Abstract
Objective: Adipocyte and hepatocyte endoplasmic reticulum (ER) stress response is activated in dietary and genetic models of obesity in mice. We hypothesized that ER stress was also activated and associated with reduced insulin sensitivity (SI) in human obesity.
Research Design and Methods: We recruited 78 healthy, nondiabetic individuals over a spectrum of body mass index (BMI) who underwent oral and iv glucose tolerance tests, and fasting sc adipose and muscle biopsies. We tested expression of 18 genes and levels of total and phosphorylated eukaryotic initiation factor 2α, c-jun, and c-Jun N-terminal kinase 1 in adipose tissue. We compared gene expression in stromal vascular and adipocyte fractions in paired samples from 22 individuals, and tested clustering on gene and protein markers.
Results: Adipocyte expression of most markers of ER stress, including chaperones downstream of activating transcription factor 6, were significantly correlated with BMI and percent fat (r > 0.5; P < 0.00001). Phosphorylation of eukaryotic initiation factor 2α but not of c-Jun N-terminal kinase 1 or c-jun was increased with obesity. ER stress response (as elsewhere) was also increased with obesity in a second set of 86 individuals, and in the combined sample (n = 161). The increase was only partially attributable to the stromal vascular fraction and macrophage infiltration. ER stress markers were only modestly correlated with SI. Clustering algorithms supported ER stress activation with high BMI but not low SI.
Conclusions: Multiple markers of ER stress are activated in human adipose with obesity, particularly for protective chaperones downstream of activating transcription factor 6α.
Studies of subcutaneous adipose tissue endoplasmic reticulum stress in healthy individuals shows marked elevation of chaperone gene expression and increased phosphorylation of initiation factor EIF2α with increasing obesity.
Despite the strong epidemiological linkage between obesity and type 2 diabetes, the pathophysiological mechanisms of that linkage remain uncertain. Considerable data suggest that obesity is a state of chronic inflammation, and that this inflammation in turn results in reduced insulin sensitivity (SI) (1), and recent genetic analyses point to inflammatory and immune pathways in insulin resistance, glucose intolerance, and metabolic syndrome (2,3). Whether inflammation is the proximal cause of diabetes or the bystander of another process such as oxidative stress (4) is unknown.
Recently, two studies suggested that increased endoplasmic reticulum (ER) stress may represent the proximal cause of the association between obesity, hepatic, and adipocyte insulin resistance, and type 2 diabetes (5,6). The ER is contiguous with the nuclear membrane, and is the location for synthesis and folding of membrane and secretory proteins. Conditions that impair protein folding, nutritional deficiency such as hypoglycemia, altered ER homeostasis such as induced by chemicals thapsigargin (altered calcium homeostasis), or tunicamycin (blocked glycosylation) all result in ER stress response (ERSR) and the elaborate unfolded protein response (UPR) (7). More recently fatty acids also induced ER stress in some cell lines (8,9).
The ERSR is mediated by three proximal transmembrane proteins that in the absence of unfolded proteins or ER stress are held inactive by the chaperone heat shock protein A5 (HSPA5) (also known as GRP78 or BiP). ER stress activates pancreatic eukaryotic initiation factor (eIF) 2α kinase [phospho-extracellular signal-regulated kinase (PERK), encoded by EIF2AK3], which by phosphorylating EIF2α, blocks most translation but favors translation of pro-apoptotic activating transcription factor (ATF) 4 and transcription of pro-apoptotic factors C/EBP homologous protein (CHOP) and ATF3. Activation of inositol-requiring enzyme 1 (IRE1) (encoded by human gene ERN1) acts as an endonuclease to splice transcription factor X-box binding protein 1 (XBP1), which in turn activates downstream chaperones. Upon activation, ATF6α is transported to the Golgi, where it is further processed by two site-specific proteases (specificity proteins 1 and 2) to release the cytosolic fragment, which is a transcriptional coactivator that in turn increases transcription of downstream chaperones, including HSPA5 (10,11,12). Recent studies suggest that ATF6α activity is critical to up-regulation of protective chaperone proteins, including HSPA5, HYOU1, CALR, DNAJC3 (p58IPK), and HSP90B1 (GRP94) (13,14), as well as increased ER-associated protein degradation (encoded in part by gene EDEM1) (13). When up-regulation of protective chaperones in not successful, apoptotic pathways predominate (11,12).
Ozcan et al. (5) demonstrated increased markers of ER stress in both liver and adipose tissue from mice with either genetic (ob/ob) or dietary (high fat diet induced) models of obesity, including increased phosphorylation of PERK and eIF2α, increased transcript levels of HSPA5 (GRP78), and increased c-Jun N-terminal kinase (JNK) activity (c-jun phosphorylation). XBP1 heterozygous null mice were both insulin resistant and glucose intolerant (5). Nakatani et al. (6) similarly showed increased ER stress markers in the livers of diabetic db/db mice. Conversely, overexpression of chaperone proteins such as HYOU1 (ORP150) (6) or the use of small molecule chaperones (15) appears to reduce ER stress, improve SI and secretion, and improve diabetes control.
Despite convincing data supporting a role for ERSR in the pathogenesis of diabetes in obese mice and cell culture, the role of this pathway in human tissues is unclear. Two other recently published studies support an increase in ERSR in human obesity (16,17). We tested the hypothesis that ER stress is activated in human sc adipose tissue with obesity independent of hyperglycemia, and in turn reduces whole body insulin action. We report on expression of both proximal sensors and downstream chaperones in sc adipose and muscle from individuals over a wide range of body mass index (BMI) and SI.
Subjects and Methods
Experimental subjects
The primary study was conducted in 78 individuals who were recruited by advertisement for good general health, age between 18 and 55 yr, BMI between 19 and 42.5 kg/m2, and either self-determined European American or African-American ancestry. All participants had a screening visit, at which time they had measurement of height, weight, waist and hip measures, body fat determination by dual x-ray absorptiometry (DXA) scan, fasting blood samples for lipid measures, and a standard 75-g oral glucose tolerance test with measurement of glucose and insulin at baseline and 30-min intervals for 2 h. Participants with diabetic glucose tolerance tests were not enrolled, but 10 subjects had impaired glucose tolerance. Subjects returned for a second visit, generally within 2 months. Premenopausal women were studied in the follicular phase of the menstrual cycle. After initiating iv lines in both arms, adipose and muscle biopsies were performed using a Bergstrom needle under local (lidocaine) anesthesia. Biopsy samples were immediately rinsed in normal saline, cut, and quick frozen in liquid nitrogen. After the biopsy an insulin-modified (0.04 U/kg), frequently sampled iv glucose tolerance test (FSIGT) was performed as described previously (18). The initial 62 participants had fasting insulin obtained before (two baseline) and after (two additional baseline samples) the biopsies to exclude altered SI induced by the biopsy. Pre- and post-biopsy insulin levels did not differ significantly (data not shown).
