Abstract
The differences among individual bile acids (BAs) in eliciting different physiological and pathological responses are largely unknown because of the lack of valid and simple analytical methods for the quantification of individual BAs and their taurine and glycine conjugates. Therefore, a simple and sensitive LC-MS/MS method for the simultaneous quantification of 6 major BAs, their glycine, and taurine conjugates in mouse liver, bile, plasma, and urine was developed and validated. One-step sample preparation using solid-phase extraction (for bile and urine) or protein precipitation (for plasma and liver) was used to extract BAs. This method is valid and sensitive with a limit of quantification ranging from 10 to 40 ng/ml for the various analytes, has a large dynamic range (2500), and a short run time (20 min). Detailed BA profiles were obtained from mouse liver, plasma, bile, and urine using this method. Muricholic acid (MCA) and cholic acid (CA) taurine conjugates constituted more than 90% of BAs in liver and bile. BA concentrations in liver were about 300-fold higher than in plasma, and about 180-fold higher in bile than in liver. In summary, a reliable and simple LC-MS/MS method to quantify major BAs and their metabolites was developed and applied to quantify BAs in mouse tissues and fluids.
1. Introduction
Bile acids (BAs) are synthesized in hepatocytes from cholesterol by pathways involving at least 17 different enzymes. In humans, there are 2 primary BAs: cholic acid (CA) and chenodeoxycholic acid (CDCA). In rodents muricholic acid (MCA) is also a primary BA, whereas ursodeoxycholic (UDCA) is a primary BA of bears. The terminal step in BA synthesis before excretion into bile involves BA conjugation with amino acids, specifically taurine or glycine [1].
In the intestine, primary BAs are deconjugated and converted by microflora to secondary bile acids, mainly deoxycholic acid (DCA) and lithocholic acid (LCA). Most of the BAs (95%) in the intestine are reabsorbed into the portal circulation [2]. Once taken up by hepatocytes, BAs are conjugated and excreted into bile to complete the enterohepatic circulation. The majority of BAs are contained in the enterohepatic circulation, and very little exists outside this system under normal conditions [3].
BAs are well known for their role in fat absorption. BAs form micelles with phospholipids, forming an emulsion to aid in the intestinal absorption of fat and fat-soluble vitamins [4]. Recently, BAs have been recognized as signaling molecules, which regulate the expression of several target genes through nuclear receptor biosensors, such as the farnesoid-X-receptor (FXR) and pregnane-X-receptor (PXR) [5,6]. However, BAs are also cytotoxic [7], tumor promoters [8], and cause hepatotoxicity [3].
Individual BAs vary markedly in their physiological and pathophysiological activities, therefore, the nature and extent of these responses are affected by both the concentration and the composition of the BA pool. For example, LCA is the most toxic BA and a potent PXR ligand [9], whereas CDCA is less cytotoxic and a potent FXR ligand [7,10]. The differences among individual BAs in eliciting different physiological and pathological responses in vivo are largely unknown and overlooked, in large part because of the lack of valid analytical methods for the quantification of individual BAs and their conjugates, to link specific BAs to the observed physiological or pathological responses. Separation and quantification of BAs and their conjugates are challenging, because of marked differences in physicochemical properties, the presence of isomeric forms, and their relatively low concentrations in biological samples.
Enzymatic methods using bile acid hydroxysteroid dehydrogenase are widely used for routine analysis of physiological fluids due to their simplicity [11,12]. However, differentiation of individual BAs is not possible using these methods because total BAs, rather than individual BAs, are quantified. Therefore, chromatographic techniques represent the method of choice for detailed analysis of bile acid profiles. Gas chromatography and gas chromatography-mass spectrometry (GC-MS) have been applied for the qualitative and quantitative analysis of bile acids [13–15]. GC-MS provides high sensitivity and resolution of isomeric bile acids. However, this technique is limited by complex sample preparation, derivatization, and most importantly requires the hydrolysis of conjugated BAs into their unconjugated form prior to their analysis [16]. Therefore, despite the lower chromatographic resolution compared to GC, which might not resolve all BA isomers, high-performance liquid chromatography (HPLC)-based assays are more desirable.
HPLC-UV assays have been applied for the determination of bile acids in biological materials [17–21]. The main disadvantages of these methods are the limited sensitivity and specificity of UV detection in complex biological matrices, such as tissues [16]. HPLC methods, coupled with more sensitive and selective detection techniques such as fluorescence, have also been used. However, these methods are limited by the complexity of sample derivatization with fluorescing chemicals [22,23].
The persistent need for rapid and sensitive methods has motivated efforts to exploit the high sensitivity, specificity, and sample preparation-convenience of HPLC-tandem mass spectrometry (LC-MS/MS) for bile acid analysis in biological fluids. Methods using fast atom bombardment (FAB-MS) [24] and electrospray ionization (ESI-MS) were developed and applied for bile acid analysis [24–26]. FAB-MS, however, has inferior quantitative capabilities and yields less intact ions (more in-source fragmentation) compared to ESI [27]. Therefore, ESI-LC-MS remains a powerful technique for direct quantitative analysis of BAs in biological matrices.
Several methods have been developed and used to quantify bile acids in biological matrices using ESI-LC-MS [27–32]. These methods provided valuable data, which deepened our understanding of the various roles of BAs in biological systems. However, most of these methods were not validated for reliability and reproducibility [29,31–34], were performed utilizing calibration curves prepared in solution rather than the biological matrix to be analyzed [27], used single MS detection rather than the more specific and sensitive MS/MS [31–34], had relatively high limits of quantification [34], had limited dynamic range [19,30], were designed to quantify BAs only in plasma or serum [19,27,29–31], only quantify conjugated BAs [32], and/or had very long run times [19,31]. In addition, methods based on direct injection into MS with no chromatography were used, which does not allow the differentiation of isomeric BAs [25,35].
In summary, there is currently no simple, sensitive, direct, and valid method available for investigators to quantify individual BAs and their conjugates in various tissues and fluids. Therefore, the purpose of this study is to develop and validate a sensitive and simple LC-MS/MS method for the simultaneous quantification of the major BAs, as well as their glycine and taurine conjugates in mouse liver, bile, plasma, and urine. The method presented in this paper is selective, sensitive with limits of quantification ranging from 10 to 40 ng/ml for various analytes, has a large dynamic range (from 10 ng/ml to 100 µg/ml for the various analytes), and was validated to have high precision and accuracy (< 15%). This method was applied to quantify BAs in mouse fluids and tissues.
2. Experimental
1.1. Chemicals and Reagents
Cholic acid (CA), chenodeoxy cholic acid (CDCA), deoxycholic acid (DCA), lithocholic acid (LCA), ursodeoxycholic acid (UDCA), tauro-cholic acid (T-CA), tauro-chenodeoxycholic acid (T-CDCA), tauro-deoxycholic acid (T-DCA), tauro-lithocholic acid (T-LCA), tauro-ursodeoxycholic acid (T-UDCA), glyco-cholic acid (G-CA), glyco-chenodeoxycholic acid (G-CDCA), glyco-deoxycholic acid (G-DCA), glyco-lithocholic acid (G-LCA), and glyco-ursodeoxycholic acid (G-UDCA) were purchased from Sigma-Aldrich (St Louis, MO). Activated charcoal was also obtained from Sigma-Aldrich. β-muricholic acid (β-MCA), α muricholic acid (α -MCA), tauro-β-muricholic acid (T-β-MCA), and tauro-α-muricholic acid (T-α-MCA) were purchased from Steraloids, Inc. (Newport, Rhode Island). 2H4-G-CDCA and 2H4-CDCA were purchased from C/D/N ISOTOPES, INC. (Pointe-Claire, Quebec, Canada). HPLC-grade methanol, acetonitrile, water, ammonium acetate, ammonium formate, ammonium hydroxide, formic acid, and acetic acid were obtained from Fisher Scientific (St Louis, MO). Oasis HLB SPE cartridges were purchased from Waters (Milford, MA).
