Abstract
Recent studies have shown that highly simplified interaction surfaces consisting of combinations of just two amino acids, Tyr and Ser, exhibit high affinity and specificity. The high functional levels of such minimalist interfaces might thus indicate small contributions of greater amino acid diversity seen in natural interfaces. Toward addressing this issue, we have produced a pair of binding proteins built on the fibronectin type III scaffold, termed “monobodies”. One monobody contains the Tyr/Ser binary-code interface (termed YS) and the other contains an expanded amino acid diversity interface (YSX), but both bind to an identical target, maltose binding protein (MBP). The YSX monobody bound with higher affinity, a slower off rate and a more favorable enthalpic contribution than the YS monobody. High-resolution x-ray crystal structures revealed that both proteins bound to an essentially identical epitope, providing a unique opportunity to directly investigate the role of amino acid diversity in a protein interaction interface. Surprisingly, Tyr still dominates the YSX paratope and the additional amino acid types are primarily used to conformationally optimize contacts made by tyrosines. Scanning mutagenesis showed that while all contacting Tyr side-chains are essential in the YS monobody, the YSX interface was more tolerant to mutations. These results suggest that the conformational, not chemical, diversity of additional types of amino acids provided higher functionality and evolutionary robustness, supporting the dominant role of Tyr and the importance of conformational diversity in forming protein interaction interfaces.
Keywords: Protein Engineering, Molecular Recognition, Scanning Mutagenesis, Antibody Mimic, Binding Hot Spot
Introduction
In natural protein-protein interfaces, combinations of all 20 genetically encoded amino acids are used to create the structural and chemical complementarity required for interaction. This high level of amino acid diversity has generally been considered essential for generating highly functional interfaces. However, recent studies have shown that high-affinity, specific protein-protein interactions can be generated using much smaller sets of amino acids.
Fellouse et al. have demonstrated that high-affinity antibody antigen-binding fragments (Fabs) can be generated using only a subset of the twenty amino acids in the complementarity determining regions (CDRs).1; 2 In the most extreme case, binary sequences of only Tyr and Ser residues in four CDRs were sufficient to produce tight and specific Fabs to a number of targets.1 The effectiveness of Tyr/Ser (YS) binary interfaces is general to even non-immunological proteins, as we have demonstrated using a single-domain β-sandwich scaffold, the tenth fibronectin type III domain of human fibronectin (FNfn10).3 Using this scaffold we have generated binary binding proteins, which we call “monobodies”. 4; 5 These YS monobodies recognize targets with remarkably high affinity and specificity, despite using only two amino acid types at as few as 16 positions in just two loops.
Given the high level of functionality exhibited by YS-binary paratopes (following a convention in the antibody field, the target side of the interface will be referred to as the epitope and the antibody/monobody side as the paratope), the role played by higher levels of amino acid diversity in natural protein-protein interfaces is unclear. Notably though, while Tyr often plays a dominant role in antigen recognition by natural antibodies and it is highly enriched in the antibody paratopes as well as in the naïve immune repertoire, the remaining fraction is broadly distributed over other amino acid types.6; 7; 8; 9 This amino acid distribution suggests an important role for amino acid diversity in constructing effective Tyr-dominated paratopes.
In this work, we investigate the role of amino acid diversity in protein-protein recognition using monobodies as a model system. We produced a pair of monobodies that use different degrees of amino acid diversity in their paratopes to recognize an identical epitope on their target protein, maltose binding protein (MBP). This unique scenario allows us to investigate how the properties of the protein-protein interface change as a function of paratope amino acid diversity. Importantly, because the epitope is held effectively constant, it is possible to isolate paratope amino acid diversity as the dominant variable differentiating the properties of the two interfaces. Here we dissect the binding kinetics and thermodynamics of the YS and YSX monobodies. High-resolution x-ray crystal structures of both monobodies provide rationales for observed functional differences of the two paratopes. Further, we examine the energetics of the YS and YSX interfaces using scanning mutagenesis. This in-depth analysis provides new insights into the role that amino acid diversity plays in fabricating the architecture and functionality of protein-protein interfaces.
Results
Selection of YSX Monobodies
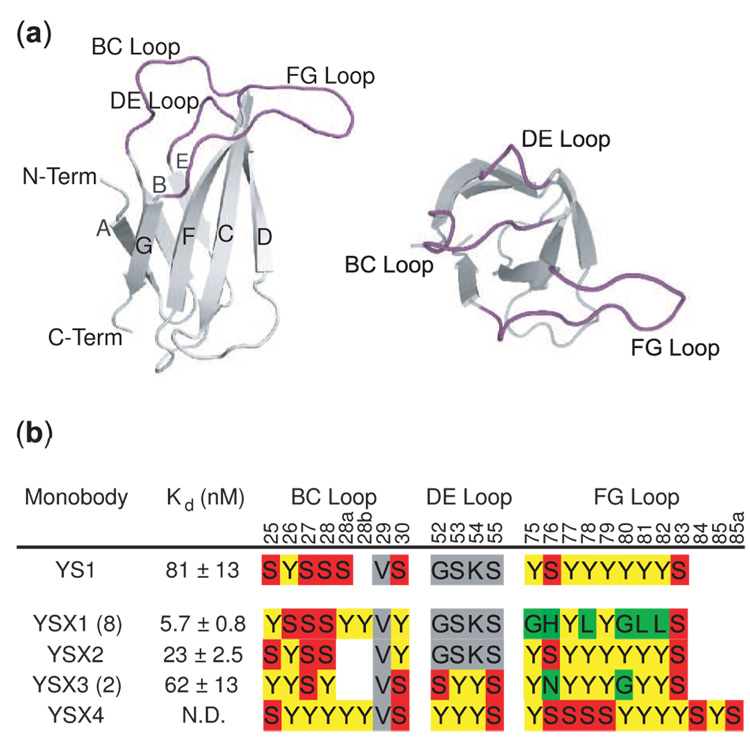
Previously, we produced monobodies with YS-only binding motifs to a number of targets.3 YS-only monobodies to MBP have been particularly well characterized both structurally and functionally. Here, we isolated monobodies to MBP from a combinatorial phage display library in which three loops of the FNfn10 scaffold were diversified in length and sequence using a mixture of 40% Y, 20% S, 10% G, and 5% each of R, L, H, D, N, A (Figure 1A). The fractions of Y and S were maintained at a high level because of their proven functionality.1; 3; 10 However, the amino acid palette was expanded by adding Gly to increase conformational diversity, Arg and His to provide positive charges, Asp to provide negative charge, Asn to provide polar non-charged surface and Ala and Leu to provide non-polar surface. This diversified but restricted amino acid mixture allowed for the sampling of representative chemical characters while keeping the library size reasonably small. After cycles of library selection for binding to MBP, we identified four unique clones (YSX1-4, Figure 1B). All four binding sequences were heavily enriched in Tyr and Ser and two clones (YSX2 and YSX4) were exclusively YS, evidencing the efficacy of the YS binary code in forming binding motifs. Interestingly, the highest affinity clone, YSX1, contained six non-YS amino acids in the FG loop, and its sequence is distinctly different from those of the YS-only monobodies. Furthermore, its affinity (Kd = 5.7 nM, measured by yeast surface display) surpassed the highest affinity for a YS-only monobody to MBP, showing that for MBP, the added amino acid diversity of the YSX library facilitated degrees of affinity not attainable using the YS-binary code.
