Abstract
Bacteria have evolved numerous mechanisms for cell–cell communication, many of which have important consequences for human health. Among these is conjugation, the direct transfer of DNA from one cell to another. For Gram-negative bacteria, conjugation requires thin, flexible filaments (conjugative pili) that are elaborated by DNA donor cells. The structure, function, and especially the dynamics of conjugative pili are poorly understood. Here, we have applied live-cell imaging to characterize the dynamics of F-pili (conjugative pili encoded by the F plasmid of Escherichia coli). We establish that F-pili normally undergo cycles of extension and retraction in the absence of any obvious triggering event, such as contact with a recipient cell. When made, such contacts are able to survive the shear forces felt by bacteria in liquid media. Our data emphasize the role of F-pilus flexibility both in efficiently sampling a large volume surrounding donor cells in liquid culture and in establishing and maintaining cell–cell contact. Additionally and unexpectedly, we infer that extension and retraction are accompanied by rotation about the long axis of the filament.
Keywords: conjugation, conjugative pilus, extension, retraction
Among Gram-negative bacteria, conjugative DNA transfer is mediated by multicomponent type IV secretion systems commonly encoded by large plasmids (1, 2). By virtue of such activity, these plasmids rapidly spread within bacterial populations, contributing to the dissemination of antibiotic resistance (3) among other traits pertinent to human health (2, 4–6).
A hallmark of Gram-negative bacterial cells capable of acting as DNA donors is the presence of surface filaments collectively designated conjugative pili (7). F-pili, characteristic of Escherichia coli carrying the conjugative plasmid F, are helical polymers of one subunit, F-pilin (8). Conjugation commences when the tips of F-pili make contact with recipient cells (9, 10). F-pili tips also bind filamentous DNA bacteriophages, such as M13, whereas icosohedral RNA bacteriophages such as R17 bind along the filament sides (11).
Indirect evidence has suggested that F-pili are dynamic structures, capable of both extension and retraction. F-pilus retraction as an essential stage of DNA transfer was first proposed by Marvin and Hohn (12) and by Curtiss (13). Various lines of indirect evidence since then have supported the retraction hypothesis (14–18), although fundamental uncertainties remain. For example, retraction was initially regarded as a triggered event, occurring only when a recipient cell or bacteriophage bound to the filament tip (12). Later it was proposed that F-pili normally undergo cycles of extension and retraction (14, 17, 19). The idea that DNA can be conducted from donor to recipient through extended F-pili (10, 20) has also received experimental support (21, 22).
Here, we have applied laser-scanning confocal microscopy to study F-pilus dynamics with living cells. Our results clarify some of the uncertainties surrounding F-pilus function and suggest a hypothesis for F-pilus function that emphasizes the mechanical properties of the filament.
Results
Visualization of F-pili on Live Cells by Fluorescent R17 Labeling.
We showed that fluorescent R17 bacteriophage binding to fixed E. coli F+ cells could be used to identify F-pili (23). We have now extended this approach to visualize filament dynamics on live cells. In developing this approach, we made two related assumptions. The first was that, as is true of R27 conjugative pili (24), F-pili extend by subunit addition at the cell-proximal base of the filament. The second was that bacteriophage R17 binding to already extant (cell-distal) F-pili segments would have no significant effect on basal F-pilus extension. Given these assumptions, we hypothesized that fluorescent R17 could be used to follow F-pilus dynamics with live cells.
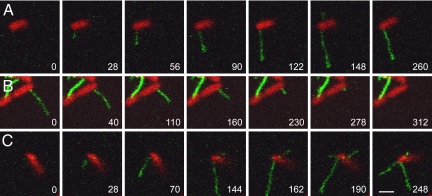
We added green fluorescent R17 to E. coli strain HfrH expressing a red fluorescent cytoplasmic marker (DsRed Express) while observing the cells by laser-scanning confocal microscopy. We found that newly assembled F-pili became visible as R17 bound along their length. The bacterium shown in Fig. 1A extended two pili during the 260-s period of observation. The first pilus reached a length of >5 μm by 148 s and then began to retract, as seen in the final frame. A second pilus grew briefly then retracted; it is visible only in frame 148 of Fig. 1A. The complete time series may be viewed as supporting information (SI) Movie S1.