Adipose tissue cDNA was available for a second sample of 86 individuals who participated in several studies using a similar protocol, as described in detail elsewhere (19,20). FSIGT studies were available on 83 African-American or European American subjects. Table 1 shows the characteristics of the primary (sample 1) and replication (sample 2) groups. Of the 80 subjects included in the analysis, 34 had impaired glucose tolerance.
Table 1.
Study population characteristics
| No. | Sample 1 | No. | Sample 2 | P value | |
|---|---|---|---|---|---|
| Male/female | 28/50 | 14/72 | |||
| Ethnicity (white/Black/Hispanic) | 54/24/0 | 72/13/1 | |||
| Age (yr) | 78 | 42.3 (8.8) | 86 | 42.7 (10.0) | NS |
| BMI (kg/m2) | 78 | 28.7 (5.6) | 86 | 31.0 (4.8) | 0.005 |
| PFAT (%) | 77 | 32.9 (10.3) | 86 | 38.4 (7.6) | 0.0002 |
| Waist circ (cm) | 77 | 95.2 (13.5) | 74 | 96.4 (14.4) | NS |
| Waist to hip ratio | 77 | 0.879 (0.072) | 74 | 0.886 (0.085) | NS |
| Systolic BP (mm Hg) | 78 | 124.2 (13.2) | 75 | 116.2 (21.4) | 0.006 |
| Diastolic BP (mm Hg) | 78 | 77 (10) | 75 | 75.7 (14.4) | NS |
| Total cholesterol (mmol/liter) | 78 | 4.58 (0.93) | 75 | 4.91 (1.05) | 0.05 |
| HDL cholesterol (mmol/liter) | 78 | 1.38 (0.83, 2.29) | 75 | 1.31 (0.76, 2.28) | NS |
| LDL cholesterol (mmol/liter) | 78 | 2.49 (1.37, 4.56) | 69 | 2.69 (1.45, 5.00) | 0.11 |
| Triglycerides | 78 | 1.08 (0.39, 3.00) | 75 | 1.32 (0.45, 3.87) | 0.02 |
| SI [×10−4 min−1 (μU/ml)−1] | 77 | 3.16 (1.08, 9.26) | 83 | 2.40 (0.69, 8.39) | 0.004 |
| AIRg (mg-min/dl) | 77 | 370 (70.0, 1960) | 83 | 328 (63, 1697) | NS |
| DI | 77 | 1169 (236, 5805) | 83 | 768 (160, 3690) | 0.001 |
The traits shared between the primary study population (sample 1) and replication population of previously examined individuals ascertained under other studies (sample 2) are shown. Skewed variables are shown as mean (95% CIs); normal variables are mean (sd). P values are from a two-tailed t test comparing the two populations. Note that the numbers of subjects used in subsequent tables and figures removed some individuals who were not of Caucasian or African-American ancestry, were studied under a different protocol, or could not be modeled for FSIGT data. For conversion to mass units (mg/dl) divide cholesterol by 0.0259 and triglycerides by 0.0113. BP, Blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein; NS, not significant; PFAT, percent fat from DXA; Waist circ, waist circumference.
All study participants provided written, informed consent under protocols approved by the University of Arkansas for Medical Sciences or Central Arkansas Veterans Healthcare System Institutional Review Boards, and the Central Arkansas Veterans Affairs Research and Development Committee.
Laboratory measurements
Insulin levels were measured using an immunochemiluminometric assay (Molecular Light Technology, Wales, UK) and plasma glucose by a glucose oxidase assay. Standard clinical assays (lipids, glucose) were performed at LabCorp, Inc. (Burlington, NC).
Gene expression
Total RNA was isolated from adipose using the RNAeasy Lipid Tissue Mini Kit (QIAGEN, Inc., Valencia, CA), and from muscle using the Ultraspec RNA kit (Biotecx Laboratories, Inc., Houston, TX). The adipocyte fraction from 22 samples was separated from the stromal vascular fraction after collagenase digestion using the method of Rodbell (21). The 22 individuals had similar BMI and SI to the full sample (data not shown). Total RNA was isolated from the adipocyte fraction using the RNeasy Lipid Tissue Mini Kit, and from the stromal-vascular fraction using the RNAqueous kit (Ambion, Inc., Austin, TX).
The quantity and quality of the isolated RNA were determined by UV spectrophotometry and electrophoresis using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA), respectively, and 1 μg was reverse transcribed using random hexamer primers with TaqMan reverse transcribed reagents (Applied Biosystems, Foster City, CA). All RNA samples from a single study population and tissue were reverse transcribed with the same kit on the same day (see supplemental Table 1S for details, which is published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org). The standard curves were generated using pooled RNA from the samples assayed. Primers were designed to capture most known splice variants and such that the amplicon spanned an intron, or one primer spanned two contiguous exons (supplemental Table 1S). The correlation between replicate runs (sample 1) was more than 0.98 for HSPA5, STC2, and 18S RNA (P < 10−50).
Quantitative Western blots
Protein was isolated from 100–200 mg sc adipose by lysis with a tissue homogenizer, sonication, and centrifugation (see supplemental data for detailed methods, which are published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org). Protein content was estimated by the Bradford method (22). Equal amounts (40–50 μg) of total protein were separated on 8% sodium dodecyl sulfate-polyacrylamide gels, and transferred onto Trans-Blot nitrocellulose membrane (Bio-Rad Laboratories, Inc., Hercules, CA), and hybridized with the primary and secondary antibodies as described in the supplemental data. Blots were scanned with the ChemiDoc XRS Image Analysis System (Bio-Rad Laboratories), and quantified by densitometry with Quantity One version 4.6.3 Image Analysis software (Bio-Rad Laboratories). To facilitate comparison across multiple gels, approximately equal numbers of lean and obese individuals were loaded on each gel, including some samples in common between gels, and protein levels were normalized to β-actin.