1.2. Instrumentation
A Waters ACQUITY ultra performance LC system (Waters, Milford, MA) was used throughout. The mass spectrometer was a Waters Quattro Premier XE triple quadrupole instrument with an ESI source (Waters, Milford, MA). The entire LC-MS system is controlled by MassLynx 4.1 software. All chromatographic separations were performed with an ACQUITY UPLC C18 column (1.7 µm, 100 × 2.1 mm I.D.) equipped with an ACQUITY UPLC C18 guard column (Waters, Milford, MA).
1.3. Liquid chromatographic and mass spectrometric conditions
The mobile phase consisted of 5% acetonitrile (ACN) in methanol (MeOH) (mobile phase A) and 7.5 mM ammonium acetate, adjusted to pH 4 using 10 M acetic acid (mobile phase B), at a total flow rate of 0.3 ml/min. The effect of mobile phase on chromatography and MS signal, and the gradient profile for the LC pumps under the final chromatography conditions are illustrated in Figure 1. A built-in switching valve was used to direct the LC flow to the MS instrument from 3–18 min during each run and to waste for the rest of the run time and between runs. The injection volume of all samples was 10 µl.
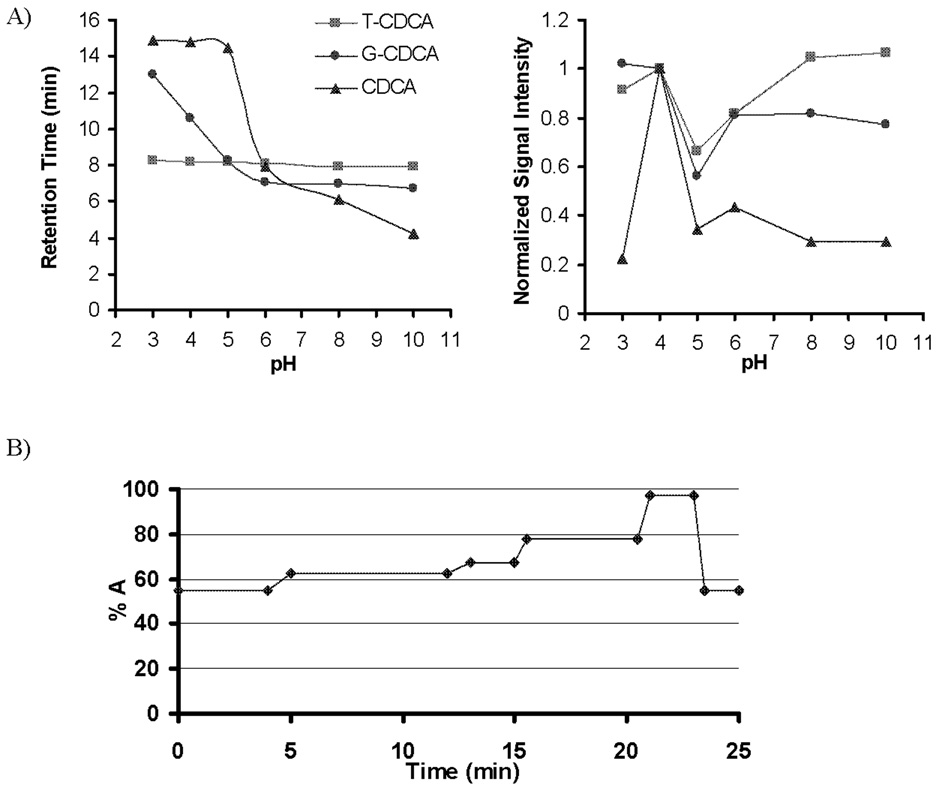
Figure 1.
A) Effect of mobile phase pH and composition on the retention time and signal sensitivity. Ammonium formate was used at pH 3 and 5, ammonium acetate at pH 4 and 6, ammonium bicarbonate at pH 7 and 8, and ammonium carbonate at pH 9 and 10, at 7.5 mM each. B) The solvent gradient profile of the HPLC pumps at the final chromatography conditions. The mobile phase consisted of 5% acetonitrile (ACN) in methanol (MeOH) (mobile phase A) and 7.5 mM ammonium acetate adjusted to pH 4 using 10 M acetic acid (mobile phase B) at a total flow rate of 0.3 ml/min.
The mass spectrometer was operated with the source and desolvation temperatures set at 120 and 375°C, respectively. Glycine conjugates (G-BAs) were detected in the positive ionization mode, whereas taurine conjugates (T-BAs) and unconjugated BAs were detected in the negative mode. The capillary, extractor, and RF voltages were 3000 V, 4 V, and 0 V, respectively. The source and desolvation gases (nitrogen) were set at a flow rate of 650 and 75 L/h, respectively. The cone voltages were 20, 65, and 90 V for G-BAs, T-BAs, and unconjugated BAs, respectively. Collision energies were 15, 70, and 15 eV for glyco-, tauro-, and unconjugated BAs, respectively. The multiple reaction monitoring (MRM) transitions are shown in Figure 2. 2H4-G-CDCA and 2H4-CDCA were used as internal standards (ISs) for analytes in the positive (G-BAs) and in the negative (T-BAs, BAs) modes, respectively.
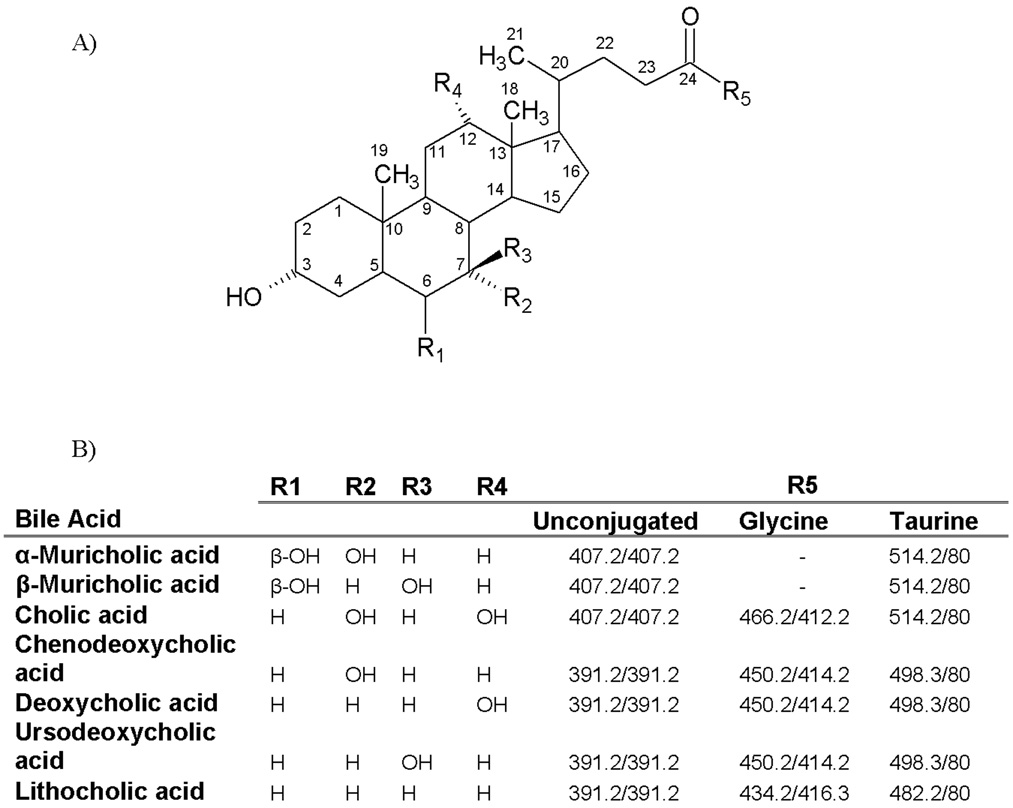
Figure 2.
A) Backbone and side chain structures of the 6 major BAs, as well as their glycine and taurine conjugates in mice. B) Parent and fragment masses used for quantification of BAs, their conjugate metabolites, and internal standards using tandem MS.