Fig. 1.
Extension and retraction of F-pili. (A) Pilus extension. The extension of a long pilus is seen in the first six frames; retraction is evident in the final frame. A second short pilus is visible only in the sixth frame. Note that during extension, the distal portion of a pilus is more brightly labeled than the cell-proximal portion, suggesting that the filament elongates from the base. (B) Pilus retraction. This 4-μm pilus retracted completely; during retraction fluorescence was uniform along the filament. (C) Independent regulation of F-pili on the same cell. This bacterium extended three pili, which grew and retracted asynchronously. For each bacterium, the complete time series may be seen as a movie: (A) Movie S1; (B) Movie S2; (C) Movie S4. Time is shown as seconds after the first frame. (Scale bar, 2 μm; all images are at the same magnification.)
The cell-proximal portion of an extending filament was always weakly labeled, as expected if new F-pilin monomers added at the base of the pilus faster than the rate of R17 binding. This observation implies that R17 binding itself does not induce retraction, for if it did, we should not have seen any elongating filaments.
F-pilus Retraction.
Fig. 1B and Movie S2 show an example of a retracting F-pilus. This pilus, which was 4 microns in length when observation began, retracted completely by 312 s. The different fluorescence intensities at the cell-distal and proximal pilus segments disappeared as this and all other filaments we observed underwent retraction. Indeed, a clear indication that a pilus had switched from extension to retraction mode was equivalent fluorescence along the entire length of the filament.
The mean retraction rate was less than half the mean extension rate of 40 nm/sec (Table 1). The extension rate was likely unaffected by R17 because newly extended, cell-proximal filament lacked bound bacteriophage. However, the retraction rate may have been slowed by bound R17, as suggested for retraction in the presence of metabolic poisons (15). Thus, shorter pili retracted completely, whereas longer, more highly fluorescent filaments were seen to initiate retraction, but then gradually slow or stop (Movie S3). A field of bacteria invariably contained many cells with brightly labeled, static pili, which may have arisen in this manner.
Table 1.
Rates of F-pilus extension and retraction
| Event | nm/sec | |||
|---|---|---|---|---|
| N | Mean rate | Range | SD | |
| Extension | 14 | 39.5 | 27.2–52.9 | 8.2 |
| Retraction | 13 | 15.8 | 8.2–23.0 | 5.4 |
Rates were determined from movies of extension or retraction events using NIH ImageJ software or Zeiss LSM-5 imaging software. Zero times were taken as the appearance of a filament beyond the cell body (extension) or the first frame where an extended filament began to retract. Total extension and retraction lengths were about 1.5–5 μm (about 30–100 frames at 2 sec/frame). N = sample size; SD = standard deviation.
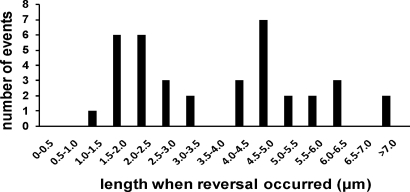
We have also observed more than three dozen examples in which extending F-pili reversed and began to retract. Invariably, the transition from extension to retraction occurred abruptly, without any pause detectable at our level of resolution (∼2-s intervals). These observations show that F-pili normally undergo a switch from extension to retraction absent a triggering event. We could reliably observe such reversals at filament lengths less than approximately 10 μm. Below this limit, reversals occurred preferentially at 1.5–2.5 μm (32% of total reversals) and at 4–5 μm (27%) (Fig. 2). Retraction seemed to be highly processive because we never observed a retracting filament switch to extension.
Fig. 2.
Switching of F-pili from extension to retraction. The figure shows the distribution of filament lengths when an extending filament switched and began to retract between two frames (2-s interval).
