Rev is an important HIV-1 regulatory protein that binds the Rev responsive element (RRE) within the env gene of HIV-1 RNA genome; the binding of Rev to RRE is essential for the expression of the structural genes gag-pol and env, and for HIV replication.1 The Rev-RRE interactions are routinely characterized by gel-shift and filter-binding-based assays;2, 3 however, these traditional methods are usually labor-intensive and time-consuming. In recent years, several other technologies have been developed for the assay of RRE-Rev interactions, such as surface plasmon resonance,4 acoustic wave,5 isothermal calorimetry,6 stopped-flow fluorescence,7, 8 and fluorescence resonance energy transfer (FRET).9 Among these methods, fluorescence-based assays are especially attractive due to their high sensitivity, good reproducibility, and real-time monitoring of binding interactions. Organic dyes are usually used in the fluorescence-based assays;7-9 however, due to their narrow absorption, red-tailed emission spectra, and susceptibility to photodegradation, the applications of fluorescent dyes in the RNA-peptide interaction studies usually involves complex experimental designs, and considerable effort must be put into addressing the problems of perturbing native interaction which result from labeling with organic dyes in/near active sites of RRE IIB RNA.7, 8 Alternatively, quantum dots (QDs) have broad excitation and size-tunable photoluminescence spectra with narrow emission bandwidth (full-width at half-maximum of ∼25−40 nm), exceptional photochemical stability and relative high quantum yields.10, 11 QDs have been widely used as fluorescent markers in genomic analysis, immunoassay, fluorescence imaging and drug delivery.12, 13 Recently, QDs have also been used as FRET donors in biosensors to detect DNA, RNA and proteins.14-17 Here we explore the potential of QD-based FRET for in vitro evaluation of RNA-peptide interactions, and develop a QD-based nanosensor for sensitive RRE-Rev interaction assay.
Stem-loop IIB of RRE containing a purine-rich internal bulge and small peptides containing a 17 amino acid arginine-rich region of Rev (amino acids 34−51) were used as a model system for this in vitro Rev-RRE interaction study.7, 8 Biotin was attached to the 5’ end of RRE RNA (Figure 1a), and Cy5 was attached at the N-terminal of Rev (Figure 1b); the positioning of biotin and Cy5 at these sites did not alter the native interaction between RRE and Rev, or even the interaction of RRE with its inhibitors.7, 8 The streptavidin-coated QDs functioned as both a nano-scaffold and a FRET donor in this QD-based nanosensor. The binding of Rev to RRE formed a biotinylated Cy5-labeled Rev-RRE complex; the subsequent addition of 605QDs resulted in the formation of 605QD/Rev-RRE/Cy5 assemblies through specific streptavidin-biotin binding (Figure 1c). Upon excitation with a wavelength of 488nm, FRET occurred between 605QDs and Cy5s in the assemblies; the fluorescence signals of 605QD and Cy5 were observed simultaneously (Figure 1c). The 605QD is an excellent energy donor with Cy5 for a number of reasons: no cross-talk between the emission spectra of 605QD and that of Cy5, and no direct excitation of Cy5 at the wavelength of 488 nm. Moreover, 605QD has a high quantum yield (∼0.6), Cy5 has a high extinction coefficient (∼250,000 M−1cm−1), and a single 605QD can efficiently couple to multiple Cy5-labeled Rev-RRE complexes around its surface;15, 17-19 thus efficient FRET can be achieved even at distance approaching 2×R0 for this 605QD/Cy5 FRET pair.17
Figure 1.
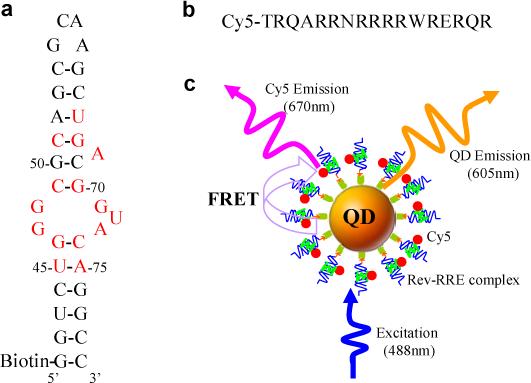
The principles of the QD-based nanosensor for Rev-RRE interaction assay. (a) Secondary structure of biotinylated RRE IIB RNA. Nucleotides identified as important for Rev binding are shown in red. (b) Sequence of Cy5-labeled Rev peptide. (c) Conceptual scheme of the QD-based nanosensor for Rev-RRE interaction assay based on FRET between 605QD and Cy5. The binding of a Cy5-labeled Rev to a biotinylated RRE IIB RNA formed a Rev-RRE complex, which was caught on the surface of a 605QD to form a 605QD/Rev-RRE/Cy5 assembly through specific streptavidin-biotin binding. FRET occurred between the 605QD and Cy5 upon illumination of the 605QD/Rev-RRE/Cy5 assemblies with an excitation wavelength of 488nm.