1.4. Preparation of Standard Solutions and Calibration Curves
Ten mg/ml stock solutions of BAs and ISs were individually prepared in water: MeOH (1:1). Liver, bile, plasma, and urine were collected and each pooled from 5 untreated mice. Livers were homogenized in deionized water (1:2 w/v). Homogenized liver, plasma, bile, and urine were incubated with 100 mg/ml activated charcoal for 1 hr to strip these matrices of endogenous BAs. Mixtures were centrifuged at 13,000 g for 10 min, and the supernatants were filtered. The filtrates of these stripped matrices were used to construct the calibration curves, each in the corresponding biological matrix to be analyzed. Fixed volumes of these stripped matrices were spiked with 20 µl of the appropriate standard solution containing ISs to construct a calibration curve with the range of 0.04–100, 0.02–50, 0.01–25 µg/ml for T-BAs, unconjugated BAs, and G-BAs, respectively. The concentration of the 2H4-G-CDCA and 2H4-CDCA ISs were 4 and 2 µg/ml, respectively.
1.5. Sample Extraction
Several solid-phase extraction and protein-precipitation techniques were investigated for sample clean-up. Solid-phase extraction (SPE) using C18, C2, Oasis-HLB, and Oasis-MCX cartridges were investigated. For plasma samples, simple protein precipitation using ice-cold ACN was used. 1 ml of ice-cold ACN was added to 100 µl serum-spiked with 20 µl ISs, vortexed, and centrifuged at 11,000 g for 10 min. The supernatant was aspirated, evaporated under vacuum, and reconstituted in 100 µl of 50% MeOH. Liver samples were extracted by protein precipitation using alkaline ice-cold ACN (5% NH4OH in ACN). Approximately 100 mg of liver was homogenized in 2 volumes of 50% MeOH. 300 µl of liver homogenate was spiked with 20 µl ISs, 2 ml of ice-cold alkaline ACN was added, vortexed, shaked continuously for 1 hr, and centrifuged at 11,000 g for 10 min. The supernatant was aspirated and precipitant was extracted with another 1 ml of ice-cold alkaline ACN. Supernatants from the 2 extraction steps were pooled, evaporated, and reconstituted in a 100 µl of 50% MeOH.
For bile samples, Oasis-HLB SPE cartridges were used for sample extraction. Bile samples were diluted 100 fold with deionized water, 100 µl of diluted bile samples were spiked with 20 µl ISs, vortexed, and loaded onto SPE cartridges pre-conditioned with 1 ml MeOH, followed by 1 ml H2O. Loaded cartridges were washed with 2 ml H2O and eluted with 2 ml MeOH. The eluate was evaporated under vacuum and reconstituted in 100 µl of 50% MeOH. Urine samples (50 µl) were spiked with 20 µl of ISs and prepared similarly as bile samples. For all samples, evaporation was under vacuum and at room temperature. In average, samples took 4–5 hours to evaporate to dryness.
Extraction recoveries were determined for each quality control (QC) point in each matrix as from the ratio of the analyte peak area in samples spiked before extraction compared to the corresponding peak area in untreated samples prepared in neat solution.
1.6. Method Validation
The method was validated using 5 QC points for each calibration curve. Five replicates of each QC point were analyzed each day to determine the intra- and inter-day accuracy and precision. This process was repeated 3 times over 3 days in order to determine the inter-day accuracy and precision using freshly prepared calibration curves. Intra-day accuracy and precision were calculated from the % bias [% (Measured – Theoritical)/Measured concentrations] and relative standard deviation [%RSD = % Standard Deviation/ Mean], respectively, for the 5 replicates of each QC point. Inter-day accuracy and precision were calculated similarly for the 15 replicates of each QC point pooled from the 3 validation runs. The concentrations of the QC points were 0.04, 0.1, 5, 50, and 100 µg/ml for T-BAs, 0.02, 0.05, 2.5, 25, and 50 µg/ml for unconjugated BAs, and 0.01, 0.025, 1.25, 12.5, and 25 µg/ml for G-BAs.
1.7. Animal Studies
Eight-week-old male C57BL/6 mice were purchased from Charles River Laboratories Inc (Wilmington, MA). Animals were housed in a temperature-, light-, and humidity-contolled environment. Mice were fed Laboratory Rodent Chow W (Harlan Teklad, Madison, WI) ad libitum. To obtain urine samples, mice (N=5) were placed in metabolic cages and urine were collected for 24 hrs. The same set of mice was anesthetized i.p.using ketamine (100 mg/kg)/midazolam (5 mg/kg) and the common bile duct was cannulated with a 30-gauge needle attached to PE-10 tubing. Bile was collected from the cannula for 2 hrs at 15-min intervals. Another set of mice (N=5) were anesthetized, a PE-10 cannula was placed in the carotid artery, and approximately 700 µl of blood was collected into heparanized tubes from each mouse. Blood samples were centrifuged at 1000 g for 5 min, and plasma collected. Livers were harvested from the same animals, gallbladders were removed, and livers were washed, frozen in liquid nitrogen, and stored at −80 ° C until time of analysis.
3. Results and Discussion
3.1. Method Development
Figure 2 shows the chemical structure, as well as parent, and fragment masses of the 6 major BAs, as well as their glycine and taurine conjugates. T-BAs and unconjugated BAs were analyzed using the negative mode, whereas G-BAs were analyzed in the positive mode. The major G-BA fragments were formed by (H2O)n loss (n=3 for tri-hydroxy, =2 for di-hydroxy, =1 for mono-hydrox G-BAs). SO3− was the major fragment for all T-BAs, whereas unconjugated BAs did not yield any major fragment, and therefore the same mass was monitored for both parent and fragment ions (Figure 2).
Hydrophilicity and therefore retention on a reversed-phase C18 column is influenced by both the BA nucleus and side chain structures. Therefore, tri-hydroxy BAs (CA, MCA) elute earlier than di-hydroxy BAs (CDCA, and DCA), which in turn elute earlier than the mono-hydroxy BA (LCA). However, retention time is also determined by the position and stereochemistry of hydroxyl groups, where the UDCA (di-hydroxy BA) elutes earlier than CA (tri-hydroxy BA). The di-hydroxy BAs elute in the order of UDCA, CDCA, followed by DCA. This elution behavior may be attributed to the orientation of the hydroxyl substitutions and their ability to form intra-molecular H-bonding [36].
To optimize the chromatographic separation, various mobile phases with various counter-ions in the pH range of 3–10 were evaluated (Figure 1.A). T-BAs (pKa ~ 1.5) are always ionized in the pH range of 3–10 used in chromatographic analysis, whereas G-BAs are primarily in the unionized form at pHs lower than the pKa of ~ 4.5. Unconjugated BAs have a pKa of approximately 6 [37]. Therefore, decreasing the pH of the mobile phase markedly increases the retention of G-BAs and unconjugated BAs, which improves their chromatographic separation with minimal influence on T-BAs. The nature of the counter-ion, i.e. formate vs. acetate vs. carbonate also plays a role in the effect of mobile phase on BA chromatographic and MS behaviors.
Ionization efficiency and therefore signal intensity in ESI is strongly dependent on mobile phase constituents and pH (Figure 1.A). Using ammonium acetate in the HPLC buffer yielded a higher signal than ammonium formate. Ammonium bicarbonate and carbonate buffers decreased the signal intensity for G-BAs and unconjugated BAs, but slightly increased the signal intensity for T-BAs. Ammonium acetate buffer at pH 4 yielded the highest MS signal intensity and chromatographic retention for all analytes. Furthermore, under these conditions, all isomeric BAs of interest were resolved from each other in less than 20 min except α and β isomers of T-MCA (Figure 3). α and β isomers of T-MCA could be separated by using less organic in the mobile phase, but the run time becomes longer than 40 minutes (data not shown). Figure 4 shows a representative chromatogram of mouse bile analyzed under the final extraction and chromatography conditions.