Figure 1.
The FNfn10 scaffold and loop sequences of MBP-binding monobodies. (a) A schematic of the FNIII scaffold. The seven β-strands are labeled A–G and three loops used for interface design (BC, DE and FG) are labeled. (b) The amino acid sequences of the loop regions of YS-only monobody YS1 and monobodies from the YSX library. For the latter group, the number of times each sequence was recovered is indicated in parentheses. The dissociation constant for MBP as measured by yeast display is also given. Unmutated residues originating from the mutagenesis template are colored gray.
Monobody YSX1 shows distinct loop sequences and lengths compared to one of the highest affinity (yeast display Kd = 81 nM) YS-only binders to MBP, a monobody which we refer to here as YS1 (Figure 1B). YS1 was referred to in previous work as MBP-74.3 We consider YS1 to be representative of the optimum YS-only monobody to MBP because of its clear dominance in a number of independent selection trials that exhaustively sampled the encoded diversity of the YS-only library (J. Wojcik, AK and SK, unpublished data). Although a limited number of YS-only binders exhibited higher affinity than YS1 as measured using yeast surface display (e.g. YSX2, Figure 1B), characterization of two of these clones, including YSX2, as free proteins revealed complicated binding kinetics and low apparent affinity. Denaturant titrations confirmed that each of these proteins was properly folded with stability comparable to the functional YS1 and YSX1 monbodies (data not shown), demonstrating that the low affinity was not due to protein instability. Because of their complex behavior, we did not pursue these clones further. Moreover, the FG loop motif found in YS1 persists among these few observed YS-only monobodies of higher affinity, suggesting that the binding mode of YS1 is conserved among high-affinity YS-only monobodies. Competitive binding experiments revealed that both YS1 and YSX1 were inhibited by β-cyclodextrin, an MBP ligand that binds to the sugar-binding pocket (data not shown). As described below, x-ray crystal structures of the YS1 and YSX1 complexes with MBP reveal that the two monobodies bind to nearly identical epitopes. This shared epitope makes YS1 and YSX1 a particularly interesting pair for analyzing how different levels of amino acid diversity affect molecular recognition.
Comparison of Binding Kinetics and Thermodynamics
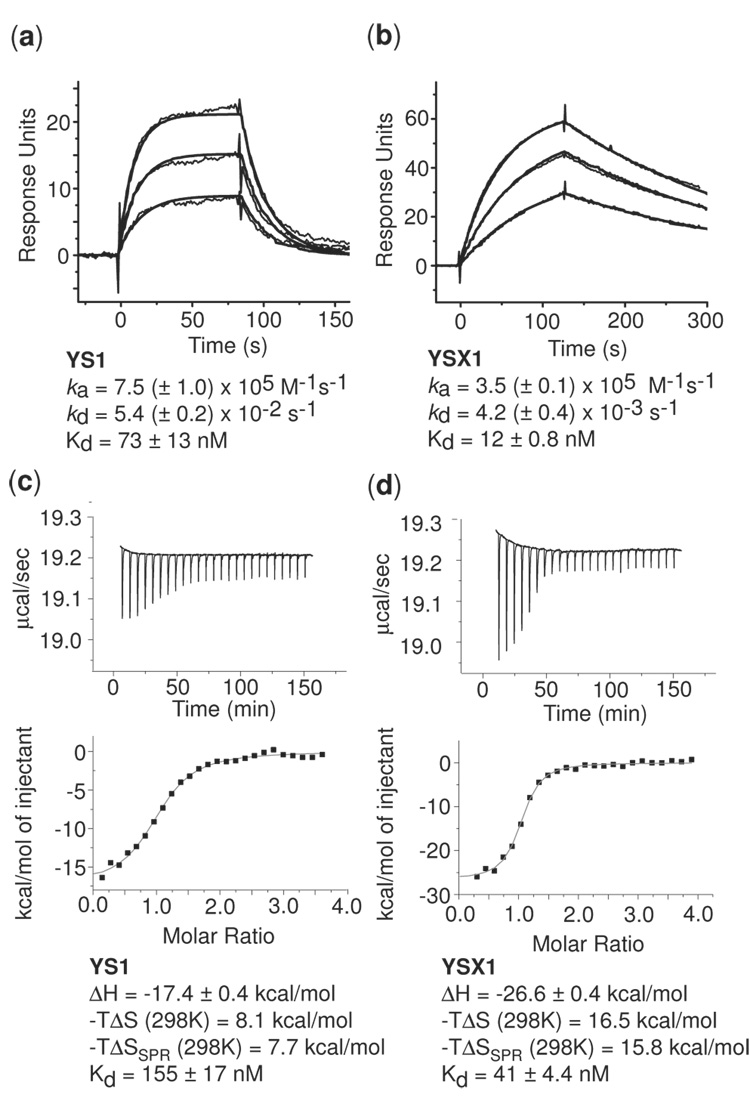
We studied binding kinetics using surface plasmon resonance (SPR) (Figure 2A), and the Kd values for YS1 and YSX1 were measured as 73 nM and 12 nM, respectively. These values were in good agreement with those measured by yeast display (81 nM and 5.7 nM; Figure 1B). In addition to their affinity differences, YS1 and YSX1 exhibited distinctly different kinetic parameters for binding to MBP. The off rate of YSX1 was more than 10-fold slower than that of YS1, and the on rate was ~2-fold slower.
Figure 2.
Binding kinetics and thermodynamics of the YS1 and YSX1 monobodies. (a) SPR traces for YS1 (left) and YSX1 (right). Kinetic parameters are also given. (b) Isothermal titration calorimetry data for YS1 (left) and YSX1 (right). The top panels show the measured heat as a function of time, and the bottom panels show the integrated heats plotted as a function of the molar ratio of MBP to monobody. The curves shown in the bottom panels are fits of a 1:1 binding model. Corresponding thermodynamic parameters are also given including –TΔS values inferred using dissociation constants measured by SPR.
