Abstract
Evidence that the androgen receptor (AR) is not only important in androgen-dependent prostate cancer, but also continues to play a role in tumors that become resistant to androgen deprivation therapies, highlights the need to find alternate means to block AR activity. AR, a hormone-activated transcription factor, and its coactivators are phosphoproteins. Thus, we sought to determine whether inhibition of specific cell signaling pathways would reduce AR function. We found that short-term inhibition of p42/p44 MAPK activity either by a MAPK kinase inhibitor, U0126, or by depletion of kinase with small interfering RNA caused target gene-specific reductions in AR activity. AR enhances histone H3 acetylation of target genes that are sensitive to U0126 including prostate-specific antigen and TMPRSS2, but does not increase histone H3 acetylation of the U0126-resistant PMEPA1 gene. Thus, although AR induces transcription of many target genes, the molecular changes induced by AR at the chromatin level are target gene specific. Long-term treatment (24–48 h) with U0126 causes a G1 cell cycle arrest and reduces AR expression both through a decrease in AR mRNA and a reduction in AR protein stability. Thus, treatments that reduce p42/p44 MAPK activity in prostate cancer have the potential to reduce AR activity through a reduction in expression levels as well as by target gene-selective inhibition of AR function.
ALTHOUGH THE IMPORTANCE of the androgen receptor (AR) and androgens in prostate cancer has long been recognized, several recent findings underscore the contribution of AR in primary tumors and in recurrent cancers subsequent to androgen ablation. Tomlins et al. (1) have found that a very high percent of prostate tumors contain a somatic genetic rearrangement, which results in a fusion between the promoter region of the androgen-regulated TMPRSS2 gene and the coding region of an Ets factor. Thus, the expression of this fusion is dependent on AR action. There is increasing evidence that recurrent prostate tumors remain AR dependent. In comparisons of androgen-dependent tumors with androgen-independent tumors from humans and from xenografts, the most consistent difference is an elevation in AR expression in the androgen ablation-resistant tumors (2). Moreover, the recurrent tumors reexpress many AR-regulated targets including prostate-specific antigen (PSA). Eliminating AR expression or activity in AR-positive androgen-independent prostate cancer cells blocks cell proliferation and eliminates PSA expression (3,4), supporting the concept that tumors develop alternate means for activating AR in the absence of normal levels of androgens. Consequently, understanding the aberrant activation of AR and developing means to inhibit its activity are high priorities. A number of potential mechanisms for reactivation of AR have been proposed, including increased AR expression, altered steroid metabolism resulting in elevated levels of androgens in tumors, and altered cell signaling that results in the activation of AR in the absence of normal levels of androgens (5). Several studies have implicated HER2 signaling in regulating AR protein stability, sensitizing AR to low levels of androgens, and even in inducing hormone-independent activation (6,7). Typical of most growth factor signaling, multiple signaling pathways are activated by HER2 heterodimers. The downstream kinase(s) responsible for regulating AR expression and activity have not been identified although Akt has been eliminated as a candidate (7). Other studies show that AR protein is more stable in androgen-independent than in androgen-dependent cell lines and that the androgen-independent cells are hypersensitive to low levels of androgens (8). The hypersensitivity to androgens can be induced by expressing a Ras effector mutant that selectively activates Raf, leading to activation of p42/p44 MAPK (also called ERK1/ERK2) (9). Interestingly, elevated p42/p44 MAPK signaling has been detected in advanced prostate tumors (10). Our studies show that blocking p42/p44 MAPK activity recapitulates the observed reductions in PSA expression and AR stability caused by inhibition of HER2 (7). Studies to date have relied on measuring AR activity using AR-responsive reporters or endogenous PSA. However, we find that inhibition of p42/p44 MAPK reduces AR activity in a target gene-specific manner. Using mammalian two-hybrid assays, we found that kinase inhibition reduces AR amino-carboxyl terminal interactions as well as the interaction between AR and the p160 coactivator, steroid receptor coactivator (SRC)-1. Consistent with this, we found that target genes that exhibit increased histone H3 acetylation in response to androgen are sensitive to the MEK inhibitor, whereas those with no hormone-dependent increase in histone H3 acetylation are resistant. Long-term inhibition of MEK activity causes a G1 arrest and reduces AR stability. Thus, treatments that reduce p42/p44 MAPK activity in prostate cancer have the potential to reduce AR activity through a reduction in expression levels as well as by target gene-selective inhibition of AR function.
RESULTS
Inhibition of MEK Reduces AR Transcriptional Activity in a Promoter-Specific Manner
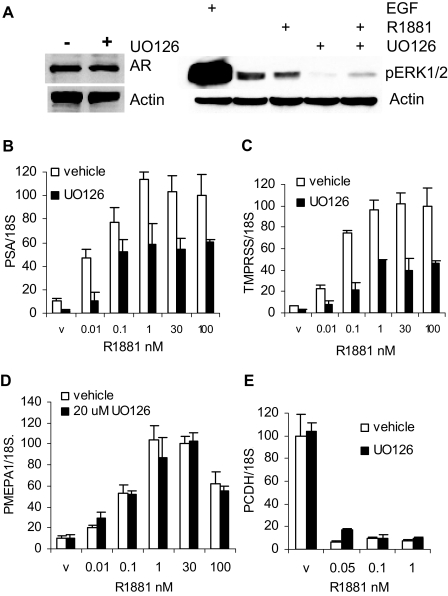
To examine the impact of MAPK signaling on AR transcriptional activity, we treated LNCaP cells with U0126, a MEK inhibitor that blocks activation of p42/p44 MAPK. At 12 h the level of AR in LNCaP treated with the synthetic androgen, R1881, was unchanged by U0126 treatment whereas phosphorylation of MAPK was inhibited (Fig. 1A). Induction of two well-known AR target genes, PSA and TMPRSS2, was reduced (Fig. 1, B and C). In contrast, another AR-activated target gene PMEPA1 (11,12) and an AR-repressed gene PCDH11 (13) were insensitive to MAPK inhibition at the 12-h time point (Fig. 1, D and E). Thus, the requirement for p42/p44 MAPK signaling is target gene dependent.
Figure 1.
MAPK Signaling Is Required for Optimal AR Transcriptional Activity
A, LNCaP cells were preincubated for 24 h in medium supplemented with 10% charcoal stripped serum (sFBS), 1 nm R1881, and vehicle (DMSO) or 20 μm UO126 was added to the medium and cells incubated for 12 h; except 100 ng/ml of EGF was added where indicated only for the last 10 min. Cells were harvested and AR, phospho-ERK1/2, and actin expression was measured by Western blotting. B–E, LNCaP cells were treated with the indicated amount of R1881 and either DMSO as a vehicle or 20 μm UO126 for 12 h. Total RNA was prepared as described in Materials and Methods and analyzed for PSA (B), TMPRSS2 (C), PMEPA1 (D), and PCDH11 (E) expression by quantitative RT-PCR. All expression values were normalized to 18S RNA expression, each point was done in triplicate, and mean and sd were calculated. The experiments were repeated three times, and a representative experiment is shown. v, Vehicle.