To characterize the FRET in the 605QD/Rev-RRE/Cy5 assembly, both 605QD and Cy5 fluorescence were measured as a function of the ratio of Cy5-labeled Rev-RRE complex to 605QD (Figure 2). As the complex-to-605QD ratio increased, the 605QD fluorescence decreased, while the Cy5 fluorescence increased. At the ratio of 30/1, the fluorescence of both 605QD and Cy5 saturated, suggesting that the number of Cy5-labeled Rev-RRE complexes assembled on the 605QD surface had reached its maximum. Assuming three available biotin binding sites per streptavidin after conjugation to QDs, in principle, up to ∼36−45 biotinylated Rev-RRE complexes could be captured by a single 605QD based on the assumption that each QD conjugated with 12−15 streptavidins. Therefore, only two thirds of available biotin binding sites around a 605QD were occupied; this probably results from the complex-complex steric hindrance. The Förster distance (R0) is 69.4 Å for 605QD/Cy5 FRET pair, and the average QD-Cy5 separation distance was calculated to be 107.0 ± 4.9 nm according to the formalism described in previous papers.17-19 Figure 2 demonstrates that the increase of the Cy5-labeled Rev-RRE complex-to-605QD ratio can significantly improve FRET efficiency, and consequently the detection sensitivity of the RRE-Rev interaction assay.
Figure 2.
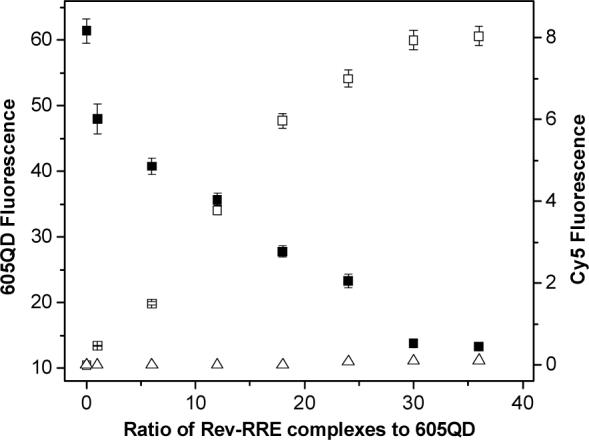
Characterization of FRET in the 605QD/Rev-RRE/Cy5 assemblies. The 605QD fluorescence (■) and Cy5 fluorescence (□) were measured as a function of the Rev-RRE complex-to-605QD ratio. The direct excitation of equivalent amounts of Cy5-labeled Rev without the presence of 605QD (Δ) was shown for comparison. 605QD concentration: 38 nM; Rev-RRE complex concentration varied with the ratio of Rev-RRE complex to 605QD as shown. Error bars, S.D. (n=3).
The binding of Rev to RRE was directly monitored by titrating a fixed concentration of RRE with Rev. The 605QD fluorescence decreased in a saturable fashion as the Rev concentration increased, while Cy5 fluorescence increased with the Rev concentration (Figure 3a). The 605QD fluorescence data was plotted and fitted with equation 1 (see Supporting Information). The dissociation constant (Kd) of the Rev-RRE complex was calculated to be 11 ± 4 nM. The dissociation constant was also calculated by fitting Cy5 fluorescence data using equation 2 (see Supporting Information). A Kd of 14 ± 5 nM was obtained. The Kd values determined in this study agree well with the Kd of 15 ± 5 nM that was obtained by the fluorescence anisotropy method20 and with the Kd of 20 ± 7 nM that was obtained by monitoring the fluorescence intensity of 2-aminopuring 2’-O-methylriboside which incorporated in position 72 of RRE RNA.8 It is worth noting that the Kd of 11 ± 4 nM obtained from the 605QD fluorescence data is in excellent agreement with the Kd of 14 ± 5 nM obtained from the Cy5 fluorescence data, since Kd was obtained from FRET between 605QD and Cy5; this further confirms the accuracy and sensitivity of this QD-based nanosensor for RRE-Rev interaction assay.
Figure 3.