We next compared the thermodynamic parameters of YS1 and YSX1 binding using isothermal titration calorimetry (ITC) (Figure 2B). The titration data for both monobodies fit exclusively to a one site binding mechanism, and by this method, the Kd of YSX1 was measured as 41 nM and that of YS1 as 156 nM. These Kd values are somewhat larger than those obtained by either yeast display or SPR methods. This discrepancy is likely due in large part to error introduced by undersampling the binding isotherm at low molar ratios. Instrument sensitivity limitations, however, precluded further optimization of the ITC experiments to determine more accurate Kd values. Experiments revealed a 9.2 kcal/mol more favorable enthalpy change upon binding for YSX1 compared to YS1, and an 8.4 kcal/mol less favorable entropy change (TΔS at 25 °C). Using the Kd values determined from by SPR, which may be more accurate than those measured by ITC, changed the difference in TΔS only slightly to 8.1 kcal/mol, indicating that the inaccuracies in the ITC-derived Kd are quite small compared to the thermodynamic differences between YS1 and YSX1. These large enthalpy and entropy differences mostly compensate, but the more favorable enthalpy change resulted in higher affinity of YSX1 relative to YS1.
High-Resolution Crystal Structures of the YSX1 and YS1 Monobodies
To understand the structural origins of the enhanced affinity and distinct binding kinetics and thermodynamics of YSX1 compared with YS1, we determined the crystal structures of the YS1/MBP and YSX1/MBP complexes at 1.8 Å and 2.0 Å, respectively (Table 1). Both complexes were crystallized as fusion proteins of the monobody and MBP, a strategy that we previously developed for maintaining 1:1 stoichiometry in the crystal lattice and facilitating the formation of higher order structure.3 The model of the YS1/MBP complex reported here is improved compared to the previously reported model at 2.35 Å resolution.3
Table 1.
Crystallographic Information
| YS1(PDB ID: 3CSG) | YSX1(PDB ID: 3CSB) | |
|---|---|---|
| Data Collection | ||
| Space Group | P41 | P41212 |
| Cell Parameters | a = b = 68.58 Å, c = 108.00 Ǻ α = β = γ = 90° | a = b = 98.82 Å, c = 134.62 Ǻ α = β = γ = 90° |
| Wavelength | 1.0 Å | 1.0 Å |
| Resolution | 50.00 − 1.80 (1.86 − 1.80) Å | 50.00 − 2.00 (2.07 − 2.00) Å |
| Unique Reflections | 46,107 | 45,823 |
| RMerge | 0.051 (0.620) | 0.058 (0.692) |
| Completeness | 99.7 % (99.9 %) | 100.0 % (100.0 %) |
| Redundancy | 3.3 (3.3) | 9.9 (9.9) |
| I/σ(I) | 28.1 (1.7) | 29.9 (3.4) |
| Refinement Statistics | ||
| Resolution Range | 20.00 − 1.80 Å (1.85 − 1.80 Å) | 20.00 − 2.00 Å (2.05 − 2.00 Å) |
| Unique Reflections | ||
| Working Set | 41,432 | 41,103 |
| Free Set | 4,615 | 4,569 |
| Rcryst | 0.187 | 0.195 |
| Rfree | 0.235 | 0.235 |
| Overall Mean B Values | 37.94 Å2 | 37.55 Å2 |
| Number of Amino Acid Residues | 458 | 464 |
| Number of Water Molecules | 285 | 213 |
| Matthews Coefficient | 2.31 (water content 46.8%) | 2.99 (water content 58.9%) |
| RMSD from Ideal Values | ||
| Bonds/Angles | 0.02 Å / 1.7° | 0.02 Å / 1.5° |
| Estimated Overall Coordinate Error Based on Maximum Likelihood | 0.1 Å | 0.2 Å |
| Estimated Overall Error for B Values Based on Maximum Likelihood | 6.7 Å2 | 6.5 Å2 |
| Ramachandran Plot Statistics | ||
| Residues in Most Favored Regions | 91.1 % | 90.9 % |
| Residues in Additional Allowed Regions | 8.6 % | 8.6 % |
| Residues in Generously Allowed Regions | 0.3 % | 0.3 % |
| Residues in Disallowed Regions | 0.0 % | 0.3 % |
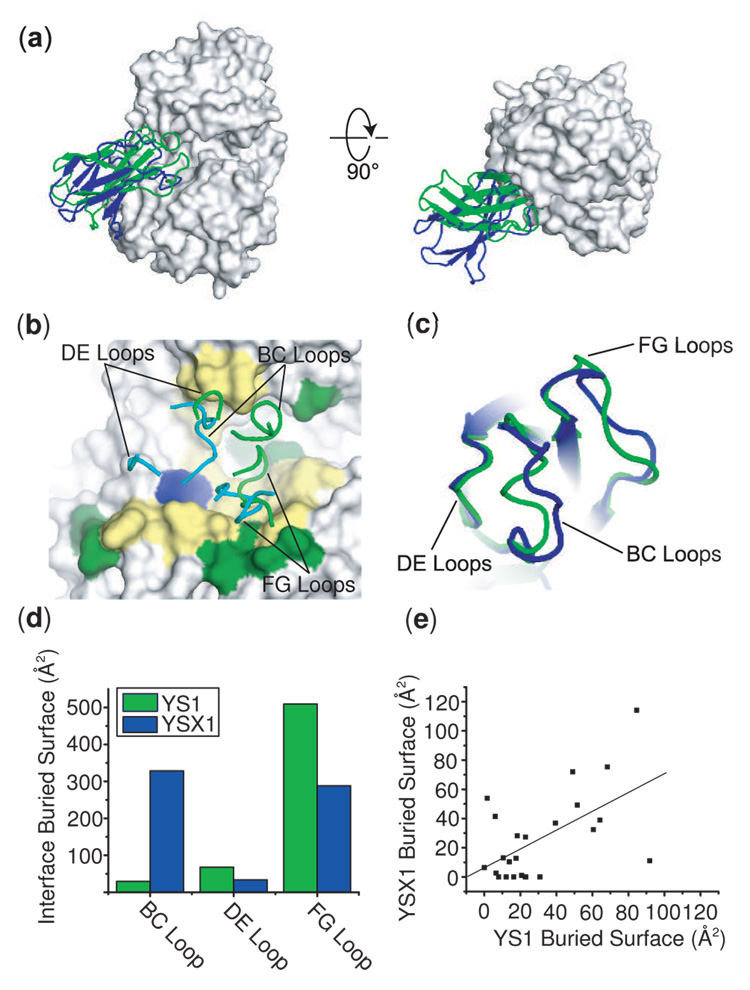
As inhibition by β-cyclodextrin indicated, the crystal structures show that both YS1 and YSX1 bind to the sugar-binding pocket of MBP. Both YS1 and YSX1 bind by insertion of a convex surface into the pocket cavity (Figure 3A). This epitope convergence despite the absence of any specific selective pressure to target monobodies to the pocket is a strong example of a previously noted preference of small antibody-like proteins to bind to concave surfaces.11; 12 The MBP molecule is in an open conformation similar to that observed in the MBP/β-cyclodextrin complex (PDB ID 1DMB), and maintains an effectively identical conformation in both monobody complexes with a Cα RMSD of 0.76 Å. The scaffold portions of the monobodies also superimpose quite well (Cα RMSD = 1.2 Å), indicating that the sequence variation in the binding loops has not perturbed the overall structure of either molecule. Although both YS1 and YSX1 occupy the sugar-binding pocket, their precise binding modes and binding loop orientations with respect to the pocket surface are distinct (Figure 3B). Consistent with their dissimilar amino acid sequences, the backbone conformations of the YS1 and YSX1 binding loops are also different (Figure 3C).
