixture of 25% glycerol and buffer. Crystals were flash-frozen in liquid nitrogen prior to diffraction measurements.
Table 4.
RMSD for Cα atoms of PSAM aromatic ladder mutants relative to their respective wild-type scaffold
| Mutant | N-term domain (Å) |
C-term domain (Å) |
All (Å) |
|---|---|---|---|
| Large scaffold | |||
| (YK)4 | 0.493 | 0.132 | 0.281 |
| (YY)4 | 0.745 | 0.399 | 2.738 |
| Small scaffold | |||
| Y3 | 0.092 | 0.138 | 0.170 |
Fig. 4.
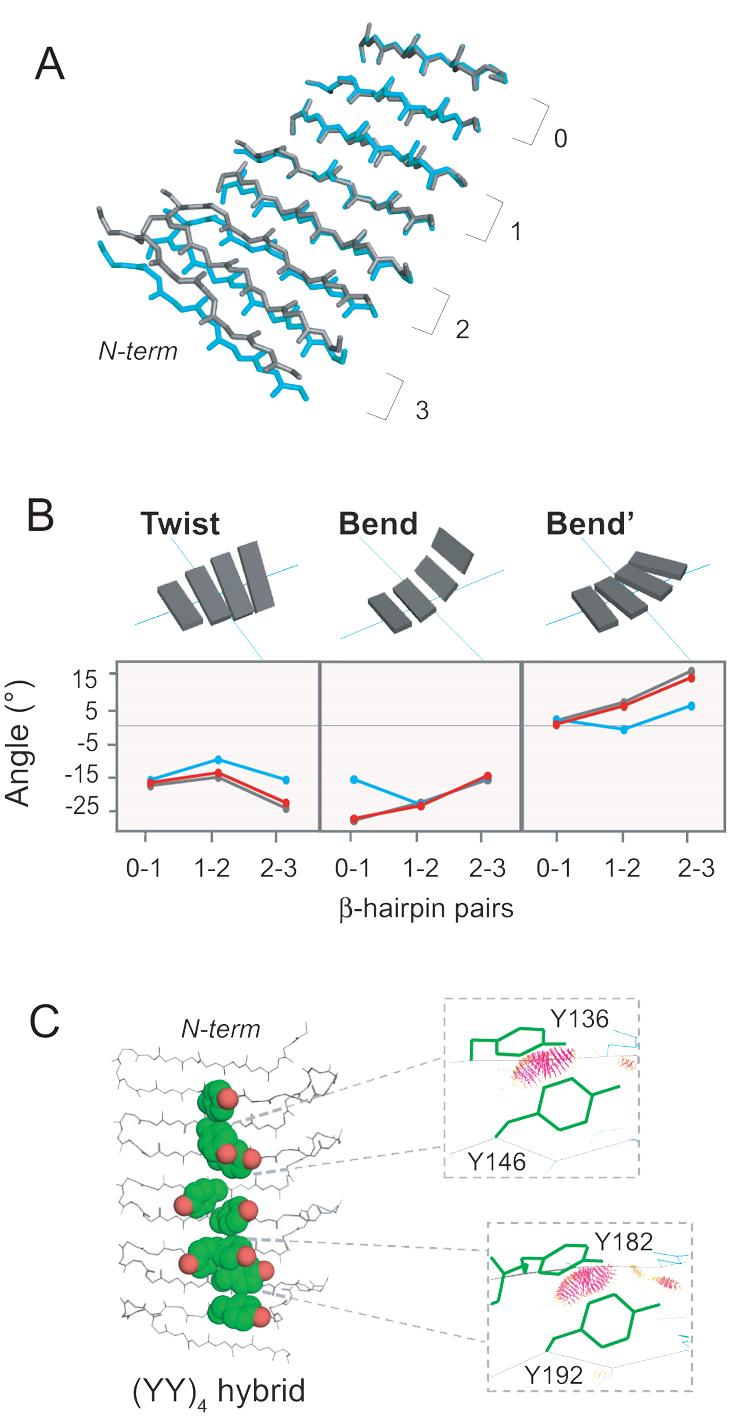
Perturbations of backbone conformation introduced by the Tyr ladders. (A) Each PSAM mutant was aligned using the most C-terminal β-hairpin of the SLB, denoted as 0 (residues 187-209). Wild-type (EK)4 is shown in grey and (YY)4 in blue. (YK)4 overlays almost perfectly with wild-type, and its structure is not depicted. The N- and C-termini along with the turn regions are omitted for clarity. The backbone of the β-strands are shown as sticks. (B) Values of the three parameters Twist, Bend, Bend' describing the 3-dimensional rotations of each β-hairpin in the SLB. Zero values define a perfectly flat, rectangular β-sheet. Wild-type (EK)4 is shown in grey, (YY)4 in blue, and (YK)4 in red. (C) Over-packing of Tyr ladders revealed by hybrid analysis. Each strand from (YY)4 was aligned separately with its homologous strand in wild-type (EK)4 using all backbone atoms. The adjusted coordinates of the aromatic side-chains were then grafted onto the wild-type (EK)4 backbone to form the (YY)4 hybrid. Severe steric conflicts (shown as red dots in the inserts) were judged using Probe.32 Y182 has two side chain conformers, and the conflicts were observed for its gauche- conformer but not for its trans conformer.