ERK1 and ERK2 Differentially Contribute to AR Activity
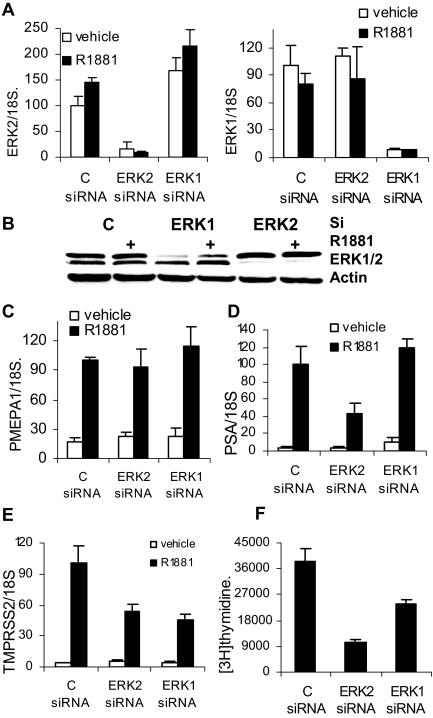
MAPK kinase (MEK) inhibition prevents the direct activation of two kinases, ERK2 (MAPK1/p42) and ERK1 (MAPK3/p44), which are both expressed in LNCaP cells (14). To eliminate the possibility of off-target effects of U0126 and to determine whether there is downstream kinase specificity, we reduced expression of each of these kinases separately using small interfering RNA (siRNA). As shown in Fig. 2, A and B, ERK1 and ERK2 expression and protein levels were effectively reduced by their respective siRNAs, and there was no androgen dependence of kinase mRNA or protein expression (Fig. 2, A and B). Consistent with the effect of UO126 on AR target gene expression, PMEPA1 expression was not affected by a reduction in either kinase (Fig. 2C). PSA expression was sensitive to the reduction in ERK2 only (Fig. 2D), whereas TMPRSS2 required both ERK1 and ERK2 (Fig. 2E) for optimal expression. The proliferation of LNCaP cells, measured using [3H]thymidine incorporation, was inhibited by reducing either ERK1 or ERK2; however, ERK2 seems to have a more profound effect on proliferation (Fig. 2F).
Figure 2.
Individual Contributions of ERK1 and ERK2 to AR Signaling
Two million LNCaP cells were electroporated with 800 pmol of either noncoding control siRNA (Dharmacon), MAPK1 (ERK2) smart pool siRNA (Dharmacon), or MAPK3 (ERK1) siRNA (Dharmacon) using an Amaxa electroporator and R Kit (Amaxa). A, Cells electroporated with the indicated siRNAs were plated on polylysine-coated plates at 260,000 cells per well in six-well plates and after 24 h were treated with 1 nm R1881 or vehicle for 12 h. Total RNA was extracted and analyzed for ERK2 (left panel), ERK1 (right panel), and 18S expression by quantitative RT-PCR. ERK1 and ERK2 expression levels were normalized for 18S expression. B, LNCaP cells were transfected and treated in parallel with panel A and cells were harvested and analyzed for ERK1, ERK2 and actin expression by Western blotting. C–E, RNA from panel A was analyzed for PMEPA1 (C), PSA (D), and TMPRSS2 (E) expression by quantitative RT-PCR, and their expression levels were normalized to 18S expression. F, LNCaP cells electroporated with control, ERK2, or ERK1 smart pools (Dharmacon) were plated at a concentration of 5000 cells per well. Cells were treated 24 h after transfection with 1 nm R1881 for 12 h, and cell proliferation was compared using a [3H]thymidine incorporation assay. Each point was done in triplicate, average and sd were calculated, and experiments were performed four times. C, Control.
U0126 Treatment Reduces AR Protein Interactions
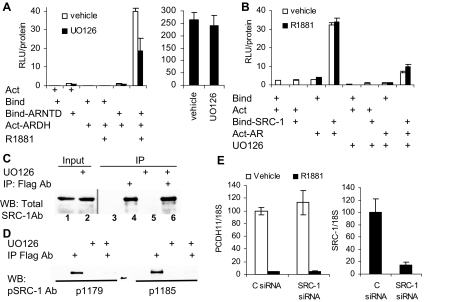
Because both SRC-1 and AR are phosphoproteins and SRC-1 is a direct target of MAPK (15), we used mammalian two-hybrid assays to measure effects of MEK inhibition on AR protein-protein interactions, including the AR amino/carboxyl (N-C) terminal and AR SRC-1 interactions, which are required for optimal induction of many target genes (16). Using a plasmid expressing the AR amino terminus and DNA binding domain linked to the VP16 activation domain and a plasmid expressing the AR DNA binding domain and hormone binding domain linked to a Gal-DNA binding domain, we observed a significant reduction in interaction of the two receptor fragments in the presence of UO126 (Fig. 3A, left). In contrast, the activity of a constitutively active VP16 activation domain linked to a Gal-DNA binding domain was unaffected by treatment (Fig. 3A, right), indicating that it is the interaction between the portions of the AR that is reduced.
Figure 3.
MAPK Signaling Affects Inter- and Intramolecular AR Interactions
A, PC-3 cells were transfected with 400 ng 17mer-Luc, and the indicated combinations of 100 ng of pBind (Promega) or pBind-AR-NTD, 100 ng of pAct (Promega) or pAct-AR-DH as described in Materials and Methods. Cells were treated with either vehicle or 10 nm R1881 with or without 20 μm U0126 overnight and assayed for luciferase activity, which was then normalized for the level of total protein. On the right, PC-3 cells were transfected with 400 ng of 17mer-Luc reporter and 10 ng of pBind-VP16 expression plasmid. Cells were then treated with either DMSO or 20 μm UO126 overnight, harvested, and assayed for luciferase activity and total protein. B, PC-3 cells were transfected with 400 ng 17mer-Luc, the indicated combinations of 100 ng of pBind or pBind-SRC-1a, and 100 ng of pAct or pAct-AR. Cells were treated overnight with either vehicle (empty bars) or 10 nm R1881 (solid bars), and either DMSO or 20 μm UO126 as indicated. Cells were harvested and assayed for luciferase activity and protein concentration. In experiments A and B, each point was done in triplicate, the experiments were repeated three times, and a representative experiment is shown. C and D, COS1 cells were transfected with flag-SRC-1 expression plasmid. After 24 h cells were treated with either DMSO or 20 μm UO126 for 12 h and harvested, and protein was extracted in the presence of protease and phosphatase inhibitors. Protein from each treatment group (1.5 mg) was used for immunoprecipitation (IP) with SRC-1-specific antibody. In panel C, lanes 1 and 2 are 50 μg of the input protein extract treated with either DMSO or 20 μm UO126; lanes 3–6 are 10% of immunoprecipitated material from cells treated with DMSO (lanes 3–4) or UO126 (lanes 5–6), using beads alone (lanes 3 and 5) or with flag antibody (lanes 4 and 6) were resolved on 6.5% PAGE and analyzed by Western blotting with total SRC-1 antibody. In D, 40% of the immunoprecipitated material was resolved on two separate 6.5% gels and analyzed by Western blotting using either pThr1179 or pSer1185 phospho-specific antibody. E, LNCaP cells were electroporated with either control or SRC-1 siRNA and plated in medium supplemented with 10% sFBS. Cells were treated 24 h later with either control or 1 nm R1881 for another 24 h. RNA was isolated and analyzed for 18S, PCDH11, and SRC-1 expression. Expression in the cells transfected with control (C) siRNA and treated with vehicle was designated as 100%, and all other values were adjusted proportionally. Each point was done in triplicate, and the experiment was repeated three times. Ab, Antibody; Act, actin; Bind, binding; RLU, relative light units; WB, Western blotting.