Figure 3.
X-ray crystal structures of the YS1 and YSX1 monobodies in complex with MBP. (a) Overall views of YS1 (green) and YSX1 (blue) bound to MBP (surface representation) in two different orientations. The two structures were superimposed using the MBP molecules. (b) Orientations of the binding loops of YS1 (green) and YSX1 (cyan) with respect to the sugar binding pocket of MBP. Yellow surfaces show the shared epitopes of YS1 and YSX1, green the epitope unique to YS1, and blue unique to YSX1. (c) The backbone conformations of YS1 (green) and YSX1 loops (blue). The scaffold portions of the monobodies are aligned. (d) The buried surface area contributions of each of the binding loops to the interface. (e) A correlation plot of the interface area contributed by the epitope residues in the YS1 and YSX1 complexes. The line shows a linear fit of the data, which gives a correlation value of 0.65.
The extent to which each of the binding loops contacts MBP differs between YS1 and YSX1. Both monobodies use the FG loop to interact with the “bottom” lobe of MBP, but YS1 relies on this contact much more heavily, resulting in an effectively single-loop mediated interaction (Figure 3D). In contrast, YSX1 also uses the BC loop to form a large portion of the interface. The DE loops, which retain the template sequence in both monobodies, make only minor contributions to the interface.
The most striking feature of this pair of structures is the extent of similarity in the epitopes for binding to YS1 and YSX1. The surface areas buried by the two monobodies in the interface are similar (733 Å2 for YS1 and 699 Å2 for YSX), although this comparison is complicated due to a portion of the YS1 interface made by residues in the scaffold, which previous work suggested to be an artifact of crystal packing.3 This size similarity, however, does indicate that the increased affinity of YSX1 is not due to a larger interface. In addition to size similarity, more than 90% of the MBP residues in the YSX1 epitope are also in the YS1 epitope. The all-atom RMSD of the shared epitope residues is 1.6 Å with the majority of variation localized to a few side chains, indicating that the epitopes are essentially identical both conformationally and chemically. Remarkably, there is also a significant correlation in the extent to which epitope residues are buried at the interfaces for YS1 and YSX1. This indicates that the shared epitope residues are even contacted to a similar extent (Figure 3E).
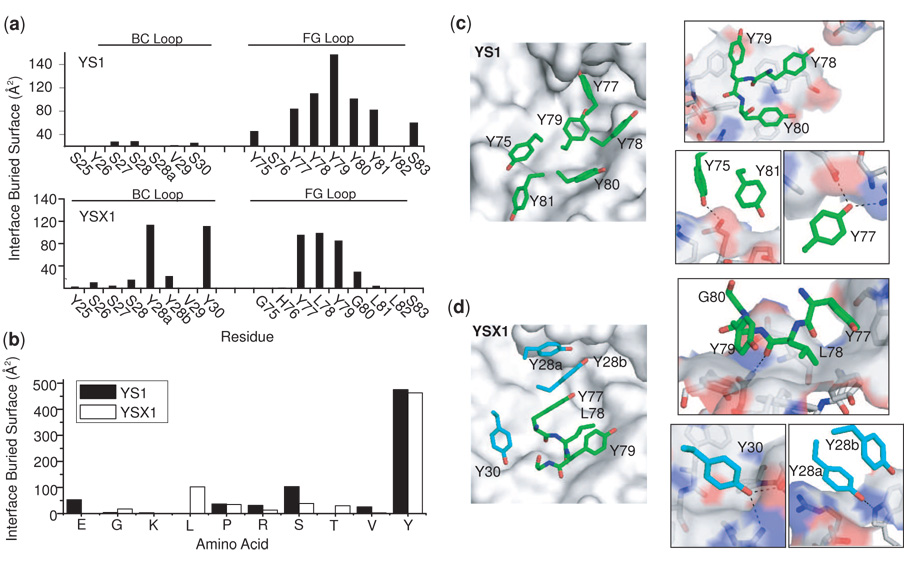
Although YS1 and YSX1 have clearly disparate binding loop sequences and additional amino acids were available in the selection of YSX1, both monobodies rely essentially on Tyr to contact their shared epitope (Figure 4A and 4B), However, this chemically similar paratope is organized differently in the two interfaces (Figure 4C and D). In the YS1 interface, Tyr side chains are splayed out flat along the surface, making stacking interactions with aromatic side chains of MBP and contacting predominantly non-polar surface. In the YSX1 interface, in contrast, Tyr side chains form more edge-plane contacts with aromatics and also interact with charged Arg side-chains. Interestingly, the YSX1 paratope results in a more tightly packed interface as measured by several metrics. The surface correlation (SC) value13 of an interface reflects how well the two surfaces complement each other geometrically, and it ranges from 0 to 1 with 1 indicating perfect shape complementarity with no gap. The YSX1 interface has a surface correlation (SC) value of 0.74 while the YS1 interface has a significantly lower value of 0.64. When considering only the packing of Tyr side chains, YSX1 gives a value of 0.78 while YS1 gives just 0.68. The better packing of the YSX1 interface is also reflected by its higher ligand efficiency (LE).14 For small molecules, the LE is defined as the free energy of binding divided by the number of atoms in the molecule. Here, the parameter is defined similarly, but with the number of atoms restricted to contacting atoms (those with epitope atoms within 4 Å or less). The LE for YSX1 is measured as 0.23 kcal mol−1 atom−1 whereas that of YS1 is 0.18. YSX1 also buries more surface area per contacting atom than does YS1 (15.2 Å2 versus 13.8 Å2). Because the YS1 interface includes scaffold contacts which are likely an artifact of crystal packing,3 inclusion of this surface in interface analysis may not accurately reflect the properties of the engineered interface. Likewise, the YSX1 interface includes some scaffold contribution. However, omission of these scaffold contacts from the calculations still results in a YSX1 SC of 0.73 versus 0.66 for YS1, a YSX1 LE of 0.29 kcal mol−1 atom−1 versus 0.23 kcal mol−1 atom−1 for YS1 and a YSX1 buried surface/contact atom of 17.6 Å2 versus 14.13 Å2 for YS1. Taken together, these measures indicate that the increased amino acid diversity of YSX1 has allowed for a more efficient packing of the interface, particularly of Tyr residues. This conformational role of the additional amino acid diversity is exemplified by two Gly residues in the FG loop of YSX1. One adopts a positive phi angle and the other is buried to the α-carbon, and neither configuration is achievable with other amino acids. These two positions provide clear examples of how the expanded diversity of the YSX library has been exploited to conformationally optimize the interface.