Statistical analysis
SI was estimated from the insulin and glucose data using the MINMOD Millennium program (23), and acute insulin response to glucose (AIRg) and disposition index (DI) were taken from the program output. SI from the oral glucose tolerance test was calculated using the homeostatic index (24). Gene expression levels were normalized to 18S RNA, and the ratio was used in all calculations. Data were assessed for normality separately within each study sample and within the combined data set, and nonnormal variables natural logarithmically transformed to normality. Partial correlation measures of obesity or SI with gene expression were calculated controlling for age, gender, and ethnicity. General linear regression models were examined with age, gender, ethnicity, and study identity in combined data sets as covariates. Obesity measures were evaluated as covariates in models with gene expression levels as the dependent variables. SI was assessed as the dependent variable with gene expression as a covariate to test the effects of ER stress on SI, with BMI as an additional covariate to control for obesity effects on SI. No interactions of study identifier with gene expression levels or measures of obesity were found. Protein levels were compared across gels after taking the protein or phosphoprotein to β-actin ratio and using both the two-tailed Student’s t test, and to avoid problems with nonnormal data, the Mann-Whitney U test. Results were similar; we report only the Mann-Whitney U results. Analyses were performed in SPSS for Windows version 12.0 (SPSS, Inc., Chicago, IL). Values are reported as means with sd values for normal data, or geometrical means and 95% confidence intervals (CIs) for data not normally distributed.
We performed the unsupervised hierarchical clustering for gene expression and phosphoprotein levels using PermutMatrix version 1.9.3 software (25). Euclidian distance matrix between each individual was generated using log-normalized values and seriation under multiple fragment heuristics. The hierarchical clustering-based tree was generated using McQuitty’s criteria (26).
Results
ER stress markers correlate with total adiposity
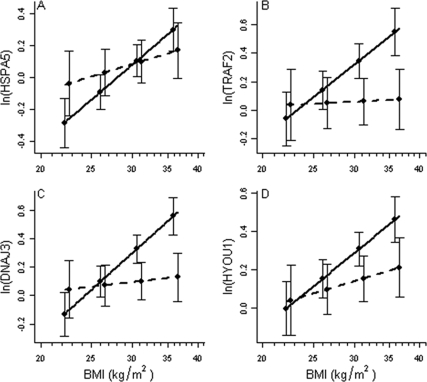
Characteristics of the study population are shown in Table 1. Among the primary study (sample 1) of 78 individuals (28 men, 50 women; BMI 19–42.5 kg/m2) recruited specifically for this study, key proximal ER stress markers HSPA5 (encoding GRP78), ATF6α, and EIF2AK3 (encoding PERK), and downstream chaperones and response genes DNAJC3, HYOU1, HSP90B1, CALR, STC2, EDEM1, and NRF2 were strongly correlated with both BMI and DXA-determined percent body fat (r > 0.45; Tables 2 and 3). Transcripts downstream of ATF4 (indirectly activated by PERK phosphorylation of EIF2α), including markers of apoptosis, were more variably associated with obesity measures. Thus, ATF3 (r > 0.4) was strongly correlated with obesity measures, factors ATF4, TRAF2, and GADD34 modestly correlated (r > 0.3), and primary CHOP showed no activation with increasing obesity. Neither ERN1 (encoding proximal sensor IRE1) nor total XBP1 transcript levels were correlated with obesity measures. Selected regression lines predicting average expression for HSPA5, HYOU1, DNAJC3, and TRAF2 given BMI by ethnicity are shown in Fig. 1. Gender and measures of obesity (BMI, waist circumference, percent fat) were significant determinants of gene expression for most markers in general linear regression models. Ethnicity was not significant as a main effect, but ethnicity and measures of total obesity showed a significant interaction to determine gene expression levels (HSPA5, ethnicity vs. BMI, P = 0.01; Fig. 1). The relationship of BMI and gene expression was reduced (lower slope) in African-American compared with European American participants. Furthermore, measures of fat distribution (waist to hip ratio, trunk to leg fat from DXA) were less correlated (r < 0.3) than obesity measures with transcriptional markers of ERSR (Tables 2 and 3).
Table 2.
Correlation of metabolic traits with ERSR in sample 1
| Gene type trait | r/p | Proximal ER sensors
|
sp XBP1 | Protein degradation
|
|||
|---|---|---|---|---|---|---|---|
| HSPA5 | ATF6α | EIF2AK3 | EDEM1 | TRAF2 | |||
| BMI | r | 0.51 | 0.48 | 0.48 | 0.43 | 0.45 | 0.38 |
| p | 6 × 10−6 | 2 × 10−5 | 2 × 10−5 | 0.0001 | 0.00008 | 0.001 | |
| Waist circ | r | 0.46 | 0.43 | 0.44 | 0.39 | 0.38 | 0.31 |
| p | 5 × 10−5 | 0.0002 | 0.0001 | 0.0007 | 0.0009 | 0.009 | |
| Hip circ | r | 0.47 | 0.44 | 0.44 | 0.43 | 0.43 | 0.35 |
| p | 4 × 10−5 | 9 × 10−5 | 0.0001 | 0.0001 | 0.00015 | 0.002 | |
| Waist to hip ratio | r | 0.25 | 0.21 | 0.23 | 0.12 | 0.13 | 0.1 |
| p | 0.037 | 0.07 | 0.05 | 0.32 | 0.26 | 0.4 | |
| PFAT | r | 0.55 | 0.53 | 0.52 | 0.46 | 0.52 | 0.42 |
| p | 6 × 10−7 | 2 × 10−6 | 3 × 10−6 | 5 × 10−5 | 2 × 10−6 | 0.0002 | |
| Trunk: leg | r | 0.23 | 0.25 | 0.214 | 0.2 | 0.19 | 0.17 |
| p | 0.06 | 0.037 | 0.07 | 0.1 | 0.12 | 0.16 | |
| SI | r | −0.21 | −0.29 | −0.18 | −0.26 | −0.17 | −0.16 |
| p | 0.08 | 0.01 | 0.14 | 0.027 | 0.14 | 0.16 | |
| AIRg | r | 0.05 | 0.018 | −0.01 | 0.03 | 0.05 | 0.059 |
| p | 0.68 | 0.89 | 0.93 | 0.79 | 0.66 | 0.62 | |
| DI | r | −0.1 | −0.19 | −0.14 | −0.16 | −0.07 | −0.05 |
| p | 0.42 | 0.1 | 0.24 | 0.19 | 0.56 | 0.656 | |
| Fasting glucose | r | 0.2 | 0.25 | 0.19 | 0.19 | 0.18 | 0.1 |
| p | 0.09 | 0.032 | 0.11 | 0.11 | 0.12 | 0.39 | |
| 2-h glucose | r | 0.04 | 0.11 | 0.07 | 0.11 | 0.11 | 0.12 |
| p | 0.75 | 0.33 | 0.54 | 0.36 | 0.36 | 0.32 | |
| Insulin AUC | r | 0.11 | 0.19 | 0.1 | 0.15 | 0.15 | 0.09 |
| p | 0.36 | 0.1 | 0.39 | 0.21 | 0.22 | 0.44 | |
| Ins/glu AUC | r | 0.08 | 0.15 | 0.07 | 0.16 | 0.12 | 0.04 |
| p | 0.5 | 0.2 | 0.55 | 0.18 | 0.33 | 0.73 | |
Correlations of traits with expression of ER stress genes in sc adipose. Underlined cells are statistically significance. The second column (r/p) gives either the partial correlation coefficient (r) or the attached significance (p). For space considerations, genes with no significant correlations are not shown: ERN1, total XBP1, and CHOP. Note that approved human gene codes are used throughout. Commonly used aliases are GRP78/BIP (HSPA5), PERK (EIF2AK3), p58IPK (DNAJC3), and ORP150 (HYOU1). circ, Circumference; Insulin AUC, insulin area under curve from oral glucose tolerance test study; Ins/glu AUC, ratio of insulin to glucose area under curve from oral glucose tolerance test visit; PFAT, percent fat from DXA; SI from MINMOD; sp XBP1, spliced XBP1 by real-time assay; Trunk:leg, ratio of mean percent fat in both legs to trunk from DXA.