SRC-1 is a histone acetyltransferase (HAT) and recruits other HATs to the promoters. In addition, MAPK-dependent phosphorylation of the p160 coactivator, SRC-1, has been shown to be necessary for IL-6 dependent potentiation of AR activity (17). To determine whether U0126 reduced interaction between SRC-1 and AR, we used a mammalian two-hybrid assay and found that U0126 greatly decreased the interaction between AR and SRC-1 (Fig. 3B). Note that the interaction is hormone independent; the major SRC-1 interaction site on AR is the hormone-independent activation function 1 (AF-1) rather than the hormone binding domain (18). Because Gioeli et al. (19,20) have reported that AR is not phosphorylated by ERK1/2, we sought to determine whether MAPK signaling directly affects SRC-1 phosphorylation by examining the level of phosphorylation on Thr1179 and Ser1185 of flag-tagged SRC-1 expressed in COS-1 cells. Overnight treatment with UO126 did not alter the level of SRC-1 (input protein in Fig. 3C) or the amounts of immunoprecipitated flag-tagged SRC-1 (IP and WB with total SRC-1 specific antibodies, lanes 4 and 6, Fig. 3C). In contrast, treatment with U0126 essentially eliminated phosphorylation of Thr1179 and Ser1185 (Fig. 3D), suggesting that SRC-1 is a direct target of MAPK signaling. Because androgen-dependent repression of PCDH11 is resistant to MEK inhibition, we asked whether SRC-1 is required for androgen-dependent repression. Depletion of SRC-1 using siRNA did not alter the ability of AR to repress PCDH11 (Fig. 3E).
U0126 Treatment Reduces the Rate of AR Recruitment to Target Gene Binding Sites
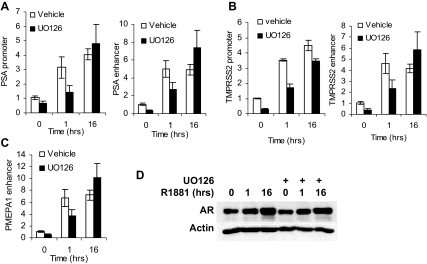
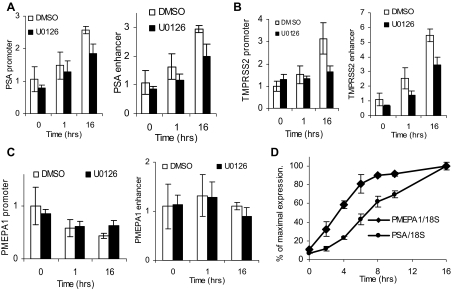
To elucidate the effects of U0126 on AR activity, R1881-dependent recruitment of AR to the PSA, TMPRSS2, and PMEPA1 target genes was measured. As shown in Fig. 4, inhibition of MEK in LNCaP cells significantly reduced recruitment of AR to the PSA promoter and enhancer (Fig. 4A), TMPRSS2 promoter and enhancer (Fig. 4B), and PMEPA1 enhancer (Fig. 4C) after 1 h of treatment with 1 nm R1881 despite the fact that there are similar amounts of AR under both conditions (Fig. 4D). After 16 h of treatment, U0126 no longer has an effect on AR binding (Fig. 4, A–C).
Figure 4.
Effect of Short-Term Inhibition of MAPK on AR Recruitment to the PSA Enhancer
A–C, LNCaP cells were placed in medium supplemented with 10% sFBS for 36 h. Cells were treated with 20 μm UO126 or vehicle (DMSO) and with either vehicle (ethanol) for 16 h, or 1 nm R1881 for 16 h or for the final 1 h. The ChIP assay was performed using an AR-specific antibody as described in Materials and Methods. DNA recovered after the ChIP assay was amplified with PSA (A) and TMPRSS2 (B) enhancer and promoter-specific TaqMan primers and probe and the PMEPA1 enhancer sequence (C). Values were divided by the corresponding input levels, and the amount of recruitment in cells treated with both vehicles was assigned a value of 1 and all others were normalized to it. The results of three independent ChIP assays were averaged and sd was calculated. D, LNCaP cells treated in parallel with panel A were harvested and analyzed for AR and actin expression by Western blotting.
Inhibition of MEK Reduces Androgen-Dependent Acetylation of Histone H3 at the PSA and TMPRSS2 Promoters and Enhancers
Activated AR recruits a series of HATs including the p160 family members and p300 to the PSA promoter and enhancer resulting in increased acetylation of histone H3 (21). To determine whether MEK inhibition reduces HAT activity at AR target genes, we examined the levels of acetyl H3 histone on the androgen-regulated promoters. Intriguingly, the level of acetylation of H3 on the promoters and enhancers of UO126-sensitive genes, PSA and TMPRSS2, increased with androgen treatment, and this increase was compromised by MEK inhibition (Fig. 5, A and B). Acetylation levels of H3 histone at the promoter and enhancer of PMEPA1 were unchanged either by androgen treatment or MEK inhibition (Fig. 5C). This suggests distinct target gene-specific roles for AR. Because AR was not required to increase acetylation of histone H3 upstream of PMEPA1 gene, we speculated that it might be primed for initiation. Thus, we tested whether genes that require histone acetylation at the promoter and enhancer are induced more slowly by androgens compared with PMEPA1, which does not require this additional step for transcriptional activation. We found that, indeed, PMEPA1 expression was induced more rapidly than PSA expression (Fig. 5D). TMPRSS2 was induced more slowly than PMEPA1 as well (data not shown).
Figure 5.
AR Target Genes Display Differential Dynamics of Histone Modification and Activation
A–C, LNCaP cells were incubated in medium with 10% sFBS for 36 h. Cells were treated with 20 μm UO126 or DMSO, and either ethanol for 16 h or 1 nm R1881 for 16 h or for the final 1 h. ChIP was performed using antiacetyl H3 antibody (Millipore) as described in Materials and Methods. Immunoprecipitated DNA was analyzed for PSA promoter and enhancer sequences (A), TMPRSS2 promoter and enhancer (B), and PMEPA1 enhancer and immediate promoter sequences (C). In panels A–C, each point was done in duplicate, experiments were repeated three times, and fold induction was averaged. D, LNCaP cells were incubated for 36 h in 10% sFBS and treated with 1 nm R1881 for the indicated lengths of time. Cells were harvested and analyzed for PSA, PMEPA1, and 18S expression. Maximal expression during 16 h was assigned as 100%, and other values were proportionally adjusted. Each point was done in triplicate and average and se was calculated. The experiment was repeated three times and a representative experiment is shown.