To identify the origin of large difference in the backbone structure between (YY)4 and (EK)4, we compared backbone dihedral angles of the PSAM structures. However, no significant changes in the (ϕ, φ) angles were found for the mutated positions in (YY)4 and (YK)4 or residues immediately adjacent to these mutations (Supplementary Fig. 3), indicating that the conformational change was not caused by localized differences around the mutation sites. To further dissect the basis of the structural variations, we analyzed the relative orientations of adjacent β-hairpin units in terms of rotations about three orthogonal axes termed Twist, Bend, and Bend', respectively, as we established previously (Fig. 4B).12 Our analysis revealed the (YY)4 ladder reduced all of these parameters, and as a result the β-sheet bent away from the face on which the (YY)4 ladder was introduced and was significantly flatter than the wild type. In contrast, (YK)4 showed marginal differences from wild-type (EK)4, as expected from its small backbone RMSD. These results suggest that the β-sheet in the (YY)4 ladder, as a whole, has a distinct conformation from those of (EK)4 and (YK)4 and that these differences originate from small differences in the backbone conformation of residues throughout these β-sheets.
Interactions involving the Tyr ladders responsible for structural and stability changes
To elucidate the structural basis for the distinct conformation of (YY)4, we then examined whether the (YY)4 side-chain conformers were compatible with the more twisted, wild-type (EK)4 backbone conformation. Tyr side-chain rotamers found in the (YY)4 ladder were grafted onto the experimentally determined wild-type (EK)4 backbone to form a “hybrid” model. This model showed steric clashes in two pairs of cross-strand Tyr residues (Fig. 4C). In contrast, the (YK)4 hybrid, constructed by grafting Tyr at every other position of the wild-type (EK)4 ladder, did not display any steric conflict. This lack of steric conflict is consistent with our experimental observation that the backbone conformation of the (YK)4 crystal structure is nearly identical to that of the wild-type. These results suggest that bulky aromatic side-chains placed in contiguous positions of a cross-strand ladder impose steric restrictions on the β-sheet backbone conformation, leading to a flat β-sheet. The lack of structural perturbation in (YK)4 may be due to “buffering” the cross-strand Tyr by less bulky Lys side-chains, which allow Tyr to be accommodated without altering backbone conformation. Taken together, these results strongly suggest that sheet flattening by a long Tyr ladder is mainly due to the avoidance of side-chain clashes.
The Tyr residues introduced in (YY)4 and (YK)4 make numerous hydrophobic contacts with neighboring side-chains (Fig. 3A), contributing an additional 336Å2 and 207Å2 of buried surface area, respectively. This result suggests that increased hydrophobic burial accounts for the increased stability of (YY)4 and (YK)4 relative to wild-type (EK)4, a view which is consistent with previous work demonstrating that hydrophobic side-chain packing is a major determinant of the stability of flat β-sheets.21 Although we do not have an atomic structure of (EY)4, we speculate that its destabilization may be caused by burial of the Glu carboxylates in a hydrophobic environment by adjacent Tyr. In contrast, in (YK)4 the amine moieties of the Lys residues are not significantly buried by the neighboring Tyr residues. Together, these results indicate Tyr ladders can substantially stabilize flat β-sheet through hydrophobic contacts, though such effects also depend on the identity of the ladder.
Smaller effects of a shorter Tyr ladder
To test whether the structural and thermodynamic effects of aromatic ladders depend on their length, we characterized a shorter Tyr ladder using a smaller PSAM scaffold. This scaffold is the same size as the natural OspA (i.e. no β-hairpin insertions) and contains three anti-parallel β-strands. These β-strands are homologous in both sequence and structure to those in the 8-stranded scaffold described above (Fig. 1C). We introduced a ladder consisting of Tyr to this three-stranded PSAM, termed Y3 (Table 1). As judged by urea denaturation, Y3 did not stabilize this small PSAM (Table 2) indicating that the thermodynamic effects of a Tyr ladder are dependent on its length.