Figure 4.
The paratope structures of the YS1 and YSX1 monobodies. (a) The surface area buried at the interface of individual residues in the BC and FG loops of YS1 and YSX1. (b) The interface buried surface area contributed by each amino type to the YS1 and YSX1 paratopes. (c, d) Structural details of the YS1 and YSX1 interfaces. The left panels show an overall view of the arrangement of major interface contacts and the right panels show close-up views of these interactions. In these illustrations MBP is shown as a gray surface/sticks and contacting monobody paratope residues are shown as sticks. The carbon atoms of the FG loop residues are colored green, and those of the BC loop residues colored cyan. Putative hydrogen bonds are indicated by dashed lines.
Analysis of Interface Energetics by Shotgun Scanning Mutagenesis
To further characterize the YS1 and YSX1 interfaces, we investigated whether these two interfaces were constructed similarly from an energetic standpoint. We first examined this qualitatively by conducting small-scale shotgun scanning mutagenesis experiments. We constructed combinatorial libraries in which the sequence of either the BC loop or FG loop of each monobody was randomized to a subset of amino acids (Table 2) while the other was held to the original sequence. This small library was then sorted for binding-competent clones, and the sequences of those clones were analyzed. While shotgun scanning mutagenesis has been previously used to quantitatively assess the energetic consequences (ΔΔGbinding) of mutation by sequencing a very large number of clones,15 our intention was to coarsely evaluate how tolerant a given position is to substitution and to what extent certain amino acids are preferred there. We report the use of site-directed alanine scanning mutagenesis to quantitatively assess the energetic importance of individual positions in the following section, which complements the shotgun analysis.
Table 2.
Amino Acid Variaiton in Shotgun Scanning Mutagenesis
| Original Amino Acid | Codona | Amino Acids Sampled in Shotgun Scanning Mutagenesis |
|---|---|---|
| Y | KHT | Y,A,D,S,V or F |
| S | KCT | S or A |
| G | GST | G or A |
| L | KYG | L,A,S,V |
| H | BMC | V,F,H,P,D,A |
K = G and T; H = A,Cand T; S = C and G; Y= C and T; B = C,G and T; M = A and C
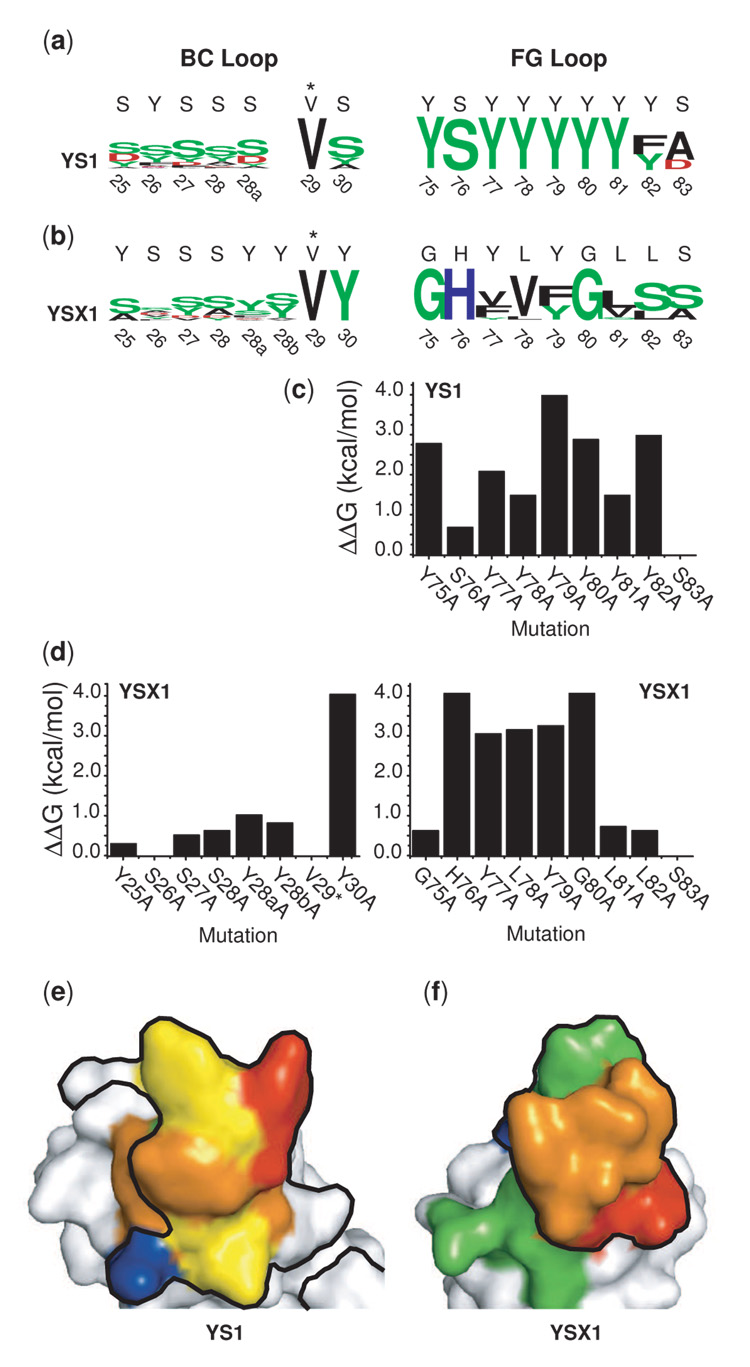
In the shotgun scanning experiments, the BC Loops for both YS1 and YSX1 were relatively robust to mutation, showing little conservation of amino acid identity at most positions (Figure 5a and 5b). Consistent with these results, the crystal structure shows that YS1 makes only minor contact through the BC loop and the observed contact is nearly exclusively backbone mediated. YSX1 however, makes much more extensive contact to the target surface through the BC loop. For instance, Tyr30 of the YSX1 BC loop is highly buried at the interface (Figure 4a). Consistent with this role, this position exhibits a strong preference for Tyr in the shotgun experiments. Interestingly, another BC loop Tyr residue, Tyr28a, is also highly buried in the crystal structure and contributes 18% of the total paratope buried surface (Figure 4a), yet this position shows little preference for Tyr, suggesting that the YSX1 interface is somehow able to accommodate the elimination of the phenolic moiety.