Inhibition of MAPK Reduces AR Stability
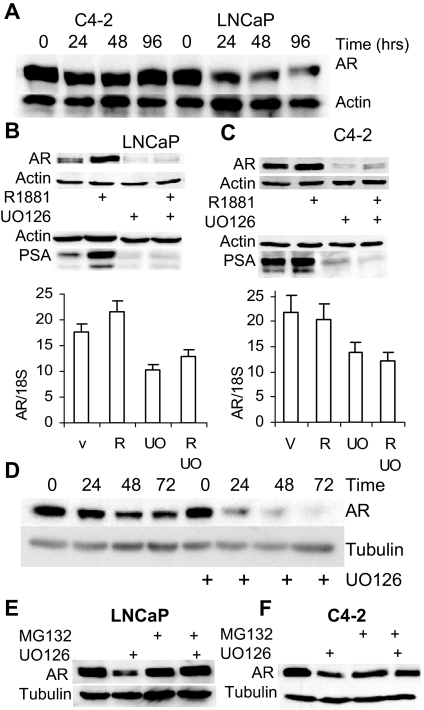
In examining AR expression in androgen-dependent LNCaP cells and in androgen independent C4-2 cells, we noted that AR expression was similar in fetal bovine serum (FBS), but that AR expression in LNCaP cells decreased upon transfer to medium containing charcoal-stripped serum (which is depleted of steroids and small hydrophobic peptides), whereas AR levels in C4-2 cells are maintained (Fig. 6A). The C4-2 cell line is an androgen-independent derivative of the LNCaP cell line (22) that expresses AR and a subset of its target genes in medium depleted of androgens; under these conditions, proliferation and PSA expression remain AR dependent (4). Others have shown that inhibition of HER2 signaling reduces AR stability through an Akt-independent mechanism (7). To test whether p42/p44 MAPK regulates AR expression, LNCaP and C4-2 cells were treated with 20 μm U0126. Remarkably, 48 h of treatment had a profound effect on the levels of AR protein in both LNCaP (Fig. 6B) and C4-2 (Fig. 6C) cells, with a modest reduction in AR mRNA (Fig. 6, B and C, lower) suggesting posttranscriptional regulation. As expected for an AR-dependent gene, PSA expression was also strongly inhibited (Fig. 6, B and C). To determine whether the decrease in AR protein was caused by increased protein turnover, C4-2 cells were treated with cycloheximide in the absence or presence of U0126, and AR expression was determined by Western blotting. As shown in Fig. 6D, AR protein was much less stable in the presence of U0126. Moreover, treating with the proteasome inhibitor, MG132, largely counteracted the effects of U0126 on AR expression in LNCaP and C4-2 cells (Fig. 6, E and F), suggesting that proteasome-mediated degradation of AR was enhanced by U0126 treatment.
Figure 6.
Inhibition of MEK Reduces AR Stability
A, LNCaP and C4-2 cells were plated in medium supplemented with 5% FBS and allowed to grow for 24 h. Cells were then rinsed with serum free medium and transferred to one supplemented with 5% sFBS for the indicated lengths of time. Cells were then harvested and analyzed for AR and actin expression by Western blotting. B, LNCaP cells grown in medium containing sFBS were treated with vehicle (V), 1 nm R1881 (R), 20 μm UO126 (UO), or 1 nm R1881 and 20 μm UO126 for 48 h. Upper, Cells were harvested, protein was extracted, and 20 μg was resolved on a 12.5% SDS-PAGE. The levels of AR, PSA, and actin were analyzed by Western blotting. Lower, Total RNA was extracted, analyzed for AR and 18S expression by quantitative RT-PCR, and AR expression was normalized to 18S RNA. For RNA analyses, each point was performed in triplicate, and the sd was calculated. C, C4-2 cells were treated and analyzed exactly as LNCaP cells in panel B. D, C4-2 cells were plated in medium with 5% sFBS and treated with 10 μg/ml cycloheximide and either DMSO or 20 μm UO126. At the indicated time points, cells were harvested, and protein was extracted and analyzed for expression of AR and tubulin. E and F, LNCaP (E) and C4-2 (F) cells plated in full serum were pretreated with 10 μm MG132 for 0.5 h and then with 20 μm UO126 as indicated. After 24 h, cells were harvested, and protein was extracted and analyzed for AR and tubulin expression by Western blotting.
MAPK Activity Is Necessary for Optimal LNCaP and C4-2 Cell Growth
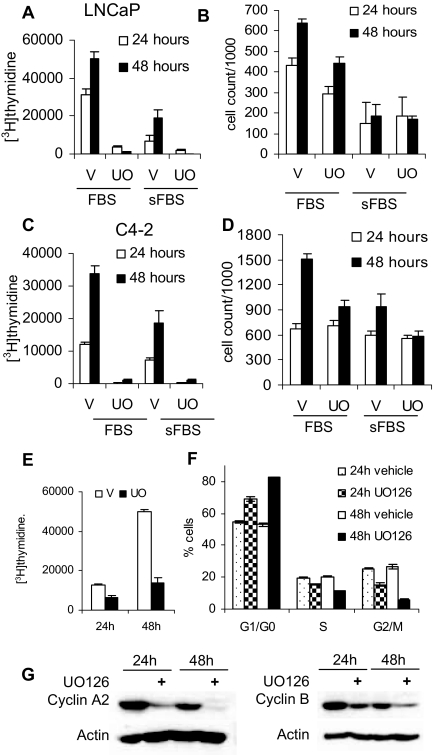
Both the androgen-dependent LNCaP and the androgen-independent C4-2 cells require AR for proliferation (3,4). We have shown that ablation of either ERK1 or ERK2 using siRNA led to decreased proliferation of the LNCaP cells (Fig. 2F). To assess the effect of MEK inhibition on LNCaP and C4-2 cell proliferation, cells were treated with UO126 and [3H]thymidine incorporation, and cell number was measured after 24 and 48 h of treatment. As shown in Fig. 7A in medium supplemented with either stripped or full FBS [3H]thymidine incorporation, a measure of cells passing through S phase, was dramatically reduced in LNCaP cells with UO126 treatment, consistent with the recent report of Carey et al. (23). Cell number was modestly reduced by treatment of cells in FBS (Fig. 7B). Consistent with previous reports, the LNCaP cells didn’t grow significantly in charcoal-stripped FBS (sFBS) (Fig. 7B) because they are androgen dependent; U0126 did not further reduce the number of cells. U0126 strongly inhibited [3H]thymidine incorporation in C4-2 cells (Fig. 7C), almost eliminated any increase in cell number in FBS, and eliminated growth in sFBS (Fig. 7D). To determine whether the MAPK dependence of cell growth in prostate cancer cells is AR dependent, we treated AR-negative PC-3 cells with either dimethylsulfoxide (DMSO) or UO126. PC-3 proliferation was reduced by U0126 treatment (Fig. 7E), demonstrating that growth regulation by MAPK signaling also has an AR-independent component. The decrease in proliferation of the UO126-treated cells was due to an accumulation of cells in G1/G0 phase (Fig. 7F) that was most evident after 48 h of treatment. Chen et al. (24) have reported that AR stability is regulated by cyclin-dependent kinase (CDK)1. Typically, CDK activity is limited by the levels of activating partner cyclins. In agreement with the change in cell cycle distribution, expression of the CDK1 partners, cyclin B and cyclin A2, was decreased in UO126-treated cells compared with those treated with vehicle (Fig. 7G). Because U0126 treatment causes reductions in AR protein only after prolonged treatment, we suggest that this reduction may be a consequence of the reduction in CDK1 activity.
Figure 7.