We determined the crystal structures of Y3 at a 1.30 Å resolution (Fig. 3B). A comparison of this structure with the previously determined structure of the wild-type OspA scaffold22 revealed virtually no effect on the β-sheet conformation (Table 4). In the Y3 crystal structure, the Tyr side-chains splay into regions that cause minimal side-chain contacts (Fig. 3B). This contrasts with the structure of (YY)4, where the splaying of Tyr side-chains is hindered by the conformational restrictions imposed by a longer aromatic ladder.
Discussion
The PSAMs in conjunction with “ladder mutations” have provided a unique system to gain insights into both the thermodynamic and structural perturbations introduced to peptide self-assemblies by mutations in the underlying peptide unit. We have experimentally demonstrated that Tyr cross-strand ladders flatten a self-assembly-like β-sheet through steric hindrance of the Tyr side-chains. The increase in the side-chain packing due to a Tyr ladder also contributes to enhanced rigidity and stability of the α-sheet. As Tyr has been predicted to be energetically primed for the formation of flat α-strands,10 and is known to affect the structure of fibril-forming peptides,23 our work provides insights into the molecular mechanism by which aromatic residues influence self-assembly morphology and stability.
The effects of introducing Tyr in cross-strand ladders were complex and dependent on the context and length of ladders. The introduction of four Tyr residues in different positions of the same host ladder (i.e. (YK)4 versus (EY)4) had dramatically different impacts on stability (Table 2). Similarly, the short Y3 ladder had little influence on stability, while (YY)4 significantly stabilized the β-sheet. These differences indicate the difficulty of predicting effects of mutations by extrapolating results from a simpler system and illustrate the importance of analyzing mutational effects on peptide self-assembly in an appropriate context.
Previous research on the amino acid determinants of peptide self-assembly has focused on the inherent chemical and structural properties of aromatic amino acids, such as their high β-sheet propensity18 and tendancy to form stabilizing π-stacking interactions.13; 14 However, a lack of high-resolution information precluded this type of research from addressing structural contributions of aromatics beyond those that can be easily predicted from the generic “cross-β” architecture. While these properties are undoubtedly important, the present work establishes an additional rationale for aromatic amino acids that is uniquely observed in the context of cross-strand ladders. Our description of the flattening and rigidifying effects of aromatic ladders may be important for rationalizing the prevalence of aromatic residues among self-assembling peptides. To laminate and form multi-layer β-sheet assemblies, each β-sheet layer must be devoid of strong twist.12 Also a high degree of flexibility poses a large entropic penalty for lamination. Thus the abundance of aromatics in self-assembling peptides may therefore be partly due to their ability to flatten and rigidify sheet conformation.
Computational studies have suggested that single-layer α-sheets are important intermediates in forming multi-layer assemblies, though they are often inaccessible to experimental observation.24 Nussinov's group has previously studied Asn ladders in a single-layer sheet by molecular dynamics simulations, finding that stacking and inter-strand hydrogen-bonding within the ladder contributed to an energetic “bottleneck” in peptide oligomerization.25 Quantitative thermodynamic data and atomic structures produced by the PSAM approach may be useful for evaluating and refining such computational studies.
Recent crystal structures of small fibril-forming peptides have provided atomic insights into the factors stabilizing laminated self-assemblies, showing that the side-chains of self-assembling peptides form a dry interdigitating “steric zipper” between β-sheet layers.7; 8 It is interesting that aromatic residues are often found in the “wet interface” of these crystal structures. Although the current PSAMs only recapitulate a solvent-exposed interface, our results provide a complementary view of how repetitive sequences of aromatic amino acids at the single-layer level can stabilize β-sheets and are likely to be relevant to the molecular mechanism underlying the formation of multi-layer self-assemblies. Together, these results highlight the importance of both inter-sheet and intra-sheet interactions in the overall stability of self-assemblies.
Conclusions
The PSAM system is effective at addressing a variety of questions related to β-rich peptide self-assembly at atomic resolution. This work has revealed structural roles for Tyr in self-assembly, indicating a long Tyr ladder both flattens and rigidifies a β-sheet layer. Knowledge gained from this work provides new insights into the sequence determinants of peptide self-assembly and suggests guidelines for designing flat, self-assembling nanomaterials. The PSAM approach can be readily adapted to study ladders composed of a variety of amino acids, which will further clarify the fundamental mechanisms of β-rich peptide self-assembly and interactions of self-assemblies with small molecules.