Figure 5.
Interface energetics of the YS1 and YSX1 monobodies. (a) Results of shotgun scanning mutagenesis shown in the sequence logo format.28; 29 In this format, the overall height of the stack of letters at each position reflects the level of consensus and the height of any one letter reflects the prevalence of that amino acid in the dataset. Amino acids are colored according to their chemical properties. Green for polar amino acids (G,S,T,Y,C,Q,N), Blue for basic (K,R,H), Red for acidic (D,E) and black for hydrophobic (A,V,L,I,P,W,F,M). Above each position in the sequence logo is the original amino acid at that position. (b) Free energy changes (ΔΔGbinding values) for paratope Ala mutants as measured by SPR. (c) A heat map representation of alanine scanning results projected onto the YS1 and YSX1 monobody surfaces. Residues for which Ala mutations increased the dissociation constant by less than 10-fold are colored green, 10 to 100-fold: yellow, 100 to 1000-fold: orange, and greater than 1000-fold: red. Those which do not significantly affect the dissociation constant are colored blue. The black outline encompasses contacting residues, defined as those that bury <5 Å2 of surface in the interface.
In contrast to its highly mutable BC loop, the FG loop of YS1 is extremely sensitive to mutation (Figure 5a). The original amino acid is completely conserved at every position except the last two, where a conservative Phe substitution is tolerated at position 82 and mutation of Ser to either Ala or Asp is favored at position 83. Remarkably, none of the other YS1 FG loop tyrosines could be replaced by Phe while maintaining binding. In contrast, the FG loop of YSX1 shows significant tolerance for substitution even at positions that make extensive contact with the target (Figure 5b). For example, the central YLY motif that forms the majority of the interface contacts is able to accommodate several other amino acid types and even accommodates mutations at all three of these positions simultaneously. However, another three residues (Gly75, His76 and Gly80) were completely conserved, and interestingly these were all newly introduced amino acid types in the YSX library.
Analysis of Binding Energetics by Alanine Scanning Mutagenesis
Complimenting shotgun scanning analysis, we performed alanine scanning mutagenesis for a quantitative assessment of the energetic importance of residues in each of the binding interfaces. Because the crystal structure and shotgun scanning experiments suggested that the BC loop of YS1 did not significantly contribute to binding, it was not investigated here.
In both monobodies, Ala mutations at a majority of positions in the FG loops resulted in significant energetic effects (Figure 5c–f). YS1 was especially sensitive: seven of the nine Ala mutations resulted in affinity decreases of greater than 10-fold, including positions which made no contact with the target surface. In contrast, the FG loop of YSX1 exhibited several positions which produced only mild negative effects on mutation, and critical residues were focused to the central YLY motif which forms the bulk of the contacting surface.
The BC loop of YSX1 was robust to alanine mutation, with the exception of Tyr30 which, as mentioned earlier, is heavily buried in the interface. Interestingly, the alanine scanning experiments again indicated that mutation of Tyr28a had very little effect on binding affinity despite the original residue’s heavy contribution to the interface in the crystal structure.
Discussion
Here we present functional and structural studies of two monobodies that use different amino acid sets to bind to the same epitope, thereby providing an unparalleled opportunity to elucidate the role that amino acid diversity plays in the construction of protein-protein interfaces. Because epitope composition is held constant in both interfaces, it is possible to isolate amino acid diversity of the monobody paratope as the dominant source of the functional and structural differences in the two interfaces.
The YSX1/MBP complex possesses a Tyr-rich paratope in spite of an expanded amino acid sequence space. High-resolution structural information revealed that YSX1’s enhanced affinity compared to YS1 originates from an optimized packing of this Tyr-rich chemistry facilitated by non-YS amino acids. Such optimization is reflected by several metrics of interface packing, including surface correlation values and ligand efficiency values.13; 14 These results support an important role for amino acid diversity in conformational optimization of a subgroup of amino acids that are well suited for forming contacts at protein-protein interfaces (e.g. Tyr). Related to our findings, Fellouse et al. recently reported that, for synthetic Fab libraries, expansion of the amino acid repertoire beyond the YS-binary code increased library performance.16 Structural analysis indicated that in spite of this expanded diversity, Tyr was still sufficient to form most of the Fab’s contacts to its target, human vascular endothelial growth factor (hVEGF). However, the relatively low resolution of this structure and the absence of a YS-binary counterpart precluded in-depth analysis. Also, a more discrete analysis of synthetic Fab libraries revealed that the addition of Gly to the YS-binary code led to a highly functional library yielding highly specific binders with Kd values in the subnanomolar range.17 These results again emphasize the efficacy of Ser and Tyr in forming functional interfaces, but also highlight the improvement of functionality facilitated by the addition of conformational diversity into the minimalist context. Our present results suggest that expanding the conformational diversity beyond the YSG-ternary code further improves the functionality of synthetic interfaces.
The more tightly packed interface of the YSX1/MBP complex rationalizes the reduced off rate and increased enthalpic gain upon binding observed for this complex compared with the YS1/MBP complex. Optimization of van der Waal’s interactions increases enthalpic favorability and stabilizes the bound state leading to a slowed off rate. Interestingly, however, we observe a large compensatory effect for YSX1 in the form of an increased entropic penalty upon binding and a slower association rate. This compensation leads to a small, six-fold difference in the Kd values for YS1 and YSX1. The differences in binding thermodynamics for YS1 and YSX1 suggest a mechanism for affinity increase through expansion of amino acid diversity. Both YS1 and YSX1 bury similar amounts of surface area with similar levels of hydrophobicity (699 and 733 Å2 with 43% and 41% non-polar atoms respectively). Therefore, it is expected that the solvent entropy component to binding is roughly equal in the two binding reactions. This narrows the source of entropic difference to monobody and target conformational entropy. Increased amino acid diversity may, as observed in this case, lead to a more effective complementation of a target surface and a tighter packed interface, which, while enthalpically beneficial, restricts side chain and backbone freedom to a greater extent. Furthermore, addition of flexible residues such as Gly to the monobody paratope may broaden the conformational space in the unbound state. This could simultaneously allow for better target surface complementation on binding, but also lead to less favorable entropy loss on binding and a higher-energy transition state leading to slower kinetics. Further studies will define the structural origins of the thermodynamic differences of these two binding reactions.