UO126 Treatment Reduces Cell Proliferation and Induces G0/G1 Accumulation
A, LNCaP cells were plated at 1.5 × 105 cells per well in six-well plates in medium supplemented with 10% FBS. Cells were allowed to attach overnight and rinsed, and medium was substituted for one supplemented with either 10% sFBS or 10% FBS. Cells were then treated with either vehicle (V) (DMSO) or 20 μm UO126 (UO); 24 and 48 h later, cell proliferation was evaluated using [3H]thymidine incorporation. B, LNCaP cells were plated at 1.5 × 105 cells per well and treated in parallel with panel A and harvested at either 24 or 48 h and counted using the Coulter counter. C, C4-2 cells were plated at 1.5 × 105 cells per well in six-well plates. Cells were allowed to attach overnight, rinsed with serum free medium, and placed in a medium with either 5% sFBS or 5% FBS treated with DMSO or UO126 (UO) for 24 or 48 h, and proliferation was examined using [3H]thymidine incorporation. D, C4-2 cells were plated at 1.5 × 105 cells per well and treated in parallel with panel C. Cells were counted at the indicated time points using a Coulter counter. E, PC-3 cells were plated at 50,000 cells per well in six-well plates, treated with either DMSO or 20 μm UO126, and incubated for 24 or 48 h. Cell proliferation was measured using a [3H] thymidine incorporation assay. F, LNCaP cells were treated with either vehicle (DMSO) or 20 μm UO126 for 24 and 48 h. Cells were harvested, fixed with ethanol, stained with propidium iodide, and used for fluorescence-activated cell sorting analysis to determine cell cycle distribution. G, LNCaP cells treated in parallel with F were used to analyze cyclin A2 and B expression by Western blotting.
DISCUSSION
The importance of AR in prostate cancer and the frequent failure of androgen deprivation therapies have led to efforts to identify alternate means for inhibiting AR activity. Indeed, primary tumors, recurrence, and failure of androgen blockade are frequently detected by rising levels of serum PSA, an androgen-regulated gene. Although AR action is growth stimulatory in the epithelial prostate cancer cells, in normal prostate epithelial cells it participates in production of tissue-specific secreted proteins. It is the stromal cell AR that is needed for development of the prostate and growth of the epithelial cells (25). Selective elimination of AR in mouse prostate epithelial cells increases proliferation (26). The activities of AR in prostate cancer cells are a combination of acquired autocrine growth-stimulatory actions and activities of normal prostate epithelial cells including the induction of secreted proteins such as the kallikreins and regulation of genes to limit proliferation. For example, AR action inhibits the production of PCDH11, an activator of Wnt signaling (13) that stimulates proliferation and is up-regulated in advanced prostate cancer (27). Moreover, we have shown that androgens repress expression of transcriptional intermediary factor (TIF)2, a p160 coactivator important both for AR-dependent and AR-independent cell growth (28). Thus, some AR actions are beneficial, and a way to selectively inhibit AR actions potentially is more useful than elimination of all AR activity.
In contrast to most steroid receptors the transcriptional activities of which are strongly dependent on the activation function in the hormone binding domain (AF-2), the amino-terminal AF-1 is dominant in AR (18). Antagonists, which bind the hormone binding domain, are less effective in blocking AF-1 function. The AR and its coactivators are phosphoproteins; consequently, inhibition of specific kinases is a potential alternative or addition to use of an antagonist for inhibiting AR action. Many of the known phosphorylation sites in AR and in the coactivators contain Ser/Thr-Pro motifs, which are often substrates of mitogen-activated kinases or of cyclin-dependent kinases. Elevated p42/p44 MAPK activity presumably due to elevated growth factor receptor signaling has been reported in more advanced prostate cancers (10), and the p160 family of coactivators are all targets of these kinases (15,29,30,31). Inhibition of AR-responsive reporter activity and/or PSA expression by a MEK inhibitor has been reported previously (23,30). We sought to examine the effects of p42/p44 MAPK on the ability of endogenous AR to regulate endogenous target genes and found that the change in induction or repression was target gene specific. Treatment with U0126 for 12 h decreased hormone-dependent induction of PSA and TMPRSS2. However, expression of another primary AR target gene PMEPA1 and the AR-repressed gene PCDH11 was unchanged. To confirm these findings and to eliminate potential off-target effects of U0126, we reduced expression of ERK1 (p42) and ERK2 (p44) separately using siRNA. Expression of ERK1 alone was sufficient for optimal PSA expression, and neither kinase affected PMEPA1 expression similar to the effects of U0126. These experiments were particularly important because Gioeli et al. (20) found that stress-activated kinases reduced AR activity but that reducing expression of MEK1/MEK2, the upstream activators of ERKs, had no effect on induction of PSA. This suggested that U0126 might have some off-target effects or that there is a MEK1/MEK2 independent means to activate ERKs. However, we find that direct elimination of ERKs substantially alters AR activity and the gene-specific pattern mimics the effects of U0126.
The gene-specific effects on AR action have both clinical and mechanistic implications. Interestingly, we found that TMPRSS2 expression was sensitive to loss of either ERK2 or ERK1; this androgen-regulated promoter is linked to portions of the coding region of an Ets factor in more than 60% of prostate cancers (1), and these studies suggest that MEK inhibitors would substantially reduce expression of these gene fusions. On the other hand, our studies highlight the problem in using PSA as a proxy for AR activity. PSA is a protease normally secreted into the lumen of the prostate, but found in the serum of prostate cancer patients; the PSA level in blood is a convenient surrogate measure of prostate tumor burden. Measuring PSA levels in AR-positive prostate cancer cell lines is typically used to evaluate overall AR activity. However, our studies show that measuring PSA induction is not an accurate reflection of all AR activities. Consistent with this, Sathya et al. (32) have shown that some synthetic AR ligands that are selective AR modulators can induce prostate cancer cell growth without inducing PSA.
The target gene-specific differences in requirements for ERK1/ERK2 likely reflect different cofactor requirements for target gene regulation. U0126 treatment reduced recruitment of AR to PSA and TMPRSS2 promoters and enhancers and to the PMEPA1 promoter after a 1-h treatment with R1881, but this deficiency is overcome at later times. Genes, the regulation of which is sensitive to MEK inhibition, exhibited androgen-dependent increases in acetylation of histone H3, which changes chromatin structure making the DNA more accessible to transcription factors. This acetylation was partially inhibited by U0126 treatment. In contrast, androgen does not induce histone H3 acetylation of the MEK inhibitor-resistant gene, PMEPA1. Presumably, the histones at this locus are adequately acetylated, and AR is only needed either to recruit other coactivators or to recruit factors such as cyclin T/Cdk9, which phosphorylate Pol II-promoting transcriptional elongation. Interactions between cyclin T/Cdk9 (P-TEFb) and AR have been reported previously (33). Consistent with the idea that PMEPA1 requires fewer steps before induction of mRNA synthesis, the expression of PSA lags behind induction of PMEPA1 in a time course of androgen-dependent induction. To date, there is no evidence that AR is a direct target of the ERKs (19,20), but new studies reveal that androgen-dependent induction of subsets of target genes requires collaborating transcription factors and, in the case of PSA and TMPRSS2, GATA2 is required not only for optimal androgen-dependent induction, but also for optimal AR binding to the enhancers of these two target genes (34). Phosphorylation of GATA-2 by ERKs has been described although the sites and function have not been elucidated (35). In addition, the p160 coactivators are targets of ERK signaling. We have shown previously that both SRC-1 and TIF2 are required for optimal induction of PSA and TMPRSS2 (4,28). We found that MEK inhibition reduced the interaction between SRC-1 and AR in a mammalian two-hybrid assay and eliminated phosphorylation of Thr1179 and Ser1185 in SRC-1; these sites are important for functional cooperation between SRC-1 and CBP (36). Gregory et al. (30) found that epidermal growth factor (EGF) treatment enhanced interaction between the AR ligand binding domain and TIF2 in a mammalian two-hybrid assay. Moreover, mutation of a MAPK consensus phosphorylation site in TIF2, Ser(736), reduced this interaction (30). Our finding that the genes that undergo hormone-dependent acetylation are sensitive to MEK inhibition is consistent with concept that MEK inhibition reduces AR interaction with HATs. Although coactivators can also facilitate transcriptional repression, the requirement for specific coactivators is target gene specific. We found that SRC-1, but not TIF2, was required for AR-dependent maspin repression (4,28). In the case of PCDH11, we found that eliminating SRC-1 had no effect on the ability of AR to repress its expression.