Materials and Methods
Sample preparation
Mutagenesis, expression and purification were performed as described previously.19 All aromatic mutations are listed in Table 1. For crystallization, the mutants were transferred into the OspAsm1 scaffold as described previously.22 The sm1 scaffold contains mutations in the N- and C-terminal caps of OspA, which increase crystallization efficiency without perturbing global structure or the SLB region.
Stability measurements
Urea-induced unfolding measurements were conducted on samples without the cleavage of the histag in 10mM sodium phosphate buffer pH 6.0 containing 50mM NaCl using an Aviv CD spectrometer with a fluorescence accessory and analyzed using a three-state model as detailed in Table 2 legend.19
NMR spectroscopy
[2H, 15N]-enriched samples of the wild-type (EK)4 and (YY)4 PSAMs were prepared as described previously. Samples were dissolved in 10 mM sodium phosphate buffer pH 6.0 and 50 mM sodium chloride prepared in 90% H2O and 10% D2O. RDC measurements were performed with samples placed in stretched 3% acrylamide gels.26; 27 Two-dimensional sensitivity- and gradient-enhanced 15N TROSY spectra were acquired at 45°C on a Varian INOVA 600 MHz spectrometer equipped with a cryogenic probe and used to determine the 1H-15N dipolar couplings.28 Resonance assignments of the globular domains were obtained by comparing spectra of the (EK)4 and (YY)4 PSAMs with those of wild-type OspA and variants for which the complete backbone resonance assignments had been established.29; 30 Because of the presence of repeating sequences and extensive resonance overlaps, only those resonances that can be unambiguously assigned to residues within the globular domains were used for RDC analysis. Details of RDC analysis have been described previously.12
Crystallization and structure determination
Crystals were obtained using the hanging-drop vapor diffusion method. Crystallization conditions are listed in Table 2. X-ray diffraction data were collected using the Advanced Photon Source at the Argonne National Laboratory. Structures were determined by molecular replacement using the N- and C-terminal domains (residues 23–131 and 132–273, respectively) of the PSAM structures 2G8C and 2HKD. Model building and refinements were performed in the same manner as the wild-type (EK) PSAMs.12
Structure analysis
The parameters Twist, Bend and Bend' were determined as described.12 The alignment and RMSD calculations were performed using CCP4.31 Dihedral angles were determined using Dang (kinemage.biochem.duke.edu). All structure images were generated with PyMOL (www.pymol.org).
Accession Codes
All structures have been deposited in the Protein Data Bank under the accession codes 2OY8, 2OY7, 2OYB.
Supplementary Material
Acknowledgements
We thank V. Terechko for assistance in x-ray diffraction data collection. This work was supported in part by NIH grant R01-GM72688, NSF grant CMMI-0709079 and by the University of Chicago Cancer Research Center. MB was supported by NIH grant T90-DK070076 and the Paul K. Richter and Evalyn E. Cobb Richter Memorial Fund. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract numbers DE-AC02-06CH11357 and W-31-109-ENG-38. Use of the SER-CAT Sector 22-ID was supported by institutions listed at www.sercat.org/members.html. Use of the GM/CA CAT Sector 23-ID has been funded by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–90. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Xing Y, Higuchi K. Amyloid fibril proteins. Mech Ageing Dev. 2002;123:1625–36. doi: 10.1016/s0047-6374(02)00098-2. [DOI] [PubMed] [Google Scholar]
- 3.Scheibel T, Parthasarathy R, Sawicki G, Lin XM, Jaeger H, Lindquist SL. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci U S A. 2003;100:4527–32. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolfson DN, Ryadnov MG. Peptide-based fibrous biomaterials: Some things old, new and borrowed. Curr Opin Chem Biol. 2006;10:559–67. doi: 10.1016/j.cbpa.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–5. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 7.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–8. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–7. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 9.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci U S A. 2005;102:315–20. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai HH, Gunasekaran K, Nussinov R. Sequence and structure analysis of parallel beta helices: implication for constructing amyloid structural models. Structure. 2006;14:1059–72. doi: 10.1016/j.str.