Table 3.
Correlations of ERSR genes with metabolic traits, chaperones and apoptosis pathway genes
| Gene type trait | r/p | Chaperones
|
Apoptosis pathway
|
|||||
|---|---|---|---|---|---|---|---|---|
| HYOU1 | DNAJC3 | CALR | STC2 | ATF4 | GADD34 | ATF3 | ||
| BMI | r | 0.51 | 0.53 | 0.53 | 0.62 | 0.3 | 0.36 | 0.48 |
| p | 6 × 10−6 | 2 × 10−6 | 2 × 10−6 | 6 × 10−9 | 0.01 | 0.002 | 2 × 10−5 | |
| Waist circ | r | 0.51 | 0.47 | 0.5 | 0.62 | 0.24 | 0.35 | 0.5 |
| p | 6 × 10−6 | 3 × 10−5 | 1 × 10−5 | 8 × 10−9 | 0.045 | 0.003 | 1 × 10−5 | |
| Hip circ | r | 0.52 | 0.5 | 0.49 | 0.59 | 0.26 | 0.4 | 0.44 |
| p | 3 × 10−6 | 9 × 10−6 | 1 × 10−5 | 3.8 × 10−8 | 0.03 | 0.0006 | 9 × 10−5 | |
| Waist to hip ratio | r | 0.25 | 0.21 | 0.27 | 0.37 | 0.1 | 0.13 | 0.34 |
| p | 0.03 | 0.07 | 0.02 | 0.001 | 0.42 | 0.29 | 0.003 | |
| PFAT | r | 0.54 | 0.57 | 0.52 | 0.65 | 0.31 | 0.38 | 0.43 |
| p | 1 × 10−6 | 1 × 10−7 | 2 × 10−6 | 6.10−10 | 0.008 | 0.0009 | 0.0001 | |
| Trunk: leg | r | 0.23 | 0.23 | 0.27 | 0.37 | 0.19 | 0.4 | 0.37 |
| p | 0.06 | 0.05 | 0.02 | 0.001 | 0.11 | 0.0004 | 0.0001 | |
| SI | r | −0.2 | −0.23 | −0.25 | −0.35 | −0.21 | −0.37 | −0.26 |
| p | 0.084 | 0.05 | 0.04 | 0.002 | 0.08 | 0.001 | 0.025 | |
| AIRg | r | 0.2 | 0.06 | 0.11 | 0.21 | −0.006 | 0.052 | −0.07 |
| p | 0.083 | 0.64 | 0.36 | 0.07 | 0.96 | 0.66 | 0.54 | |
| DI | r | 0.075 | −0.1 | −0.06 | −0.02 | −0.16 | −0.21 | −0.27 |
| p | 0.53 | 0.38 | 0.6 | 0.83 | 0.18 | 0.08 | 0.02 | |
| Fasting glucose | r | 0.17 | 0.22 | 0.26 | 0.13 | 0.001 | 0.2 | 0.19 |
| p | 0.16 | 0.06 | 0.03 | 0.28 | 0.99 | 0.1 | 0.11 | |
| 2-h glucose | r | 0.07 | 0.08 | 0.11 | 0.16 | 0.13 | 0.24 | 0.15 |
| p | 0.55 | 0.51 | 0.35 | 0.18 | 0.27 | 0.038 | 0.21 | |
| Insulin AUC | r | 0.19 | 0.17 | 0.15 | 0.28 | 0.12 | 0.3 | 0.23 |
| p | 0.11 | 0.15 | 0.22 | 0.02 | 0.3 | 0.009 | 0.05 | |
| Ins/glu AUC | r | 0.19 | 0.14 | 0.1 | 0.25 | 0.1 | 0.26 | 0.21 |
| p | 0.1 | 0.23 | 0.42 | 0.04 | 0.38 | 0.03 | 0.08 | |
Correlations of traits with expression of ER stress genes in sc adipose. Underlined cells are statistically significance. The second column (r/p) gives either the partial correlation coefficient (r) or the attached significance (p). For space considerations, genes with no significant correlations are not shown: ERN1, total XBP1, and CHOP. Note that approved human gene codes are used throughout. Commonly used aliases are GRP78/BIP (HSPA5), PERK (EIF2AK3), p58IPK (DNAJC3), and ORP150 (HYOU1). circ, Circumference; Insulin AUC, insulin area under curve from oral glucose tolerance test study; Ins/glu AUC, ratio of insulin to glucose area under curve from oral glucose tolerance test visit; PFAT, percent fat from DXA; SI from MINMOD; sp XBP1, spliced XBP1 by real-time assay; Trunk: leg, ratio of mean percent fat in both legs to trunk from DXA.
Figure 1.
Gene expression vs. BMI by ethnicity. The figure shows regression plots for HSPA5 (A), TRAF2 (B), DNAJC3 (C), and HYOU1 (D). For all figures, both gene expression measures (ratio to 18S RNA) and BMI were logarithmically transformed, but BMI is shown as kg/m2. Solid lines represent means for European American participants (n = 54), and dotted lines represent means for African-American participants (n = 24). Bars represent 95% CIs.