In addition to the changes in the intrinsic activity of AR, we also found that longer term inhibition of MEK with UO126 led to reduced levels of AR protein with minimal changes in AR mRNA levels. The interactions between the amino and carboxyl termini of AR have been implicated in stabilization of the receptor (37). Overexpression of TIF2/glucocorticoid receptor interacting protein 1 increases expression of an AR mutant lacking the N/C interaction motifs either in the presence of dihydrotestosterone or EGF (30). Thus, both N/C interactions and coactivator interactions can contribute to the overall stability/expression of the protein and both are sensitive to MEK inhibition. The reduction in the N/C terminal interaction in a two-hybrid assay despite the apparent lack of MAPK phosphorylation of AR suggests that proteins such as SRC-1 and TIF2, which interact with regions in the N terminus as well as the ligand-binding domain, serve as bridges between the two regions. A recent study implicates CDK1 in stabilization of AR (24). The prolonged treatment with U0126 caused a G1 arrest and corresponding reduction in the cyclins that partner with CDK1. Thus, the reduction in CDK1 activity also should result in decreased AR stability.
Previous reports have shown that AR is more stable in androgen-independent prostate cancer cells (8), and our study shows that transfer of LNCaP and C4-2 cells to medium containing charcoal-stripped serum depleted of steroids and small hydrophobic growth factors leads to a reduction in AR protein in androgen-dependent LNCaP cells, but not in androgen-independent C4-2 cells. U0126 treatment of C4-2 cells substantially decreases AR stability, suggesting that enhanced autocrine growth factor signaling and continued progression through the cell cycle contributes to maintenance of AR expression facilitating AR activity in androgen-depeleted medium. In tumors, the combination of enhanced p160 coactivator expression (4,28,38,39) and elevated cell signaling likely contributes to increased AR levels and facilitates AR action despite a reduction in androgens. Thus, inhibition of MAPK signaling is a candidate for inhibition of AR activity and expression in androgen-independent cancers.
MATERIALS AND METHODS
Materials
UO126 was purchased from Promega Corp. (Madison, WI), R1881 from PerkinElmer Life Sciences (Boston, MA), and MG132 from EMD Biosciences (San Diego, CA), Protein A sepharose was obtained from GE Healthcare Life Sciences (Piscataway, NJ), and anti-flag antibody, ribonuclease A, and propidium iodide were from Sigma (St. Louis, MO). λ-Protein phosphatase was obtained New England Biolaboratories (Ipswich, MA). Antiacetylated histone H3 antibody was purchased from Millipore Corp. (Temecula, CA).
Cell Culture
LNCaP, PC-3, and COS-1 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). C4-2 cells were purchased from UroCor, Inc. (Oklahoma City, OK). Cells were maintained in RPMI 1640 with 10% FBS (Intergen Co., Purchase, NY) (LNCaP), DMEM/F12 with 5% FBS (PC-3), T medium with 5% FBS (C4-2), and DMEM with 5% FBS (COS-1) with penicillin and streptomycin (Invitrogen, Carlsbad, CA). All cell lines were maintained at 37 C in a humid atmosphere containing 5% CO2. Tissue culture supplies were purchased from Fisher Scientific (Pittsburgh, PA). All chemicals were reagent grade unless otherwise indicated.
Plasmids
Expression plasmids pAct and pBind were purchased from Promega. Expression plasmids pAct-AR (full-length AR), pAct-ARDH (AR DNA and hormone-binding domains), pBind-AR-NTD (AR amino terminus and DNA-binding domain) [kindly provided by Dr. Elizabeth Wilson, University of North Carolina, Chapel Hill, NC (37,40)], pCR3.1-AR, pBind-SRC-1, GRE2E1b-luciferase [an AR-responsive reporter provided by Dr. Carolyn Smith (Baylor College of Medicine, Houston, TX], and 17mer-luc were described previously (41). Expression plasmid pSVL-flag-SRC1 was kindly provided by Dr. David Moore (Baylor College of Medicine).
Transfection, Luciferase, β-Galactosidase, and Proliferation
Assays were performed as were previously described (4,41). Plasmid transfections were performed using polylysine-coupled adenovirus. The luciferase assay was performed using a Promega kit. 2-Nitrophenyl β-d-galactopyranoside (Sigma) colorimetric assay was used to determine β-galactosidase activity. As a surrogate for proliferation, we measured the incorporation of [3H]thymidine (PerkinElmer Life Sciences) (42).
Transfection of siRNA
To down-regulate SRC-1 we transfected Dharmacon smart pool siRNA specific for SRC-1 and Dharmacon noncoding control siRNA (Dharmacon, Lafayette, CO) using AMAXA electroporation system (Amaxa, Gaithersburg, MD). To down-regulate ERK1 and ERK2 we used MAPK3 and MAPK1 smart pools (Dharmacon) and noncoding control siRNA. Two million LNCaP cells were electroporated with 1 nmol of siRNA according to the manufacturer’s protocol, cells were then split into six wells of a six-well plate, treated, and used for RNA and protein analysis.
Chromatin Immunoprecipitation (ChIP) Assay
Cells plated at 2 million cells per 100-mm dish and maintained in medium supplemented with 10% charcoal stripped serum (sFBS) for 36 h were treated for a total of 16 h with vehicle or 20 μm UO126; for the 16-h R1881 time point, 1 nm R1881 was added with U0126. For the 1-h R1881 treatment, R1881 was added for the last hour of U0126 treatment. The ChIP assay was performed as described previously (28) except that 1 μg of N20 AR antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or antiacetyl-Histone H3 (Millipore) was used. The following TaqMan primer and probe sets were used to detect AR binding. The locations of the PSA enhancer and promoter have been described previously (43,44) as have the TMPRSS2 enhancer and promoter (34). The PMEPA1 enhancer was identified by Wang, Q., W. Li, Y. Zhang, X. Yuan, R. Beroukhim, H. Wang, M. Lupien, T. Wu, M. M. Regan, C. A. Meyer, J. S. Carroll, A. K. Manrai, O. A. Jänne, S. P. Balk, R. Mehra, A. M. Chinnaiyan, M. A. Rubin, L. True, M. Fiorentino, C. Fiore, M. Loda, P. W. Kantoff, X. S. Liu, and M. Brown, manuscript submitted (chr3:137538689–137953935), and the PMEPA1 promoter was identified by Masuda et al. (45). PSA enhancer: GCCTGGATCTGAGAGAGATATCATC, ACACCTTTTTTTTTCTGGATTGTTG, 6-FAM-TGCAAGGATGCCTGCTTTACAAACATCC-TAMRA. PSA promoter: TGGGCATGTCTCCTCTGC, CCTGGATGCACCAGGCC, FAM-TTGTCCCCTAGATGAAGTCTCCATGAGCTACAA-TAMRA. TMPRSS2 enhancer: TCCAGGCAGAGGTGTGGC, GCGTATGTCTCCCTGCACCA, FAM-CACCACTTCCTCACCCCTGCCCTAGTT-TAMRA. TMPRSS2 promoter: ATTAGAAAGAACCTCTCAAGTGCCC, GCACCACCGGCCAGG, FAM-CTGAGGTGTGTCCCACCACTTCCTCACTC-TAMRA. PMEPA1 enhancer: GCATTTCTTGGTAAGTCCCTGAGA, CCAGGTGCTAATTTCAGTTGGC, FAM-AAACAGACCTGCCCAATGAAAATGCACA-TAMRA. PMEPA1 promoter: CAGGGAGGGGAGGTCTCTTA, TCAAAAGGGGTATGAGCAGG, FAM-TGAACTAAAAGTACACCCCTCCTG CTCATACCC-TAMRA.