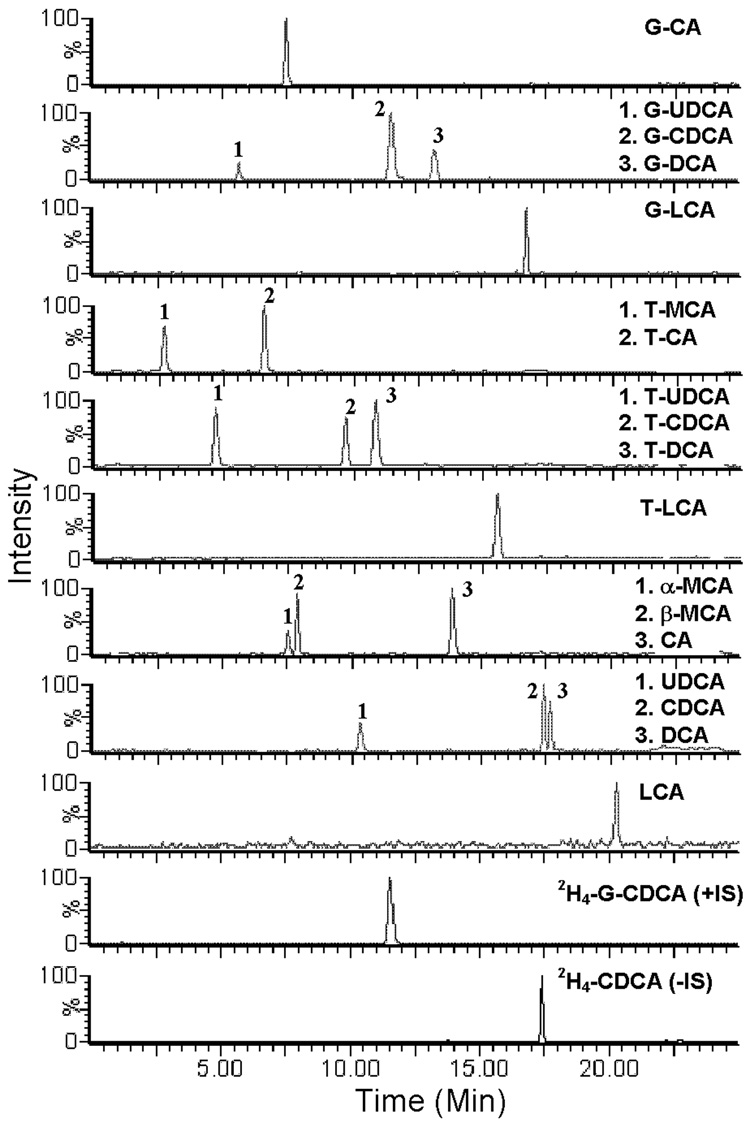
Figure 3.
Representative chromatogram at the lowest limit of quantification of a mixure of G-BAs (10 ng/ml) T-BAs (40 ng/ml), unconjugated BAs (2 0ng/ml), and the 2 ISs (+IS= 0.4 µg/ml, −IS= 4 µg/ml) under the final chromatography and detection conditions.
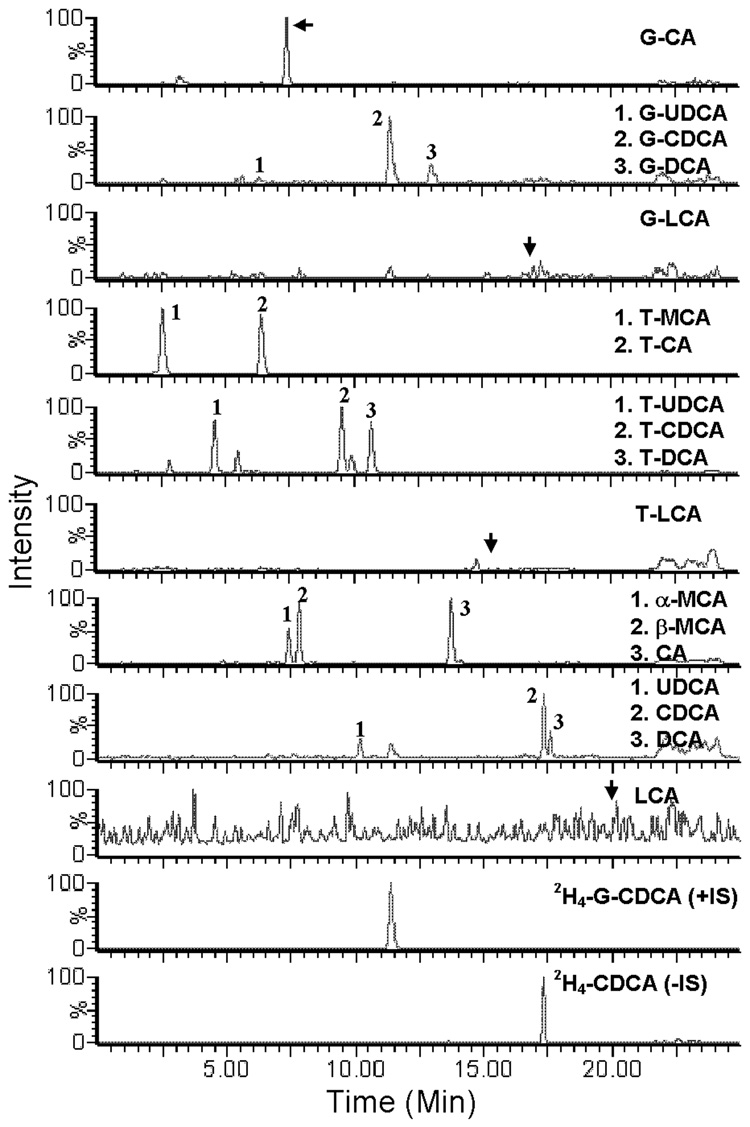
Figure 4.
Representative chromatogram of BAs in mouse bile.
To quantify BAs using ESI-MS, calibration and quality control samples were prepared and in the same biological matrices to be analyzed; this is to compensate for analytes lost during sample extraction and ion suppression/enhancement in the ESI source. Because BAs are endogenous compounds, there are no BA-free biological matrices. The method of standard addition was investigated, where calibration curves were prepared in the matrices of interest and corrected for the endogenous content of these matrices by subtracting the background in unspiked samples [38]. However, this method was neither accurate nor precise. Alternatively, stable-labeled isotopes can be used to build the calibration curve for each analyte because they have the same retention time and MS response. However, only a few of these stable-isotopes are commercially available and they are expensive. Therefore, blank matrices free of BAs were prepared by stripping BAs from the matrices of interest using activated charcoal [39]. These stripped matrices are quite different than their original counterparts, and might result in different ion suppression/enhancement effect as well as extraction recoveries of analytes. However, in our study, the 2 stable-labeled isotope internal standards have similar extraction recoveries from the stripped matrices compared to the original matrices (data not shown). Therefore, we assume this applies to all the other BA analytes in this study.
Several protein-precipitation and SPE methods were investigated to increase extraction recovery and decrease suppression effect of the matrix. The large variation in physicochemical properties between conjugated and unconjugated BAs caused different extraction efficiencies of these compounds. Table 1 shows the average extraction recoveries (5 QC points) of all G-BAs, TBAs, and unconjugated BAs in mouse plasma, bile, liver, and urine. Extraction recoveries were higher than 80% for all analytes in the 4 matrices. Furthermore, extraction recoveries were consistent throughout the calibration range.
Table 1 .