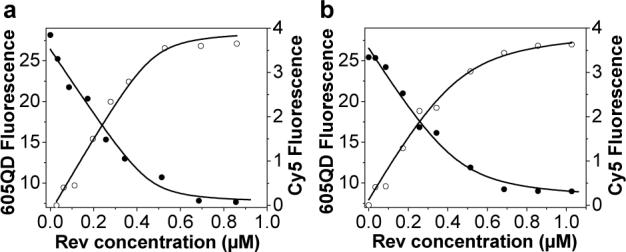
QD-based nanosensor for Rev-RRE interaction assay. (a) Titration of RRE RNA with increasing Rev concentration. (b) Titration of RRE RNA in presence of neomycin B with increasing Rev concentration. The solid curves are fits of 605QD fluorescence data (•) using equation 1, and fits of Cy5 fluorescence data (○) using equation 2. 605QD concentration: 19 nM; RRE concentration: 456 nM; Neomycin B concentration: 4.56 μM.
A previous study has demonstrated that neomycin B can efficiently inhibit Rev binding to RRE at a concentration of ∼10-fold lower than other aminoglycoside antibiotics studied.21 To test whether this QD-based nanosensor might be used to directly assess the inhibitory effect of neomycin B upon Rev-RRE binding, titration experiments with Rev in the presence of a fixed concentration of RRE and neomycin B were performed (Figure 3b). A Kd of 40 ± 9 nM for Rev peptide binding to RRE RNA in presence of neomycin B was calculated from 605QD fluorescence data with equation 1. A similar Kd of 46 ± 10 nM was obtained by fitting the Cy5 fluorescence data with equation 2. These Kd values are larger than those obtained in the absence of neomycin B, suggesting weaker binding between Rev and RRE due to the presence of inhibitor neomycin B.
In conclusion, this QD-based nanosensor offers a simple approach for sensitive Rev-RRE interaction assay due to its unique characteristics of non interference in the native interaction of Rev with RRE, improved FRET efficiency, high sensitivity and simultaneously FRET-related two-parameter detection using the simple instrumentation of a standard fluorescence spectrometer and commercially available streptavidin-coated QDs. This QD-based nanosensor can be used to study the effect of inhibitor upon Rev-RRE interaction, and may have a wide applicability in the development of new drugs against HIV-1 infection.
Supplementary Material
Acknowledgement
The authors thank Dr. R. Desamero for helpful discussions. This work was supported by NIH (GM08153).
Footnotes
Supporting Information Available: Experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gullen BR, Malim MH. Trends Biochem. Sci. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- 2.Friesen WJ, Darby MK. J. Biol. Chem. 2001;276:1968–1973. doi: 10.1074/jbc.M008927200. [DOI] [PubMed] [Google Scholar]
- 3.Clemens KR, Wolf V, McBryant SJ, Zhang P, Liao X, Wright PE, Gottesfeld JM. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 4.Van Ryk DI, Venkatesan S. J. Biol. Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 5.Tassew N, Thompson M. Biophys. Chem. 2003;106:241–252. doi: 10.1016/s0301-4622(03)00155-8. [DOI] [PubMed] [Google Scholar]
- 6.Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC. Proc. Natl. Acad. Sci. USA. 1995;92:3199–3203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeJong ES, Chang CE, Gilson MK, Marino JP. Biochemistry. 2003;42:8035–8046. doi: 10.1021/bi034252z. [DOI] [PubMed] [Google Scholar]
- 8.Lacourciere KA, Stivers JT, Marino JP. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 9.Tamilarasu N, Zhang J, Hwang S, Rana T. Bioconjugate Chem. 2001;12:135–138. doi: 10.1021/bc000104z. [DOI] [PubMed] [Google Scholar]
- 10.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 11.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 12.Medintz IL, Uyeda HT, Goldman E, Mattoussi H. Nat. Mater. 2005;4:437–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 13.Alivisatos AP. Nat. Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 14.Patolsky F, Gill R, Weizmann Y, Mokari T, Banin U, Willner IJ. Am. Chem. Soc. 2003;125:13918–13919. doi: 10.1021/ja035848c. [DOI] [PubMed] [Google Scholar]
- 15.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat. Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 16.Medintz IL, Konnert JH, Clapp AR, Stanish IM, Twigg E, Mattoussi H, Mauro JM, Deschamps JR. Proc. Natl. Acad. Sci. USA. 2004;101:9612–9617. doi: 10.1073/pnas.0403343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 18.Clapp AR, Medintz IL, Uyeda HT, Fisher BR, Goldman ER, Bawendi MG, Mattoussi H. J. Am. Chem. Soc. 2005;127:18212–18221. doi: 10.1021/ja054630i. [DOI] [PubMed] [Google Scholar]
- 19.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi HJ. J. Am. Chem. Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Hamasaki K, Rando RR. Biochemistry. 1997;36:768–779. doi: 10.1021/bi962095g. [DOI] [PubMed] [Google Scholar]
- Zapp ML, Stern S, Green MR. Cell. 1993;74:969–978. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