The shotgun scanning and alanine scanning experiments reported here for the monobody YS1 (Figure 5) represent the first such analysis of a binary interface in the context of high-resolution structural information. Interestingly, the YS1 paratope had elevated sensitivity to mutation. In most cases, even homologous amino acid replacement was not tolerated. Conversely, the functional sequence space for the YSX1 interface was broader and some positions, which are heavily buried at the interface, tolerated substitution even with non-homologous amino acids (Figure 5). The apparent fragility of the YS1 interface is also in sharp contrast with the high levels of mutation tolerance of a synthetic Fab paratope composed of four amino acids (Tyr, Ala, Asp and Ser).10 In this Fab, many of the positions including interface Tyr residues could be substituted with homologous and/or non-homologous amino acids while maintaining and, in some cases, improving binding affinity.
We speculate that the reason for the unusually high level of mutational sensitivity of the monobody YS1 is that its paratope was selected under conditions where, due to a lack of amino acid diversity, many or all of the contacting residues were unable to be conformationally optimized. This is evidenced by low surface correlation values of the YS1 interface. In such a scenario, selection for high-affinity binding may demand that a large number of such sub-optimal interactions all contribute significantly to binding in order for the molecule to “survive” in the library sorting. These results suggest that YS binary interfaces may lack mutational plasticity and exhibit a certain “brittleness” which is not observed in higher diversity interfaces.
The sensitivity to mutation in YS1 also resulted in a lack of so-called “hotspots” in the interface, as revealed by alanine scanning mutagenesis (Ala mutations at these positions reduced Kd by more than 10-fold) (Fig. 5E). Such regions are frequently identified in natural protein-protein interfaces and are characterized by their disproportionately large contribution to the binding energy with other areas contributing relatively little.18; 19 Instead of this architecture, almost all of the YS1 paratope residues contribute significantly to the binding free energy, and in a more uniform fashion (Figure 5C). In this sense, the entire paratope of YS1 can be viewed as one large hotspot. This expanded hotspot is not observed for YSX1 which exhibits a more typical localized arrangement. We have, however, observed similarly expanded hotspots in other interfaces. In a camelid heavy chain antibody (VHH) raised against RNaseA, affinity maturation by combinatorial library selection (Kd = 180 pM) resulted in an expansion in hot spot size, so that 76% of contacting residues became hot spots compared with 35% in the wild-type paratope.20 We rationalized that in order to produce such high affinity, the affinity-matured antibody was forced by selection to depend on an increasing number of residues to contribute significantly to the total binding energy so that the entire interface needed to be productively involved in binding. Analogously, in the YS1 case, with interface diversity limited to only two amino acid types, the selective pressure may have already been sufficiently high to force a similar expansion of hotspot residues. In contrast, YSX1, with an expanded set of amino acids was able to highly optimize the conformations of a subset of residues, which became hotspots. The binding energy provided by these highly optimized positions, if sufficient to survive in the library sorting, may have reduced the selective pressure on other positions to positively contribute to binding, leading to a localization of binding energy in the interface. One significant difference between YS1 and YSX1 that may partially account for the observed difference in interface architecture, however, is that YSX1 uses two loops for recognition of the target surface while YS1 uses one. Because a mutation is likely to affect another residue within the same loop to a greater extent than a residue in another loop, distributing the binding energy across two separated regions may also serve to make the interface more robust to mutation in both the shotgun and alanine scanning experiments.
Conclusions
In the case of monobodies binding to MBP, increasing amino acid diversity in the protein-protein interface facilitates a level of target affinity not achievable with the YS binary code. This improvement results not from chemical optimization of the interface, but conformational optimization of Tyr-rich chemistry. These results support the view that Tyr is capable of forming a majority of interactions required for high affinity binding as long as a sufficient degree of conformational diversity is provided. This work also revealed the evolutionary brittleness of the YS-only interfaces. The sensitivity to mutation observed in YS1 may be a general property of YS interfaces, and, as seen in the YSX1 paratope, increasing amino acid diversity may alleviate this sensitivity through increased localization of the binding energy to fewer interface residues. Our results rationalize the observed amino acid bias in protein-protein interfaces and in immune repertoires in terms of both binding function and evolutionary robustness.
Materials and Methods
Phage Display Library Construction
Library construction was conducted as described previously.3 The randomization of loop residues in the YSX library was achieved using high-efficiency Kunkel mutagenesis. The mutagenic oligonucleotides used were generated using a custom Trimer Phosphoramidite Mix (Glen Research, Sterling, VA) containing codons in the following molar ratios: 40% Y, 20% S, 10% G, 5% R, H, L, D, N, A. Sorting of the phage displayed library was carried out as described previously.3 After three rounds of sorting, the genes for monobodies in the phagemid library were transferred to a yeast surface display vector, and after one additional round of library sorting using a fluorescence activated cell sorter, individual clones were analyzed as described previously.3
Protein sample preparation
Proteins were expressed with an N-terminal His-tag and purified by high-throughput nickel affinity purification using a KingFisher automated purification system (Thermo Fisher Scientific) or by using a Ni-Sepharose affinity chromatography column (Amersham Pharmacia) as described previously.3
Kinetic Analysis by Surface Plasmon Resonance
Measurements were taken using a BIAcore2000 instrument at 298 K. Purified YS1 and YSX1 monobodies were immobilized via a His-tag to a Nickel-NTA sensor chip (Biacore) so that the response due to immobilization was approximately 30 response unit (RU). MBP was then flowed over the sensor chip at a rate of 30 µL/min and the association and dissociation kinetics were monitored. All data were fit to a 1:1 Langmuir binding model using global fitting of multiple kinetic traces with the BIAevaluation software. Data were taken at four concentrations: 0, 15, 30, and 50 nM. Zero nM traces were used for blank run subtraction. Kinetic parameters reported are the average of triplicate measurements. Buffer conditions were 10 mM HEPES, pH 7.4, 150 mM NaCl, 0.005% Tween20, 50 µM EDTA.