Real-Time Quantitative PCR and RT-PCR
Total RNA was prepared using TRIzol reagent (Invitrogen). Target genes were detected using TaqMan primer and probe sets (Applied Biosystems, Foster City, CA) using TaqMan One-Step RT-PCR master mix reagents (Applied Biosystems). The PCR was run on an ABI PRISM 7700 sequence detection system (Applied Biosystems). All data were normalized to the expression of 18S RNA. AR, PSA, PMEPA, and TMPRSS2 amplicons have been described previously (28). PCDH-11 primers and probe were: TTGTTGTCCGGGACGTACATTT, FAM-CGCGGTCCTGCTAGTATGCGTGGTG-TAMRA, TGGGCGCCAGAGTGGA.
Generation of Phosphorylation-Specific SRC-1 Antibodies
Antibodies were raised in rabbits and affinity purified by Bethyl Laboratories (Montgomery, TX). Antibody specificity was confirmed as previously described for SRC-3 antibodies (31) (see supplemental Fig. 1 and figure legend, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org, for specificity). The peptide sequence used to generate the P-Thr1179 antibody was (C)PPNYGTNPGT(PO4)PP and (C)GTPPASTS(PO4)PFSQLAA for Ser1185.
Immunoprecipitation
Ten COS-1 cell plates were plated at 106 cells per 100-mm plate and transfected with 2 μg of flag-SRC1 expression plasmid. SRC-1 was expressed for 24 h, after which cells were treated with either DMSO or 20 μm UO126 for 12 h. Cells were harvested in TEN buffer with protease and phosphatase inhibitors, lysed in FEB (50 mm Tris, pH 8; 5 mm EDTA, 1% Nonidet P-40, 0.2% sarkosyl, 0.4 m NaCl, 0.2 mm Na3VO4, 10 mm NaMoO4, 20 mm NaFl, 1 mm phenylmethylsulfonylfluoride), freeze thawed three times and cellular lysates spun for 5 min at 14,000 rpm at 4 C. Lysates were diluted 2-fold with TE (10 mm Tris, pH 8; 1 mm EDTA), incubated on ice for 10 min, and spun for 10 min at 100,000 rpm at 4 C. Clarified lysates were incubated with or without 5 μg of anti-FLAG, 12.5 μg rabbit antimouse antibody, and 75 μl of 1:1 TE-protein A sepharose suspension for 1 h at 4 C. Cells were washed twice with 1:1 mix of TE and FEB, once with TE, and extracted twice with Laemmli buffer at 100 C. Eluates were resolved by SDS-PAGE and analyzed for total SRC-1 and phospho-SRC-1 levels by Western blotting.
Western Blotting
AR (using AR441 antibody), PSA (DakoCytomation, Glostrup, Denmark), Flag (Sigma), and SRC-1 (BD Pharmigen, San Diego, CA) Western blottings were performed exactly as we previously reported (4,15). SRC-1 phospho-Thr1179, and phospho-Ser1185 were detected using antibody produced by Bethyl Laboratories. Briefly, the membrane was blocked in 5% milk in TBST (10 mm Tris HCl plus 150 mm NaCl plus 0.1% Tween 20; pH 7.5) supplemented with phosphatase inhibitors (0.2 mm Na3VO4, 20 mm NaF) for 1 h at room temperature, incubated with primary antibody in 1% milk in TBST with phosphatase inhibitors overnight at 4 C, and incubated with rabbit antimouse horseradish peroxidase-conjugated antibody in TBST for 1 h at 4 C. Cyclin A2 and cyclin B were detected using 20 μg of protein extract, resolved on 10% SDS-PAGE, and transferred to nitrocellulose membrane. The membrane was blocked in 1% milk solution in TBST, incubated with cyclin A2 antibody (Santa Cruz Biotechnology) diluted 1:400 or cyclin B antibody (BD Pharmigen) diluted 1:1000. Blots were washed three times in TBST and incubated with secondary antibody conjugated with horseradish peroxidase (GE Healthcare, Buckinghamshire, UK), and protein levels were analyzed using enhanced chemiluminescence detection reagent (Amersham Pharmacia Biotech, Piscataway, NJ).
Fluorescence-Activated Cell Sorting Analysis
C4-2 and LNCaP cells were harvested and fixed exactly as described previously (46). Cells were sorted until 104 events accumulated. The percentage of cells in each cell phase was calculated, triplicates were averaged, and se was calculated.
Supplementary Material
Acknowledgments
We thank Misty Alvarado for expert technical assistance.
Footnotes
This work was supported by National Institutes of Health (NIH) Grant DK65262 (to N.L.W. and I.U.A.); NIH Grant CA58204, the Specialized Program in Research Excellence in Prostate Cancer (to I.U.A. and N.L.W.); and NIH Grant T32DK07696 (to M.N.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online September 11, 2008
Abbreviations: AF-1, Activation function 1; AR, Androgen receptor; CDK, cyclin-dependent kinase; ChIP, chromatin immunoprecipitation; DMSO, dimethylsulfoxide; EGF, epidermal growth factor; FBS, fetal bovine serum; HAT, histone acetyltransferase; MEK, MAPK kinase; PSA, prostate-specific antigen; sFBS, charcoal-stripped FBS; siRNA, small interfering RNA; SRC, steroid receptor coactivator; TBST, Tris HCl-NaCl-Tween 20; TE, Tris-EDTA; TIF, transcriptional intermediary factor.