Extraction recoveries of BAs, T-BAs, and G-BAs in mouse fluids and tissues.
| Liver | Bile | Plasma | Urine | |
|---|---|---|---|---|
| G-CA | 93.9 ± 14.1 | 77.3 ± 14.2 | 85.4 ± 7.22 | 83.6 ± 9.51 |
| G-UDCA | 96.2 ± 7.82 | 81.1 ± 7.21 | 87.1 ± 9.13 | 85.7 ± 8.15 |
| G-CDCA | 94.3 ± 7.35 | 81.3 ± 11.1 | 88.7 ± 5.77 | 87.7 ± 3.32 |
| G-DCA | 98.6 ± 9.05 | 84.6 ± 7.23 | 88.8 ± 7.01 | 86.7 ± 13.4 |
| G-LCA | 92.1 ± 2.13 | 86.9 ± 8.36 | 90.1 ± 12.2 | 89.9 ± 7.58 |
| T-MCA | 85.2 ± 8.23 | 79.3 ± 12.3 | 82.6 ± 11.1 | 82.4 ± 9.81 |
| T-CA | 92.5 ± 12.3 | 78.9 ± 14.2 | 85.6 ± 14.2 | 84.1 ± 13.3 |
| T-UDCA | 87.1 ± 4.22 | 84.5 ± 7.41 | 88.2 ± 18.1 | 82.6 ± 3.22 |
| T-CDCA | 91.2 ± 7.61 | 89.6 ± 4.75 | 89.4 ± 17.2 | 84.5 ± 12.7 |
| T-DCA | 90.2 ± 13.5 | 90.3 ± 7.53 | 91.2 ± 9.32 | 88.1 ± 3.22 |
| T-LCA | 92.5 ± 8.45 | 92.6 ± 9.31 | 93.3 ± 13.9 | 89.5 ± 8.44 |
| αMCA | 95.2 ± 7.72 | 88.2 ± 6.56 | 90.2 ± 13.3 | 84.7 ± 12.8 |
| βMCA | 93.3 ± 13.8 | 89.6 ± 5.42 | 90.7 ± 8.37 | 84.9 ± 12.8 |
| CA | 95.1 ± 12.9 | 90.1 ± 6.11 | 93.6 ± 14.1 | 85.1 ± 9.43 |
| UDCA | 96.4 ± 18.9 | 94.3 ± 6.63 | 94.5 ± 8.88 | 87.4 ± 6.58 |
| CDCA | 97.4 ± 19.8 | 92.6 ± 6.28 | 96.2 ± 9.41 | 90.1 ± 5.49 |
| DCA | 94.6 ± 19.1 | 93.6 ± 8.51 | 95.1 ± 8.32 | 91.5 ± 7.21 |
| LCA | 95.3 ± 12.1 | 92.7 ± 12.2 | 97.7 ± 7.29 | 93.4 ± 9.32 |
Data are presented as the average of 5 QC concentrations, 5 samples each ± %RSD.
3.2. Method Validation
To ensure the method reliability and reproducibility for BA analysis, intra-day (data not shown) and inter-day accuracy and precision were determined using 5 QC concentrations distributed throughout the calibration range for each analyte in each matrix. Table 2 shows the inter-day accuracy and precision data. Precision and accuracy for all analytes were less than 15%. The limit of quantification for G-BAs, unconjugated BAs, and T-BAs in all 4 matrices were 10, 20, and 40 ng/ml, respectively. The limits of detection (signal/noise ratio=3) for the various BAs in all 4 matrices after extraction were in the range of 2–5 ng/ml. Furthermore, storage stability of stock solutions in the freezer and extracted biological samples in the autosampler were tested. BAs were stable for at least 3 months in the −20 °C freezer, and 36 hrs in the 4 °C autosampler (data not shown).
Table 2.
Summary of the inter-day accuracy and precision
| QC1 | QC2 | QC3 | QC4 | QC5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nominal Conc | 10 (ng/ml) | %RSD | 25 (ng/ml) | %RSD | 1.25 (µg/ml) | %RSD | 12.5 (µg/ml) | %RSD | 25 (µg/ml) | %RSD |
| G-CA | 10.0 | 13.7 | 23.5 | 14.2 | 1.1 | 14.2 | 12.0 | 14.1 | 24.7 | 12.2 |
| G-UDCA | 9.1 | 10.6 | 25.1 | 9.8 | 1.3 | 9.8 | 13.0 | 10.3 | 27.3 | 5.2 |
| G-CDCA | 10.0 | 9.1 | 23.3 | 8.4 | 1.3 | 8.4 | 11.7 | 9.5 | 24.8 | 12.5 |
| G-DCA | 9.6 | 11.9 | 23.9 | 10.4 | 1.3 | 10.4 | 11.8 | 8.4 | 23.1 | 7.5 |
| G-LCA | 9.5 | 9.9 | 26.8 | 6.9 | 1.4 | 6.9 | 11.9 | 10.1 | 24.8 | 9.11 |
| Nominal Conc | 40 (ng/ml) | %RSD | 100 (ng/ml) | %RSD | 5 (µg/ml) | %RSD | 50 (µg/ml) | %RSD | 100 (µg/ml) | %RSD |
| T-MCA | 42.8 | 7.1 | 102.4 | 13.2 | 5.1 | 13.2 | 47.8 | 9.8 | 98.8 | 9.2 |
| T-CA | 42.1 | 13.4 | 101.7 | 2.3 | 5.1 | 2.3 | 47.7 | 6.7 | 106.3 | 6.4 |
| T-UDCA | 44.7 | 3.27 | 116.5 | 2.85 | 5.5 | 2.85 | 55.9 | 2.45 | 103.9 | 16.03 |
| T-CDCA | 41.7 | 6.7 | 49.0 | 0.7 | 4.4 | 0.7 | 42.7 | 3.9 | 97.8 | 9.9 |
| T-DCA | 43.9 | 4.6 | 105.4 | 2.2 | 4.4 | 2.2 | 50.8 | 7.6 | 92.4 | 9.7 |
| T-LCA | 36.8 | 5.7 | 111.7 | 3.9 | 5.2 | 3.9 | 56.3 | 5 | 108.3 | 2.9 |
| Nominal Conc | 20 (ng/ml) | %RSD | 50 (ng/ml) | %RSD | 2.5 (µg/ml) | %RSD | 25 (µg/ml) | %RSD | 50 (µg/ml) | %RSD |
| αMCA | 21.7 | 2.6 | 43.9 | 3.8 | 2.3 | 3.8 | 26.2 | 9 | 47.7 | 2.2 |
| βMCA | 17.1 | 14.9 | 47.5 | 3.8 | 2.5 | 3.8 | 21.4 | 6.2 | 45.0 | 1.7 |
| CA | 18.4 | 3.7 | 46.7 | 5 | 2.7 | 5 | 24.9 | 9.9 | 49.1 | 6.4 |
| UDCA | 19.9 | 2 | 49.2 | 5.7 | 2.5 | 5.7 | 26.4 | 3.3 | 51.7 | 4.3 |
| CDCA | 21.1 | 1.8 | 50.6 | 7.7 | 2.6 | 7.7 | 24.3 | 11.8 | 52.1 | 8.1 |
| DCA | 21.2 | 8.5 | 46.9 | 8.5 | 2.6 | 8.5 | 24.1 | 1.96 | 48.7 | 12 |
| LCA | 18.7 | 11.1 | 52.5 | 3.6 | 2.5 | 3.6 | 25.6 | 12.1 | 50.8 | 8.2 |
Results are calculated from the 3 validation trials of calibration curves prepared in bile.