Isothermal Titration Calorimetry
Experiments were conducted using a VP-ITC instrument (MicroCal, Northampton, MA). Buffer conditions were 50 mM phosphate, pH 7.4, 150 mM NaCl, and all experiments were performed at 25°C. Both monobody and MBP samples were exhaustively dialyzed together against the ITC buffer. Titrations were performed using 25 10 µL injections of 30 µM MBP into 1.4 mL of 3 µM monobody. Heats of dilution were corrected for by subtracting the average of the heats measured after the reaction reached saturation. Titration curves were fit using a 1:1 binding model using the Origin 7-based ITC program supplied by the vendor. n-values were forced to 1 during fitting due to difficulty in accurate determination of monobody concentration. The data however, could be exclusively fit to a 1:1 model, and a 1:1 binding mode is supported by surface plasmon resonance, solution NMR, and x-ray crystal structural data.
Crystallization, Data Collection, and Structure Solution
Both YS1 and YSX1 were crystallized as C-terminal fusions to MBP as previously described3. The YSX1 construct included a Gly-Ser-Ser linker between the last residue of MBP and the first residue of the monobody while the YS1 construct did not contain a linker. Both proteins were crystallized by the hanging drop vapor diffusion method. Concentrated protein solution was mixed 1:1 with the well solution in a 2 µL drop volume. YS1 crystallized in 20% polyethyleneglycol-1000, 0.1 M Na/K phosphate buffer, 0.2 M NaCl, pH 6.2. YSX1 crystallized in 41% polyethelyeneglycol-400, 2% 2-methyl-2,4-pentanediol, 50 mM MnCl2, 0.1M MES pH 6.9/MES pH 4.0 in a ratio of 5:1. Data were collected at APS beamline 23 ID-D (Advanced Photon Source at Argonne National Laboratory).
Crystallographic information is given in Table 1. Diffraction data were processed and scaled using the HKL2000 package.21 The structures were solved by molecular replacement using the MOLREP program in the CCP4 program suite.22 For YS1, the previous 2.35 Å structure was used as the search model (PDB ID 2OBG). For YSX1, a multicopy search was performed using MBP and FNfn10 as the search models (PDB IDs 1DMB and 1FNA respectively). Rigid body refinement was carried out using REFMAC5 in the CCP4 program suite.23 CNS 1.1 was used for simulated annealing.24 TLS (Translation/Libration/Screw), B-factor refinement, bulk solvent parameters, final positional refinement, and the search for and refinement of water molecules was carried out using REFMAC5.23 Model building and evaluation were carried out using the Coot program25, and molecular graphics were generated using PyMOL (www.pymol.org). Surface area calculations were performed using the protein-protein interaction server26; 27.
Shotgun Scanning Mutagenesis
Shotgun scanning mutagenesis for YS1 was carried out as described previously.3 The “shaved” FNIII template3 was used in all experiments. For YSX1, eight positions in the BC loop or nine in the FG were randomized using the codons outlined in Table 2 while the wild-type sequence of YSX1 was introduced into the other loop . The DE loop of YSX1 contained the template sequence and was not investigated here. The resulting libraries were subjected to 2–4 rounds of sorting for binding to MBP as previously described.3 Twelve clones were sequenced from the FG loop library and 19 from the BC loop library.
Alanine Scanning Mutagenesis
Alanine scanning mutagenesis was carried out by introducing single Ala mutations at all positions of the YS1 FG loop and all positions of the YSX1 BC and FG loops with exception of V29 in the BC loop which has been previously shown to be important for the stability of the FNIII scaffold.3 All mutations were confirmed by sequencing. SPR experiments were performed as described above except that a flow rate of 20 µL/min was used. Initially, all mutants were screened using 300 nM MBP. If high quality traces were obtained, Kd values were calculated by fitting the kinetic traces. If binding was not detected, concentrations of 1, 5, and 10 µM were tested. If binding was then observed, the saturated portions of the kinetic trace were used to calculate the equilibrium rate constant by plotting a standard saturation curve. If binding was not observed, the affinity of the mutant was considered to be at least 1000-fold reduced compared to the wild-type (> 70 µM for YS1 and > 12 µM for YSX1) and the ΔΔG value estimated at 4 kcal/mol.
Acknowledgments
We thank Drs. V. Tereshko and K. Makabe for their assistance with x-ray structure determination, Dr. E. Solomaha for assistance with ITC experiments. This work was supported in part by NIH grants R01-GM72688, R21-CA132700 and U54 GM74946 and by the University of Chicago Cancer Research Center. RG was supported in part by T32 GM007183-32A1. We acknowledge the use of the DNA sequencing and Flow Cytometry core facilities at the University of Chicago. We thank the staff of the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA CAT) beamline at the Advanced Photon Source. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein Data Bank accession codes
The coordinates and structure factors have been deposited in the PDB with entry code 3CSG and 3CSB for the YS1 and YSX1 complexes, respectively.
References
- 1.Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J Mol Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 2.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci U S A. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc Natl Acad Sci U S A. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 6.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J. Mol. Biol. 1991;217:133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 8.Tsumoto K, Ogasahara K, Ueda Y, Watanabe K, Yutani K, Kumagai I. Role of Tyr residues in the contact region of anti-lysozyme monoclonal antibody HyHEL10 for antigen binding. J Biol Chem. 1995;270:18551–18557. doi: 10.1074/jbc.270.31.18551. [DOI] [PubMed] [Google Scholar]
- 9.Shiroishi M, Tsumoto K, Tanaka Y, Yokota A, Nakanishi T, Kondo H, Kumagai I. Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL. J Biol Chem. 2007;282:6783–6791. doi: 10.1074/jbc.M605197200. [DOI] [PubMed] [Google Scholar]
- 10.Fellouse FA, Barthelemy PA, Kelley RF, Sidhu SS. Tyrosine plays a dominant functional role in the paratope of a synthetic antibody derived from a four amino acid code. J Mol Biol. 2006;357:100–114. doi: 10.1016/j.jmb.2005.11.092. [DOI] [PubMed] [Google Scholar]
- 11.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. Embo J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 15.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, Koide S, Sidhu SS. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 18.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 19.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 20.Koide A, Tereshko V, Uysal S, Margalef K, Kossiakoff AA, Koide S. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J Mol Biol. 2007;373:941–953. doi: 10.1016/j.jmb.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enz. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Jones S, Thornton JM. Protein-protein interactions: a review of protein dimer structures. Prog Biophys Mol Biol. 1995;63:31–65. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]