We stratified the population into lean (BMI < 25 kg/m2), overweight (BMI 25–30 kg/m2), and obese (BMI > 30 kg/m2). As shown in Fig. 2, proximal sensors ATF6, EIF2AK3 (PERK), and HSPA5, and factors downstream of ATF6 (EDEM1; chaperone proteins HYOU1, HSPA5, DNAJC3, HSP90B1, CALR, and STC2) showed increased gene expression by over 60% from lean to obese individuals.
Figure 2.
Gene expression of ERSR genes by BMI group. Gene expression, calculated as real-time values normalized to 18S RNA, is shown by BMI divided into lean (BMI < 25 kg/m2; n = 21, black bars), overweight (25–30 kg/m2; n = 29, white bars), or obese (BMI > 30 kg/m2, n = 28, hatched bars). Significant differences based on ANOVA of logarithmically-transformed values are marked by asterisks; no asterisks represent no significant difference. Other levels of significance are as follows: *, <0.01; **, P < 0.001; ***, P < 0.0001; ****, P < 0.00001. The graph is shown on a linear scale.
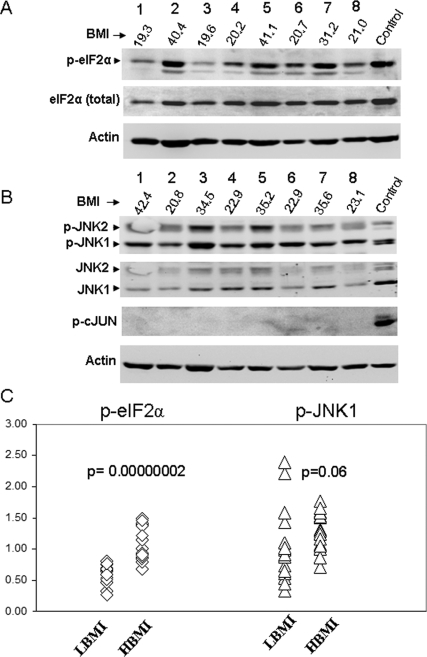
Effects on SI are proposed to result from kinase action of IRE1 (encoded by ERN1) on JNK1. Similarly, PERK (encoded by EIF2AK3) is activated by phosphorylation, and in turn phosphorylates EIF2α to halt translation. We examined total and phosphorylated JNK1, c-jun, and EIF2α in the 15 leanest (lowest BMI) and 19 most obese (highest BMI) individuals (Fig. 3). eIF2α (Fig. 3, A and C; P = 3 × 10−7) but not JNK1 or c-jun (Fig. 3B) showed increased phosphorylated in the high BMI group. The increased EIF2α phosphorylation was less significant (P < 0.05) when normalized to total EIF2α rather than β-actin.
Figure 3.
Western blot of phosphoproteins. Representative Western blots showing samples with high and low BMI. Antibodies are either total protein as noted, or phosphoproteins. A, Total and phosphorylated eIF2α, with β-actin for normalization. Control is palmitate-treated HepG2 cells, which show acute ERSR. Numbers below lane numbers 1–8 give BMI of individuals represented. B, Phosphorylated and unphosphorylated forms of JNK1 and JNK2, and phosphorylated c-jun. Control is from palmitate-treated HepG2 cells. The gel is representative of Western blots on 34 individuals, as described in the Results, with each individual measured at least twice. C, Scatterplot for 34 samples for p-eIF2α and p-JNK1. Significance (P values) is based on the Mann-Whitney U test. HBMI, high BMI group (n = 19); LBMI, low BMI group (n = 15).
Activation of IRE1 in the acute state results in activation of transcription factor XBP1 by splicing of a 24-bp intron. No splicing was observed in lean or obese subjects using gel electrophoresis standard methods (supplemental Fig. 2S, which is published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org), but spliced XBP1 transcript increased by 47% from the lean to obese subjects using a real-time PCR assay (P = 0.0002; Fig. 2).
ER stress and obesity: replication study
We examined genes HSPA5, EDEM1, DNAJC3, HYOU1, and TRAF2 in a second group of 86 individuals ascertained originally for other studies (Table 1), again categorizing individuals as lean (n = 13, BMI < 25 kg/m2), overweight (n = 16, BMI 25–30 kg/m2), or obese (n = 57, BMI > 30 kg/m2). As in sample 1, HSPA5 expression was significantly increased with obesity [means and 95% CIs 0.73 (0.63, 0.85), 0.84 (0.74, 0.95), and 1.07 (1.00, 1.16) for lean, overweight, and obese, respectively; P < 0.00001]. Chaperones DNAJC3, HYOU1, and TRAF2 were also significantly increased in obese individuals (P < 0.05), whereas EDEM1 did not reach significance. Partial correlations with obesity measures are shown in supplemental Table 3S, and results were confirmed in linear regression analyses with BMI (data not shown). When samples 1 and 2 were combined (n = 161), the increase of ERSR transcripts with obesity class was highly significant, and increased from lean to obese classes by 31% for TRAF2 (P < 0.0005) to 53% for HSPA5 (P < 1 × 10−11) (supplemental Table 2S).
ER stress and muscle gene expression
Muscle gene ERSR gene did not correlate with obesity measures (BMI, percent fat, or waist circumference; P > 0.35) for genes HSPA5, EDEM, DNAJC3, and TRAF2 examined in 77 individuals (sample 1) for whom paired adipose and muscle biopsies were available. In both sc adipose and in muscle, expression of chaperone genes downstream of the ERSR was mutually highly correlated (r > 0.60 in muscle; r > 0.9 for factors downstream of ATF6α in adipose), suggesting that all chaperones were similarly increased in adipose in response to obesity. In contrast, expression of ERSR genes in adipose tissue correlated poorly with the same gene expression levels in muscle with the exception of HSPA5, for which the correlation in expression was nominally significance (r = 0.25; P = 0.03). Interestingly, muscle ERSR gene expression tended to be higher in African-Americans, reaching significance for HSPA5 (HSPA5 to 18S ratio 0.91, CI 0.83–1.0 in European Americans; 1.11, CI 0.97–1.28 in African-Americans; P = 0.02).