References
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM 2005 Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310:644–648 [DOI] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL 2004 Molecular determinants of resistance to antiandrogen therapy. Nat Med 10:33–39 [DOI] [PubMed] [Google Scholar]
- Zegarra-Moro OL, Schmidt LJ, Huang H, Tindall DJ 2002 Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res 62:1008–1013 [PubMed] [Google Scholar]
- Agoulnik IU, Vaid A, Bingman 3rd WE, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL 2005 Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res 65:7959–7967 [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Weigel NL 2006 Androgen receptor action in hormone-dependent and recurrent prostate cancer. J Cell Biochem 99:362–372 [Google Scholar]
- Craft N, Shostak Y, Carey M, Sawyers CL 1999 A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med 5:280–285 [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Vivanco I, Kwon A, Tran C, Wongvipat J, Sawyers CL 2004 HER2/neu kinase-dependent modulation of androgen receptor function through effects on DNA binding and stability. Cancer Cell 6:517–527 [DOI] [PubMed] [Google Scholar]
- Gregory CW, Johnson Jr RT, Mohler JL, French FS, Wilson EM 2001 Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res 61:2892–2898 [PubMed] [Google Scholar]
- Bakin RE, Gioeli D, Sikes RA, Bissonette EA, Weber MJ 2003 Constitutive activation of the Ras/mitogen-activated protein kinase signaling pathway promotes androgen hypersensitivity in LNCaP prostate cancer cells. Cancer Res 63:1981–1989 [PubMed] [Google Scholar]
- Gioeli D, Mandell JW, Petroni GR, Frierson Jr HF, Weber MJ 1999 Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res 59:279–284 [PubMed] [Google Scholar]
- Xu LL, Shanmugam N, Segawa T, Sesterhenn IA, McLeod DG, Moul JW, Srivastava S 2000 A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics 66:257–263 [DOI] [PubMed] [Google Scholar]
- Anazawa Y, Arakawa H, Nakagawa H, Nakamura Y 2004 Identification of STAG1 as a key mediator of a p53-dependent apoptotic pathway. Oncogene 23:7621–7627 [DOI] [PubMed] [Google Scholar]
- Yang X, Chen MW, Terry S, Vacherot F, Chopin DK, Bemis DL, Kitajewski J, Benson MC, Guo Y, Buttyan R 2005 A human- and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res 65:5263–5271 [DOI] [PubMed] [Google Scholar]
- Hatziapostolou M, Polytarchou C, Katsoris P, Courty J, Papadimitriou E 2006 Heparin affin regulatory peptide/pleiotrophin mediates fibroblast growth factor 2 stimulatory effects on human prostate cancer cells. J Biol Chem 281:32217–32226 [DOI] [PubMed] [Google Scholar]
- Rowan BG, Weigel NL, O'Malley BW 2000 Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J Biol Chem 275:4475–4483 [DOI] [PubMed] [Google Scholar]
- He B, Lee LW, Minges JT, Wilson EM 2002 Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J Biol Chem 277:25631–25639 [DOI] [PubMed] [Google Scholar]
- Ueda T, Mawji NR, Bruchovsky N, Sadar MD 2002 Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J Biol Chem 277:38087–38094 [DOI] [PubMed] [Google Scholar]
- Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG 1999 The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol 19:8383–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioeli D, Ficarro SB, Kwiek JJ, Aaronson D, Hancock M, Catling AD, White FM, Christian RE, Settlage RE, Shabanowitz J, Hunt DF, Weber MJ 2002 Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J Biol Chem 277:29304–29314 [DOI] [PubMed] [Google Scholar]
- Gioeli D, Black BE, Gordon V, Spencer A, Kesler CT, Eblen ST, Paschal BM, Weber MJ 2006 Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol Endocrinol 20:503–515 [DOI] [PubMed] [Google Scholar]
- Jia L, Kim J, Shen H, Clark PE, Tilley WD, Coetzee GA 2003 Androgen receptor activity at the prostate specific antigen locus: steroidal and non-steroidal mechanisms. Mol Cancer Res 1:385–392 [PubMed] [Google Scholar]
- Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW 1994 Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer 57:406–412 [DOI] [PubMed] [Google Scholar]
- Carey AM, Pramanik R, Nicholson LJ, Dew TK, Martin FL, Muir GH, Morris JD 2007 Ras-MEK-ERK signaling cascade regulates androgen receptor element-inducible gene transcription and DNA synthesis in prostate cancer cells. Int J Cancer 121:520–527 [DOI] [PubMed] [Google Scholar]
- Chen S, Xu Y, Yuan X, Bubley GJ, Balk SP 2006 Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc Natl Acad Sci USA 103:15969–15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha GR, Donjacour AA, Sugimura Y 1986 Stromal-epithelial interactions and heterogeneity of proliferative activity within the prostate. Biochem Cell Biol 64:608–614 [DOI] [PubMed] [Google Scholar]
- Wu CT, Altuwaijri S, Ricke WA, Huang SP, Yeh S, Zhang C, Niu Y, Tsai MY, Chang C 2007 Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc Natl Acad Sci USA 104:12679–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Shukeir N, Potti A, Sircar K, Aprikian A, Goltzman D, Rabbani SA 2004 Up-regulation of Wnt-1 and β-catenin production in patients with advanced metastatic prostate carcinoma: potential pathogenetic and prognostic implications. Cancer 101:1345–1356 [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman III WE, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM, Weigel NL 2006 Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res 66:10594–10602 [DOI] [PubMed] [Google Scholar]
- Font de Mora J, Brown M 2000 AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol 20:5041–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CW, Fei X, Ponguta LA, He B, Bill HM, French FS, Wilson EM 2004 Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J Biol Chem 279:7119–7130 [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW 2004 Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic responses to multiple cellular signaling pathways. Mol Cell 15:937–949 [DOI] [PubMed] [Google Scholar]
- Sathya G, Chang CY, Kazmin D, Cook CE, McDonnell DP 2003 Pharmacological uncoupling of androgen receptor-mediated prostate cancer cell proliferation and prostate-specific antigen secretion. Cancer Res 63:8029–8036 [PubMed] [Google Scholar]
- Lee DK, Duan HO, Chang C 2001 Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J Biol Chem 276:9978–9984 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M 2007 A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towatari M, May GE, Marais R, Perkins GR, Marshall CJ, Cowley S, Enver T 1995 Regulation of GATA-2 phosphorylation by mitogen-activated protein kinase and interleukin-3. J Biol Chem 270:4101–4107 [DOI] [PubMed] [Google Scholar]
- Rowan BG, Garrison N, Weigel NL, O'Malley BW 2000 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol Cell Biol 20:8720–8730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E, Kemppainen JA, Wilson EM 1998 Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J Biol Chem 273:92–101 [DOI] [PubMed] [Google Scholar]
- Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM 2001 A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res 61:4315–4319 [PubMed] [Google Scholar]
- Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, Ittmann M, Tsai SY, Tsai MJ 2005 SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res 65:7976–7983 [DOI] [PubMed] [Google Scholar]
- Langley E, Zhou ZX, Wilson EM 1995 Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem 270:29983–29990 [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Krause WC, Bingman III WE, Rahman HT, Amrikachi M, Ayala GE, Weigel NL 2003 Repressors of androgen and progesterone receptor action. J Biol Chem 278:31136–31148 [DOI] [PubMed] [Google Scholar]
- Stewart LV, Weigel NL 2005 Role of insulin-like growth factor binding proteins in 1α,25-dihydroxyvitamin D3-induced growth inhibition of human prostate cancer cells. Prostate 64:9–19 [DOI] [PubMed] [Google Scholar]
- Schuur ER, Henderson GA, Kmetec LA, Miller JD, Lamparski HG, Henderson DR 1996 Prostate-specific antigen expression is regulated by an upstream enhancer. J Biol Chem 271:7043–7051 [DOI] [PubMed] [Google Scholar]
- Riegman PH, Vlietstra RJ, van der Korput JA, Brinkmann AO, Trapman J 1991 The promoter of the prostate-specific antigen gene contains a functional androgen responsive element. Mol Endocrinol 5:1921–1930 [DOI] [PubMed] [Google Scholar]
- Masuda K, Werner T, Maheshwari S, Frisch M, Oh S, Petrovics G, May K, Srikantan V, Srivastava S, Dobi A 2005 Androgen receptor binding sites identified by a GREF_GATA model. J Mol Biol 353:763–771 [DOI] [PubMed] [Google Scholar]
- Narayanan R, Edwards DP, Weigel NL 2005 Human progesterone receptor displays cell cycle-dependent changes in transcriptional activity. Mol Cell Biol 25:2885–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.