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Betts S, Haase-Pettingell C, Cook K, King J. Buried hydrophobic side-chains essential for the folding of the parallel beta-helix domains of the P22 tailspike. Protein Sci. 2004;13:2291–303. doi: 10.1110/ps.04676704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makabe K, McElheny D, Tereshko V, Hilyard A, Gawlak G, Yan S, Koide A, Koide S. Atomic structures of peptide self-assembly mimics. Proc Natl Acad Sci U S A. 2006;103:17753–8. doi: 10.1073/pnas.0606690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. Faseb J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 14.Gazit E. Mechanisms of amyloid fibril self-assembly and inhibition. Model short peptides as a key research tool. Febs J. 2005;272:5971–8. doi: 10.1111/j.1742-4658.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- 15.Jack E, Newsome M, Stockley PG, Radford SE, Middleton DA. The organization of aromatic side groups in an amyloid fibril probed by solid-state 2H and 19F NMR spectroscopy. J Am Chem Soc. 2006;128:8098–9. doi: 10.1021/ja0581898. [DOI] [PubMed] [Google Scholar]
- 16.Tartaglia GG, Cavalli A, Pellarin R, Caflisch A. The role of aromaticity, exposed surface, and dipole moment in determining protein aggregation rates. Protein Sci. 2004;13:1939–41. doi: 10.1110/ps.04663504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Lei H, Duan Y. The role of Phe in the formation of well-ordered oligomers of amyloidogenic hexapeptide (NFGAIL) observed in molecular dynamics simulations with explicit solvent. Biophys J. 2005;88:2897–906. doi: 10.1529/biophysj.104.055574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bemporad F, Taddei N, Stefani M, Chiti F. Assessing the role of aromatic residues in the amyloid aggregation of human muscle acylphosphatase. Protein Sci. 2006;15:862–70. doi: 10.1110/ps.051915806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan S, Gawlak G, Smith J, Silver L, Koide A, Koide S. Conformational heterogeneity of an equilibrium folding intermediate quantified and mapped by scanning mutagenesis. J Mol Biol. 2004;338:811–25. doi: 10.1016/j.jmb.2004.02.063. [DOI] [PubMed] [Google Scholar]
- 20.Pawley NH, Koide S, Nicholson LK. Backbone dynamics and thermodynamics of Borrelia outer surface protein A. J Mol Biol. 2002;324:991–1002. doi: 10.1016/s0022-2836(02)01146-4. [DOI] [PubMed] [Google Scholar]
- 21.Yan S, Gawlak G, Makabe K, Tereshko V, Koide A, Koide S. Hydrophobic Surface Burial Is the Major Stability Determinant of a Flat, Single-layer beta-Sheet. J Mol Biol. 2007;368:230–43. doi: 10.1016/j.jmb.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makabe K, Tereshko V, Gawlak G, Yan S, Koide S. Atomic-resolution crystal structure of Borrelia burgdorferi outer surface protein A via surface engineering. Protein Sci. 2006;15:1907–14. doi: 10.1110/ps.062246706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marek P, Abedini A, Song B, Kanungo M, Johnson ME, Gupta R, Zaman W, Wong SS, Raleigh DP. Aromatic Interactions Are Not Required for Amyloid Fibril Formation by Islet Amyloid Polypeptide but Do Influence the Rate of Fibril Formation and Fibril Morphology. Biochemistry. 2007 doi: 10.1021/bi0621967. [DOI] [PubMed] [Google Scholar]
- 24.Haspel N, Zanuy D, Ma B, Wolfson H, Nussinov R. A comparative study of amyloid fibril formation by residues 15-19 of the human calcitonin hormone: a single beta-sheet model with a small hydrophobic core. J Mol Biol. 2005;345:1213–27. doi: 10.1016/j.jmb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Tsai HH, Reches M, Tsai CJ, Gunasekaran K, Gazit E, Nussinov R. Energy landscape of amyloidogenic peptide oligomerization by parallel-tempering molecular dynamics simulation: significant role of Asn ladder. Proc Natl Acad Sci U S A. 2005;102:8174–9. doi: 10.1073/pnas.0408653102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii Y, Markus MA, Tycko R. Controlling residual dipolar couplings in high-resolution NMR of proteins by strain induced alignment in a gel. J Biomol NMR. 2001;21:141–51. doi: 10.1023/a:1012417721455. [DOI] [PubMed] [Google Scholar]
- 27.Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J Biomol NMR. 2001;21:377–82. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 28.Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc. 1998;120:10778–9. [Google Scholar]
- 29.Pham T-N, Koide S. NMR studies of Borrelia burgdorferi OspA, a 28 kDa protein containing a single-layer β-sheet. J. Biomol. NMR. 1998;11:407–414. doi: 10.1023/a:1008246908142. [DOI] [PubMed] [Google Scholar]
- 30.Koide S, Huang X, Link K, Koide A, Bu Z, Engelman DM. Design of single-layer beta-sheets without a hydrophobic core. Nature. 2000;403:456–460. doi: 10.1038/35000255. [DOI] [PubMed] [Google Scholar]
- 31.Collaborative-Computational-Project The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallography. 1994;D 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol. 1999;285:1711–33. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.