ER stress and SI
If the increased adipocyte ER stress in obese individuals contributes to insulin resistance, we hypothesized that transcriptional markers would predict whole body SI. In sample 1 (Tables 2, 3, and 4), the strongest correlations with SI were for ATF6α (r = −0.29; P = 0.01), GADD34 (r = −0.37; P = 0.001), and DNAJC3 (r = −0.23; P = 0.05). Stronger correlations were observed in sample 2 for HSPA5 (r = −0.43; P = 0.00006), and in the combined sample the correlation was highly significant (P = 0.00007; r = −0.31; Table 4). Partial regression or general linear models are standard approaches to determine which of several correlated variables is most probably driving an association. In a regression model that included HSPA5 expression, BMI, age, gender, and protocol, BMI remained highly predictive (P < 1 × 10−8), gender was nominally significant (P = 0.02), but HSPA5 expression failed to reach significance as a predictor of SI. In a further attempt to control for the strong correlations of ER stress transcripts and obesity measures, and the known negative correlation of obesity and SI, we calculated the residual for SI controlling for BMI, age, gender, ethnicity, and protocol. Again, no individual marker of ER stress (HSPA5, DNAJC3, EDEM1, GADD34, or TRAF2) was significantly correlated with residual SI (r2 < 0.01; P > 0.18). Surrogate measures of SI from the oral glucose tolerance visit were highly correlated with SI, and like SI were not significantly correlated with ERSR (data not shown).
Table 4.
Correlation of ER stress factors gene expression in adipose with SI
| Gene | Sample 1 (n = 78)
|
Sample 2 (n = 83)
|
Combined sample (n = 161)
|
|||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| HSPA5 | −0.21 | 0.08 | −0.43 | 5.9E-05 | −0.31 | 7.3E-05 |
| EDEM1 | −0.18 | 0.14 | −0.22 | 0.042 | −0.18 | 0.03 |
| TRAF2 | −0.16 | 0.16 | −0.11 | 0.33 | −0.08 | 0.29 |
| DNAJC3 | −0.23 | 0.052 | −0.16 | 0.15 | −0.15 | 0.06 |
| HYOU1 | −0.20 | 0.08 | −0.29 | 0.009 | −0.22 | 0.006 |
Correlation of SI with sc adipose in the primary study (sample 1) and replication study (sample 2), and combined sample. Note that in contrast to Table 1, Table 4 shows only individuals who had successfully completed the insulin-modified FSIGT and were either African-American or Caucasian were included. Columns ″r″ are the partial correlation coefficient with SI; columns ″p″ are the attached significance level.
ERSR in stromal vascular vs. adipocyte fraction
To exclude macrophage infiltration as the explanation for the findings of increased ERSR with obesity, we examined markers ATF6α, HSPA5, and macrophage marker CD68 in paired adipocyte and stromal vascular fractions from 22 individuals (BMI 21.6–39.9 kg/m2). Mean BMI and SI of these individuals was similar to the full study population (data not shown). In contrast to CD68 expression, which showed 7.5-fold enrichment in the stromal vascular fraction (mean 0.16 vs. 1.24; P = 3 × 10−13), HSPA5 was only 1.4-fold enriched in the stromal vascular fraction (1.30 vs. 1.82; P = 0.01), and ATF6α was not different between fractions (1.43 vs. 1.27; P = 0.3) (supplemental Fig. 3S). We reexamined the correlation of ER stress transcripts with BMI and percent body fat, controlling for CD68 gene expression by partial regression to remove further the macrophage contribution (supplemental Table 4S). Correlations with percent fat remained significant for HSPA5, EDEM1, DNAJC3, HSP90B1, ATF6, PERK, HYOU1, and STC2, ranging from r = 0.31 (EDEM1, P = 0.009) to 0.54 (STC2, P = 1 × 10−6). In contrast, significant correlations were lost for pro-apoptotic makers ATF4, ATF3, GADD34, and CHOP.
Hierarchical clustering analysis of ER stress markers
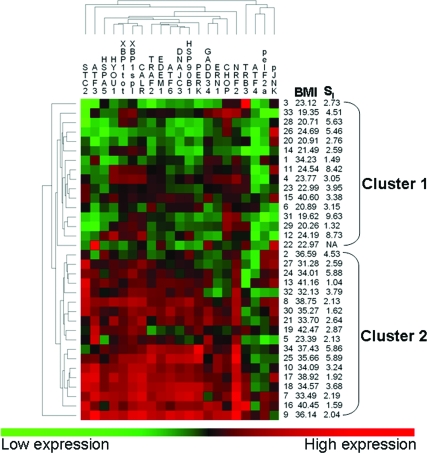
The phosphoprotein and transcript levels measured in this study represent the integrated downstream response of the three proximal sensors, and different pathways may be activated in response to different levels or substances inducing ER stress. To exclude the possibility that a single gene or phosphoprotein analysis masked individual variation, we used the program PermutMatrix to examine clustering in the 34 individuals with low and high BMI tested for protein levels. As shown in Fig. 4, ER stress markers clustered into two clear groups that clearly predicted BMI with only two exceptions. BMI between groups predicted by unbiased clustering was highly significantly different (P = 0.00001), whereas SI did not differ significantly between groups with low and high levels of ER stress markers (P = 0.15).
Figure 4.
Hierarchical clustering of gene expression for ER stress transcripts phosphorylated EIF2a and JNK1. These are data on 15 individuals selected for lowest BMI and 19 individuals selected for highest BMI from the full sample of 76 individuals, as shown in Fig. 3. Euclidian distance matrix between each individual was generated using log-normalized values of all expression measures and was used for seriation under multiple fragment heuristics.
Discussion
Induction of obesity activated the ERSR in both liver and adipose tissue, but not in muscle from mice (5). Furthermore, in mice inactivation of XBP1(5) or reduced expression of HYOU1 (ORP150) (6) in the liver resulted in insulin resistance and glucose intolerance, whereas hepatic or systemic overexpression of HYOU1 (6,27), or XBP1 overexpression in mouse embryo fibroblast cells (5) improved SI, as did chemical chaperones (15). Therefore, the implication of ER stress as a mediator of obesity induced insulin resistance and glucose intolerance in humans would have clear therapeutic implications.