3.3. Mouse BA Profiles
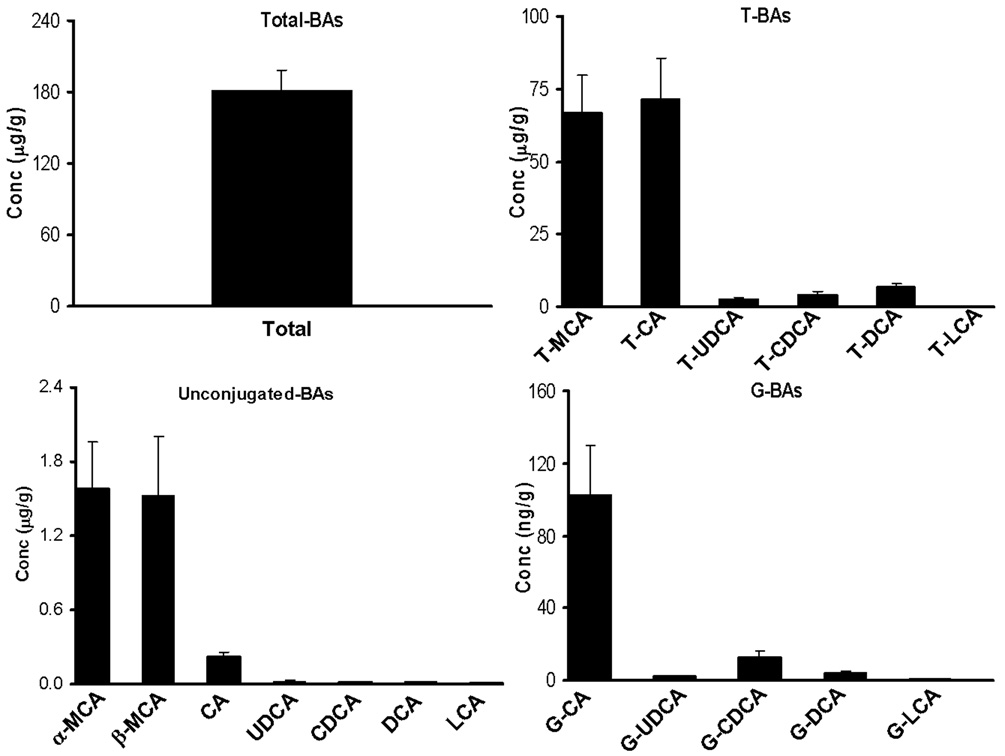
BA profiles in mouse liver, bile, plasma, and urine were characterized using this LC-MS/MS method. Total BA concentration in mouse liver was 181.3 µg/g (Fig 5). The taurine conjugates of CA and MCA constitute 44% each of the total hepatic BAs (Table 3). Of the BAs in liver, 97.7% are taurine conjugates, 2.2% are unconjugated, and less than 0.1% are glycine conjugates (Table 3). In contrast to humans and rats, the enzyme responsible for BA conjugation with amino acids in mice is specific to taurine, rather than glycine conjugation [40]. Therefore, G-BAs are major components of the BA pool in humans and rats, but not in mice. Previous studies report highly variable total BA concentrations in mouse liver, from 0.04–1.2 µmol/g (20–618 µg/g) [33,41–48]. This large variability can be attributed to the various analytical methodologies used for BA analysis, the different mouse strains studied, and the method by which livers were collected from the animals. Because BAs are at least 100-fold more concentrated in bile than liver, any contamination with gall-bladder bile during collection of liver tissues can lead to misleading BA concentrations in liver.
Figure 5.
BA concentrations in mouse liver. The results are shown as mean (N=5) ± S.E.M.
Table 3.
Percentages of various BAs in liver, bile, and plasma.
| Liver | Bile | Plasma | |
|---|---|---|---|
| G-CA | 0.064 ± 0.0192 | 0.034 ± 0.0056 | - |
| G-UDCA | 0.002 ± 0.0005 | - | - |
| G-CDCA | 0.009 ± 0.0033 | 0.004 ± 0.0032 | - |
| G-DCA | 0.003 ± 0.0009 | 0.002 ± 0.0008 | - |
| G-LCA | - | - | - |
| G-BAs | 0.077 ± 0.0017 | 0.041 ± 0.0062 | |
| T-MCA | 44.83 ± 3.8306 | 48.89 ± 2.1865 | 32.16 ± 5.545 |
| T-CA | 44.13 ± 2.9915 | 46.26 ± 2.1722 | 23.39 ± 5.313 |
| T-UDCA | 1.625 ± 0.3160 | 1.249 ± 0.1592 | - |
| T-CDCA | 2.727 ± 0.4818 | 2.302 ± 0.2934 | - |
| T-DCA | 4.405 ± 1.6352 | 0.878 ± 0.2023 | - |
| T-LCA | 0.008 ± 0.0051 | 0.011 ± 0.0052 | - |
| T-BAs | 97.72 ± 0.8740 | 99.59 ± 0.1782 | 55.55 ± 4.802 |
| αMCA | 1.045 ± 0.3714 | 0.166 ± 0.0453 | 31.35 ± 10.35 |
| βMCA | 0.967 ± 0.3042 | 0.082 ± 0.0196 | 7.822 ± 3.190 |
| CA | 0.150 ± 0.0421 | 0.113 ± 0.0388 | 5.284 ± 2.551 |
| UDCA | 0.014 ± 0.0044 | 0.001 ± 0.0007 | - |
| CDCA | 0.013 ± 0.0046 | 0.002 ± 0.0012 | - |
| DCA | 0.012 ± 0.0045 | 0.002 ± 0.0011 | - |
| LCA | - | - | - |
| BAs | 2.205 ± 0.8038 | 0.365 ± 0.0791 | 44.45 ± 4.801 |
Results are shown as the mean of BA% (N=5) ± SD. (−) is used for BAs under the limits of detection or with % < 0.0005%. Bile data are calculated by averaging data from all 8 collection intervals (15, 30, 45, 60, 75, 90, 105, and 120 min).
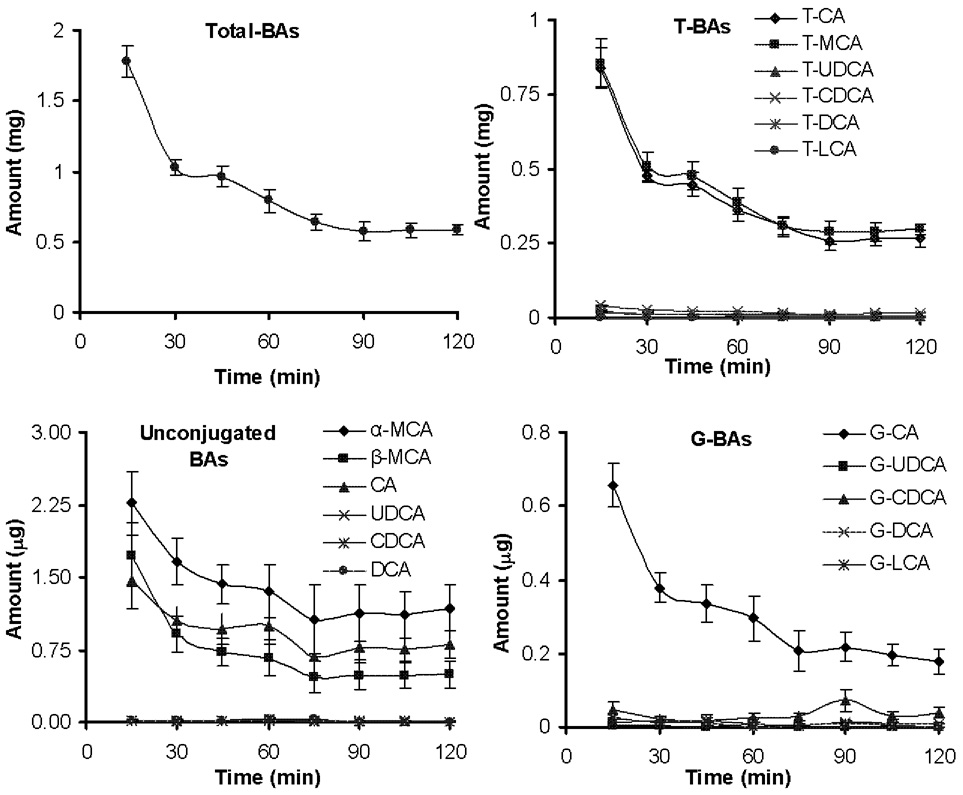
Total BA excreted into bile was 1800 µg for the first 15 min (32 mg/ml) and decreased to 589 µg by the last collection (19 mg/ml) (Figure 6). This 65% decrease in biliary excretion rate was due to both a decrease in bile flow and BA concentration (data not shown). The taurine conjugates of CA and MCA constitute 46 and 49% of total BAs in bile, respectively. In bile, 99.6% of BAs are taurine conjugates, 0.4% are unconjugated, and less than 0.1% are glycine conjugates. Table 3 shows the relative composition of BAs in mouse bile averaged for the 8 samples collected throughout the 2 hr collection period. The variation in BA composition within each collection point is small (see Fig 6). The large variation for some minor BA components, such as unconjugated DCA, UDCA, and CDCA, are not due to analysis-related factors but largely due to variation between bile samples collected at various time points throughout the 2 hrs collection period. Previous studies in mice have reported total BAs in bile from 32–200 µmole/ml (16–102 mg/ml) [33,41–43,46,47,49–53]. The large variation in these reports may be attributed to the method of bile collection (bile duct cannulation vs gallbladder bile), the different mouse strains used, as well as the analytical techniques used for BA analysis.