Marchetti et al. (28) reported induction of ER stress in human β-cells from diabetic subjects (16,17) with hyperglycemia. Two other studies have addressed the role of ERSR in human adipose. We recently reported in a limited sample that marker HSPA5 was increased in obese individuals who correspond to the replication set in the current study (16). Boden et al. (17) examined sc adipose from the thigh in six lean and six obese subjects, and reported an increase in UPR proteins calreticulin, protein disulfide isomerase A3, and glutathione S transferase P with obesity in a proteomic analysis. Additional analyses supported these studies by showing obesity related increases in spliced XBP1, calnexin, and phospho-JNK1. These published studies are considerably smaller than the present study and present a more limited picture of ER stress. Furthermore, neither study attempted to distinguish the correlations with obesity and SI. The increased JNK1 phosphorylation in obese subjects observed by Boden et al. (17) is in contrast to a study of Bashan et al. (29), which found an increase in JNK1 and p38MAPK phosphorylation with obesity in omental but not sc adipose tissue. Our larger study (Fig. 3) suggests considerable individual fluctuation. Therefore, we propose that these apparent discrepancies represent sampling fluctuations resulting from a small sample size.
We thoroughly evaluated the UPR pathways downstream of EIF2AK3, ATF6α, and ERN1 across a spectrum of BMI from lean to obese in a large sample. We confirm the findings in mice by showing increased transcription of ER chaperone genes with increasing body fat, including HSPA5 (GRP78), which is viewed as a key monitor of ER stress and the UPR (30). Particularly strongly induced were ATF6α and protective chaperones that are primarily under ATF6α control (13,14), including HYOU1 (ORP150) and DNAJC3 (p58IPK) (10,31), and stanniocalcin 2 (STC2), the most strongly up-regulated transcript and also thought to be protective (32). We also found evidence that the EIF2AK3 (PERK) pathway may be activated, based on significantly increased phosphorylation of eIF2α in obese individuals. However, other kinases are known to phosphorylate eIF2α, and because we could not demonstrate PERK phosphorylation, we cannot be certain that this increase is related to ER stress. We also observed modest increases in ATF4, which is increased when eIF2α is phosphorylated. Like Ozcan et al. (5), we found no increase in ER stress with obesity in muscle.
An unexpected finding was the much stronger correlation of UPR in adipose with obesity than with whole body SI. Separating SI from obesity measures is challenging, given the relatively strong and consistently observed negative correlation. Nonetheless, we used several standard statistical methods, including general linear and partial regression models, which consistently failed to find an association of UPR in adipose with SI once we accounted for the known effects of obesity (BMI). If activation of JNK1 is indeed the mechanism for ER stress-induced insulin resistance (5), this pathway appeared not to be activated in human sc adipose. Finally, results of our hierarchical clustering analysis strongly predicted BMI but not SI, again suggesting a poor prediction of SI by sc adipose UPR genes.
The lack of correlation of UPR with SI might reflect tissue specificity. ER stress genes in liver or visceral (omental) adipose might have been more predictive of SI. This latter possibility is suggested by higher levels of phospho-JNK1 in omental than sc adipose (29). Alternatively, our results may reflect differences in the species. Sorting out these possibilities is essential in determining whether reduction of ER stress human liver or adipose will improve SI or glucose tolerance.
Obesity is a state of mild inflammation with increasing macrophage infiltration into adipose with obesity (33). Indeed, two recent papers suggested that an inflammatory and immune response gene transcription network, particularly representing macrophages, was activated in obesity and metabolic syndrome in both mouse and human adipose (2,3). Therefore, a prominent ER stress activation in infiltrating macrophages might explain the increased ER stress markers in obesity. We found that the stromal vascular fraction partially contributed to but could not totally explain the UPR transcripts measured in whole biopsy samples. Similarly, macrophage marker CD68 explained part but not all of the strong correlation of ER stress transcripts with obesity. Interestingly, the weaker association of apoptotic pathways with obesity (ATF4, GADD34, ATF3) was lost when corrected for CD68, suggesting that these markers may derive primarily from infiltrating macrophages. Whether ER stress is the cause of the immune activation or the result cannot be determined from the present study or from available published data.
Recent studies in cell culture have distinguished the acute ERSR (cells treated with thapsigargin or tunicamycin) from the response to chronic, low-grade exposure (10,34,35). Chronic, low-grade stress up-regulated ER chaperones such as HSPA5 and presumably other compensatory chaperones such as DNAJC3 (p58IPK), HYOU1 (ORP150), and HSP90B1 (GRP94), along with increased transcription of proteins involved in degradation of unfolded proteins (EDEM1), but did not stimulate markers of apoptosis (CHOP) (10,34). Furthermore, during persistent ER stress as might be expected with obesity, ERN1 signaling was rapidly attenuated along with downstream XBP1 splicing and JNK phosphorylation (35). Recent findings suggest that ATF6α is essential to these protective responses (14). These cell culture findings mirror our observations from obese humans, in whom the most striking correlations with obesity are in chaperones that are thought to protect against ER stress-induced apoptosis. Indeed, in preliminary experiments we observe a similar pattern of ERSR transcripts when HepG2 cells were conditioned with low doses of palmitate or oleate and then challenged with doses that typically induce an acute UPR. Such findings are markedly different from the acute UPR induced by thapsigargin, tunicamycin, or palmitate. One possible interpretation of our findings is that human obesity may be a state of chronic, compensated ER stress in adipose. Perhaps those individuals who compensate adequately with high levels of protective chaperones ameliorate the metabolic consequences observed in inbred mice.
The strengths of this study include the very broad assessment of each ER stress/UPR pathway by both protein phosphorylation and gene transcription, and the replication of the key transcription findings in an independent sample. The association of ER stress factors with obesity measures is statistically convincing with P values well less than 0.0001. The findings were robust to multiple methods of statistical analysis, and are confirmed by cluster analysis in a subset of samples with extremes of BMI. SI in both the primary and replication populations was assessed directly using the insulin-modified FSIGT in addition to surrogate measures based on fasting insulin and glucose. Nonetheless, our conclusions have some necessary limitations. As noted previously, work in mice focused on hepatic ER stress (5,6), whereas a comparable study would be difficult in humans. The UPR in tissues other than sc adipose may be more predictive of SI.
In conclusion, we show that ER stress markers, mostly representing the activation of the ATF6α pathway, are increased by 50% or more in sc adipose tissue from obese compared with lean humans. Furthermore, eIF2α is phosphorylated, suggesting a possible decrease in sc adipose translation with obesity and possibly reflecting activation of the PERK pathway. JNK1 activation was not convincingly demonstrated in our study, nor could we demonstrate that the association of ER stress with SI was independent of the well-known inverse correlation of SI and obesity. Future studies are needed to explore the implications of eIF2α phosphorylation and to determine whether individuals who develop type 2 diabetes show a different pattern of adipocyte ERSR, perhaps with less compensation and increased activation of pro-apoptotic pathways.
Supplementary Material
Acknowledgments
We thank Terri Hale and Regina Dennis for assisting with subject recruitment, S. Ranganathan for insulin measurement, Cynthia Witkowski, R.N., and Carol Smith, R.N., for outstanding support of the clinical studies, and Richard Harris for assistance with database design and data management.
Footnotes
This work was supported by the Research Service of the Department of Veterans Affairs (Merit funds to S.C.E., P.A.K., and N.R.; Resource Enhancement and Protection Program funds), and in part by the General Clinical Research Center (Grant M01RR14288 from National Center for Research Resources, National Institutes of Health to the University of Arkansas for Medical Sciences).
Disclosure Statement: The authors have no conflicts to declare in relation to the material reported in this manuscript.
First Published Online August 26, 2008
Abbreviations: AIRg, Acute insulin response to glucose; ATF, activating transcription factor; BMI, body mass index; CHOP, C/EBP homologous protein; CI, confidence interval; DI, disposition index; DXA, dual x-ray absorptiometry; eIF, eukaryotic initiation factor; ER, endoplasmic reticulum; ERSR, endoplasmic reticulum stress response; FSIGT, frequently sampled iv glucose tolerance test; HSPA5, heat shock protein A5; JNK, c-Jun N-terminal kinase; IRE1, inositol-requiring enzyme 1; PERK, phospho-extracellular signal-regulated kinase; SI, insulin sensitivity; STC2, stanniocalcin 2; UPR, unfolded protein response; XBP1, x-box binding protein 1.
References
- Hotamisligil GS 2006 Inflammation and metabolic disorders. Nature 444:860–867 [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE 2008 Variations in DNA elucidate molecular networks that cause disease. Nature 452:429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K 2008 Genetics of gene expression and its effect on disease. Nature 452:423–428 [DOI] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES 2006 Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440:944–948 [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS 2004 Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306:457–461 [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, Ogawa S, Hori M, Yamasaki Y, Matsuhisa M 2005 Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem 280:847–851 [DOI] [PubMed] [Google Scholar]
- Gregor MF, Hotamisligil GS 2007 Thematic review series: adipocyte biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J Lipid Res 48:1905–1914 [DOI] [PubMed] [Google Scholar]
- Guo W, Wong S, Xie W, Lei T, Luo Z 2007 Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3–L1 and rat primary preadipocytes. Am J Physiol Endocrinol Metab 293:E576–E586 [DOI] [PubMed] [Google Scholar]
- Wei Y, Wang D, Pagliassotti MJ 2007 Saturated fatty acid-mediated endoplasmic reticulum stress and apoptosis are augmented by trans-10, cis-12-conjugated linoleic acid in liver cells. Mol Cell Biochem 303:105–113 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ 2007 That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci 32:469–476 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P 2007 Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529 [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ 2007 The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol 18:716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K 2007 Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell 13:365–376 [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ 2007 ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell 13:351–364 [DOI] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS 2006 Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313:1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Chu WS, Mondal AK, Sharma NK, Kern PA, Rasouli N, Elbein SC 2008 Effect of pioglitazone treatment on endoplasmic reticulum stress response in human adipose and in palmitate-induced stress in human liver and adipose cell lines. Am J Physiol Endocrinol Metab 295:E393–E400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, Cheung P, Merali S 2008 Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes 57:2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Chu WS, Das SK, Yao-Borengasser A, Hasstedt SJ, Wang H, Rasouli N, Kern PA 2007 Transcription factor 7-like 2 polymorphisms and type 2 diabetes, glucose homeostasis traits and gene expression in US participants of European and African descent. Diabetologia 50:1621–1630 [DOI] [PubMed] [Google Scholar]
- Rasouli N, Yao-Borengasser A, Miles LM, Elbein SC, Kern PA 2006 Increased plasma adiponectin in response to pioglitazone does not result from increased gene expression. Am J Physiol Endocrinol Metab 290:E42–E46 [DOI] [PubMed] [Google Scholar]
- Yao-Borengasser A, Rasouli N, Varma V, Miles LM, Phanavanh B, Starks TN, Phan J, Spencer III HJ, McGehee Jr RE, Reue K, Kern PA 2006 Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator-activated receptor γ activation. Diabetes 55:2811–2818 [DOI] [PubMed] [Google Scholar]
- Rodbell M 1964 Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J Biol Chem 239:375–380 [PubMed] [Google Scholar]
- Bradford MM 1976 A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN 2003 MINMOD Millennium: a computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther 5:1003–1015 [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC 1985 Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419 [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S 2005 PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21:1280–1281 [DOI] [PubMed] [Google Scholar]
- McQuitty LL 1966 Similarity analysis by reciprocal pairs for discrete and continuous data. Educ Psychol Meas 26:825–831 [Google Scholar]
- Ozawa K, Miyazaki M, Matsuhisa M, Takano K, Nakatani Y, Hatazaki M, Tamatani T, Yamagata K, Miyagawa J, Kitao Y, Hori O, Yamasaki Y, Ogawa S 2005 The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes 54:657–663 [DOI] [PubMed] [Google Scholar]
- Marchetti P, Bugliani M, Lupi R, Marselli L, Masini M, Boggi U, Filipponi F, Weir GC, Eizirik DL, Cnop M 2007 The endoplasmic reticulum in pancreatic β cells of type 2 diabetes patients. Diabetologia 50:2486–2494 [DOI] [PubMed] [Google Scholar]
- Bashan N, Dorfman K, Tarnovscki T, Harman-Boehm I, Liberty IF, Bluher M, Ovadia S, Maymon-Zilberstein T, Potashnik R, Stumvoll M, Avinoach E, Rudich A 2007 Mitogen-activated protein kinases, inhibitory-κB kinase, and insulin signaling in human omental versus subcutaneous adipose tissue in obesity. Endocrinology 148:2955–2962 [DOI] [PubMed] [Google Scholar]
- Lee AS 2005 The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35:373–381 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS 2007 The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell 18:3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Walker JR, Thompson CS, Moroz I, Lin W, Veselits ML, Hakim AM, Fienberg AA, Thinakaran G 2004 Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol 24:9456–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio GB, Yao-Borengasser A, Rasouli N, Varma V, Lu T, Miles LM, Ranganathan G, Peterson CA, McGehee RE, Kern PA 2005 Expression of CD68 and macrophage chemoattractant protein-1 genes in human adipose and muscle tissues: association with cytokine expression, insulin resistance, and reduction by pioglitazone. Diabetes 54:2305–2313 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ 2006 Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol 4:e374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P 2007 IRE1 signaling affects cell fate during the unfolded protein response. Science 318:944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.