Figure 6.
BA excretion in mouse bile. The results are shown as mean (N=5) ± S.E.M.
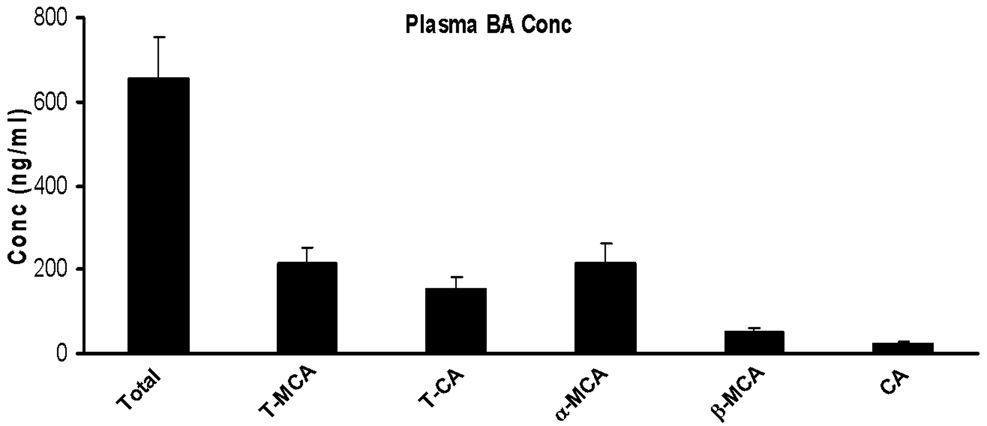
Total BA concentration in mouse plasma was 600 ng/ml (Figure 7). Previous reports of BAs in mouse plasma ranged from 0.4–15 nmol/ml ( 204–7650 ng/ml) [33,41,43,45–47,51,54,55]. The large variation in these results may be due to which BAs were targeted in the analysis. It was previously reported that additional polar forms of BAs exist in plasma, including polyhydroxylated, sulfated, and glucuronidated metabolites [56]. However, we were not able to find masses corresponding to these species (data not shown) under the detection conditions of our method. The present data is in agreement with previous reports that such BAs only appear in mouse and rat blood under abnormal liver conditions [41,48].
Figure 7.
Concentration of bile acids in plasma. The results are shown as mean (N=5) ± S.E.M.
Only trace amounts of total BAs in human and mouse urine were detected in previous reports [26,33,54,57]. However, other studies were not able to detect BAs in urine [58]. Using our method, all BA levels in mouse urine were below the lower limit of quantification (LLOQ). However, the LLOQ in our method is as low or lower than liquid chromatography-based methods in the literature. Therefore, the fact that we could not detect BAs at quantifiable levels in urine is not likely due to the lack of required sensitivity, but it might be attributed to the fact that most of the BA content in mouse urine is composed of more polar species such as poly hydroxylated BA metabolites [41], which are not included in our method. Also, longer periods of urine collection might be needed. More importantly, the method by which urine is collected can make a large difference, because contamination with feces can easily occur. In this study we were able to detect large amount of BAs in urine samples contaminated with feces. However, when metabolic cages completely separated feces from urine, BA levels were lower than the limits of quantification.
In summary, we have developed and validated a LC-MS/MS method for the simultaneous quantification of 6 major BAs, as well as their glycine and taurine conjugates in mouse liver, bile, plasma, and urine. This method is novel because it provides high sensitivity and specificity to perform quantitative profiling of 17 individual BA species in a relatively short run-time in mouse tissues and fluids using a simple single-step sample preparation. In addition this method was validated to ensure high accuracy and precision, and therefore the reliability of it’s results. This method was applied to quantify BAs in mouse fluids and tissues. The tri-hydroxy BAs, CA and MCA, in the taurine conjugate form comprised more than 95% of the total BAs in mouse liver and bile. Low concentrations of BAs were detected in plasma but none were detected in urine under the detection conditions of our method.
Acknowledgment
This work was supported by NIH grants ES-09649, ES-09716, ES013714 and COBRE grant P20-RR-021940.
Abbreviations
- LC-MS
liquid chromatography-mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- BA
bile acids
- G-BAs
glycine conjugated bile acids
- T-BAs
taurine conjugated bile acids
- MCA
muricholic acid
- CA
cholic acid
- UDCA
ursodeoxycholic acid
- DCA
deoxycholic acid
- CDCA
chenodeoxycholic acid
- FXR
farnesoid X receptor
- PXR
pregnane X receptor
- IS
internal standard
- ACN
acetonitrile
- QC
quality control
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Russell DW. Annu Rev Biochem. 2003;72:137. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 2.Danielsson H, Sjovall J. Annu Rev Biochem. 1975;44:233. doi: 10.1146/annurev.bi.44.070175.001313. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann AF. Arch Intern Med. 1999;159:2647. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 4.Gu JJ, Hofmann AF, Ton-Nu HT, Schteingart CD, Mysels KJ. J Lipid Res. 1992;33:635. [PubMed] [Google Scholar]
- 5.Chiang JY. Endocr Rev. 2002;23:443. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- 6.Zollner G, Marschall HU, Wagner M, Trauner M. Mol Pharm. 2006;3:231. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
- 7.Latta RK, Fiander H, Ross NW, Simpson C, Schneider H. Cancer Lett. 1993;70:167. doi: 10.1016/0304-3835(93)90227-z. [DOI] [PubMed] [Google Scholar]
- 8.Reddy BS, Watanabe K, Weisburger JH, Wynder EL. Cancer Res. 1977;37:3238. [PubMed] [Google Scholar]
- 9.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. Proc Natl Acad Sci U S A. 2001;98:3369. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Science. 1999;284:1365. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 11.Mashige F, Tanaka N, Maki A, Kamei S, Yamanaka M. Clin Chem. 1981;27:1352. [PubMed] [Google Scholar]
- 12.Murphy GM, Billing BH, Baron DN. J Clin Pathol. 1970;23:594. doi: 10.1136/jcp.23.7.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alme B, Bremmelgaard A, Sjovall J, Thomassen P. J Lipid Res. 1977;18:339. [PubMed] [Google Scholar]
- 14.Setchell KD, Worthington J. Clin Chim Acta. 1982;125:135. doi: 10.1016/0009-8981(82)90190-5. [DOI] [PubMed] [Google Scholar]
- 15.Stellaard F, Langelaar SA, Kok RM, Jakobs C. J Lipid Res. 1989;30:1647. [PubMed] [Google Scholar]
- 16.Roda A, Piazza F, Baraldini M. J Chromatogr B Biomed Sci Appl. 1998;717:263. doi: 10.1016/s0378-4347(98)00174-1. [DOI] [PubMed] [Google Scholar]
- 17.Goto J, Hasegawa M, Kato H, Nambara T. Clin Chim Acta. 1978;87:141. doi: 10.1016/0009-8981(78)90068-2. [DOI] [PubMed] [Google Scholar]
- 18.Labbe D, Gerhardt MF, Myara A, Vercambre C, Trivin F. J Chromatogr. 1989;490:275. doi: 10.1016/s0378-4347(00)82785-1. [DOI] [PubMed] [Google Scholar]
- 19.Lee BL, New AL, Ong CN. J Chromatogr B Biomed Sci Appl. 1997;704:35. doi: 10.1016/s0378-4347(97)00443-x. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama F, Nakagaki M. J Chromatogr. 1980;183:287. doi: 10.1016/s0378-4347(00)81708-9. [DOI] [PubMed] [Google Scholar]
- 21.Shaw R, Smith JA, Elliott WH. Anal Biochem. 1978;86:450. doi: 10.1016/0003-2697(78)90768-6. [DOI] [PubMed] [Google Scholar]
- 22.Sakakura H, Kimura N, Takeda H, Komatsu H, Ishizaki K, Nagata S. J Chromatogr B Biomed Sci Appl. 1998;718:33. doi: 10.1016/s0378-4347(98)00342-9. [DOI] [PubMed] [Google Scholar]
- 23.Kamada S, Maeda M, Tsuji A. J Chromatogr. 1983;272:29. doi: 10.1016/s0378-4347(00)86100-9. [DOI] [PubMed] [Google Scholar]
- 24.Libert R, Hermans D, Draye JP, Van Hoof F, Sokal E, de Hoffmann E. Clin Chem. 1991;37:2102. [PubMed] [Google Scholar]
- 25.Mills KA, Mushtaq I, Johnson AW, Whitfield PD, Clayton PT. Pediatr Res. 1998;43:361. doi: 10.1203/00006450-199803000-00009. [DOI] [PubMed] [Google Scholar]
- 26.Bobeldijk I, Hekman M, de Vries-van der Weij J, Coulier L, Ramaker R, Kleemann R, Kooistra T, Rubingh C, Freidig A, Verheij E. J Chromatogr B Analyt Technol Biomed Life Sci. 2008 doi: 10.1016/j.jchromb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Tagliacozzi D, Mozzi AF, Casetta B, Bertucci P, Bernardini S, Di Ilio C, Urbani A, Federici G. Clin Chem Lab Med. 2003;41:1633. doi: 10.1515/CCLM.2003.247. [DOI] [PubMed] [Google Scholar]
- 28.Ando M, Kaneko T, Watanabe R, Kikuchi S, Goto T, Iida T, Hishinuma T, Mano N, Goto J. J Pharm Biomed Anal. 2006;40:1179. doi: 10.1016/j.jpba.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Bootsma AH, Overmars H, van Rooij A, van Lint AE, Wanders RJ, van Gennip AH, Vreken P. J Inherit Metab Dis. 1999;22:307. doi: 10.1023/a:1005543802724. [DOI] [PubMed] [Google Scholar]
- 30.Burkard I, von Eckardstein A, Rentsch KM. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;826:147. doi: 10.1016/j.jchromb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Roda A, Gioacchini AM, Cerre C, Baraldini M. J Chromatogr B Biomed Appl. 1995;665:281. doi: 10.1016/0378-4347(94)00544-f. [DOI] [PubMed] [Google Scholar]
- 32.Setchell KD, Vestal CH. J Lipid Res. 1989;30:1459. [PubMed] [Google Scholar]
- 33.Stedman C, Robertson G, Coulter S, Liddle C. J Biol Chem. 2004;279:11336. doi: 10.1074/jbc.M310258200. [DOI] [PubMed] [Google Scholar]
- 34.Warrack BM, DiDonato GC. Biol Mass Spectrom. 1993;22:101. doi: 10.1002/bms.1200220202. [DOI] [PubMed] [Google Scholar]
- 35.Perwaiz S, Tuchweber B, Mignault D, Gilat T, Yousef IM. J Lipid Res. 2001;42:114. [PubMed] [Google Scholar]
- 36.Ikegawa S, Murao N, Motoyama T, Yanagihara T, Niwa T, Goto J. Biomed Chromatogr. 1996;10:313. doi: 10.1002/(SICI)1099-0801(199611)10:6<313::AID-BMC603>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 37.Scalia S. J Chromatogr B Biomed Appl. 1995;671:299. doi: 10.1016/0378-4347(95)00215-5. [DOI] [PubMed] [Google Scholar]
- 38.Alnouti Y, Li M, Kavetskaia O, Bi H, Hop CE, Gusev AI. Anal Chem. 2006;78:1331. doi: 10.1021/ac051806q. [DOI] [PubMed] [Google Scholar]
- 39.Hochberg RB. Endocr Rev. 1998;19:331. doi: 10.1210/edrv.19.3.0330. [DOI] [PubMed] [Google Scholar]
- 40.Falany CN, Fortinberry H, Leiter EH, Barnes S. J Lipid Res. 1997;38:1139. [PubMed] [Google Scholar]
- 41.Marschall HU, Wagner M, Bodin K, Zollner G, Fickert P, Gumhold J, Silbert D, Fuchsbichler A, Sjovall J, Trauner M. J Lipid Res. 2006;47:582. doi: 10.1194/jlr.M500427-JLR200. [DOI] [PubMed] [Google Scholar]
- 42.Wagner M, Fickert P, Zollner G, Fuchsbichler A, Silbert D, Tsybrovskyy O, Zatloukal K, Guo GL, Schuetz JD, Gonzalez FJ, Marschall HU, Denk H, Trauner M. Gastroenterology. 2003;125:825. doi: 10.1016/s0016-5085(03)01068-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, Salem M, Yousef IM, Tuchweber B, Lam P, Childs SJ, Helgason CD, Ackerley C, Phillips MJ, Ling V. Proc Natl Acad Sci U S A. 2001;98:2011. doi: 10.1073/pnas.031465498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masson D, Lagrost L, Athias A, Gambert P, Brimer-Cline C, Lan L, Schuetz JD, Schuetz EG, Assem M. Arterioscler Thromb Vasc Biol. 2005;25:2164. doi: 10.1161/01.ATV.0000183674.88817.fb. [DOI] [PubMed] [Google Scholar]
- 45.Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Science. 2006;312:233. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 46.Ferdinandusse S, Denis S, Overmars H, Van Eeckhoudt L, Van Veldhoven PP, Duran M, Wanders RJ, Baes M. J Biol Chem. 2005;280:18658. doi: 10.1074/jbc.M414311200. [DOI] [PubMed] [Google Scholar]
- 47.Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, Nagata K, Sinal CJ, Guo GL, Gonzalez FJ, Yamazoe Y. J Biol Chem. 2003;278:17838. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- 48.Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall HU, Trauner M. Am J Physiol Gastrointest Liver Physiol. 2006;290:G923. doi: 10.1152/ajpgi.00490.2005. [DOI] [PubMed] [Google Scholar]
- 49.Uchida K, Takase H, Nomura Y, Takeda K, Takeuchi N, Ishikawa Y. J Lipid Res. 1984;25:236. [PubMed] [Google Scholar]
- 50.Uchida K, Makino S, Akiyoshi T. Diabetes. 1985;34:79. doi: 10.2337/diab.34.1.79. [DOI] [PubMed] [Google Scholar]
- 51.Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. J Biol Chem. 2003;278:45062. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- 52.Kok T, Hulzebos CV, Wolters H, Havinga R, Agellon LB, Stellaard F, Shan B, Schwarz M, Kuipers F. J Biol Chem. 2003;278:41930. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 53.Kuipers F, van Ree JM, Hofker MH, Wolters H, In't Veld G, Havinga R, Vonk RJ, Princen HM, Havekes LM. Hepatology. 1996;24:241. doi: 10.1002/hep.510240138. [DOI] [PubMed] [Google Scholar]
- 54.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Cell. 2000;102:731. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 55.Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, Evans RM, Downes M. Proc Natl Acad Sci U S A. 2005;102:2063. doi: 10.1073/pnas.0409794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takikawa H, Otsuka H, Beppu T, Seyama Y, Yamakawa T. Digestion. 1983;27:189. doi: 10.1159/000198952. [DOI] [PubMed] [Google Scholar]
- 57.Uppal H, Saini SP, Moschetta A, Mu Y, Zhou J, Gong H, Zhai Y, Ren S, Michalopoulos GK, Mangelsdorf DJ, Xie W. Hepatology. 2007;45:422. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- 58.van Berge Henegouwen GP, Brandt KH, Eyssen H, Parmentier G. Gut. 1976;17:861. doi: 10.1136/gut.17.11.861. [DOI] [PMC free article] [PubMed] [Google Scholar]