Independent Behavior of F-pili on the Same Cell.
F-pili on the same cell extended or retracted independently (Fig. 1 A and C). Fig. 1C and Movie S4 show a bacterium that extended three pili. The first grew beyond the field of view (144- and 162- second frames), but was retracting in the last two frames. A second pilus on the other side of the cell, first seen in the 144-second frame, grew a short distance then retracted almost completely by the final frame. The cell also extended a third pilus from one pole, first seen in the 162-second frame. This pilus continued to grow throughout the remaining frames, over the same interval when the other pili were retracting. Similar behavior has been reported for nonconjugative type IV pili, which are chromosomally encoded filaments that undergo cycles of extension and retraction responsible for twitching or gliding motility in several Gram-negative bacteria (25, 26).
Cell–Cell Interactions Mediated by F-pili.
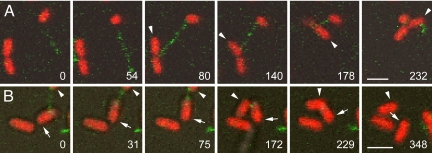
When the tip of an extending pilus made contact with another cell, the linkage strength allowed the two cells to be drawn into close contact upon pilus retraction (Fig. 3). As the two cells drew closer together during retraction, one or both of them sometimes appeared to flip or rotate (Movies S5 and S6), although our temporal resolution was not adequate to resolve the behavior clearly. Cells in contact separated shortly after complete retraction of the pilus (Fig. 3B; Movie S6), possibly because all of the cells were Hfr and therefore expressed surface exclusion (10).
Fig. 3.
Cell–cell contact resulting from pilus retraction. Two examples are shown in which the tip of a pilus extended by an HfrH cell makes contact with another HfrH cell and retraction of the pilus draws the two cells together. In A, the pilus is extending in the first two frames and makes contact with a dividing cell in the third. (An arrowhead marks the member of the dividing pair contacted by the pilus.) Retraction draws the dividing cell closer in the 140-s frame, and it has rotated and flipped upward in the 178-s frame. By 232 s, the cells are in close contact. (Movie S5). In B, a pilus links a cell marked with an arrow to another marked with an arrowhead, initially just out of the frame. The two cells draw closer in the next four frames, and the upper cell flips down into the field of view. After a period of close contact, the two cells separate. (Movie S6.) Time is shown as seconds after the first frame. (Scale bars, 2 μm.)
Do F-pili Rotate During Extension and Retraction?
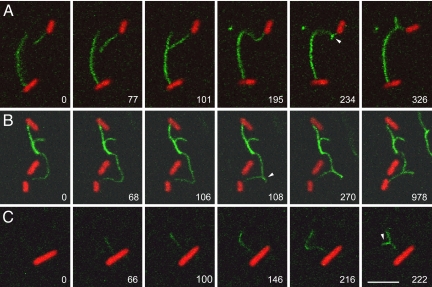
F-pili exhibit a whip-like motion, evident in Movies S1, S4, and S5 and consistent with other evidence that they are flexible filaments (7). A related property of F-pilus dynamics was suggested by three examples in which the distal portion of an F- pilus became immobilized during extension (Fig. 4 and Movies S7–S9). In Fig. 4A, the tip of a growing pilus became bound to a second F- pilus on another cell. In Fig. 4B, the growing F-pilus had already adhered over part of its length to a filament on another cell before recording began. In Fig. 4C, the tip of the growing pilus became attached to the substratum. In each case the growing filament curved in an arc owing to the restraints on its distal segment. As the pilus continued to elongate, a linear protrusion abruptly interrupted this arc (arrowheads). The transition occurred between one frame and the next, an interval of 2 sec (Fig. 4B, 106- and 108-s frames). After the transition, further growth of the pilus was reflected in elongation of the protruded segment. (Compare the 108- and 270-s frames in Fig. 4B). Also evident in the last frame of Fig. 4B is shortening of the protruded segment, presumably as the filament retracted. Given that this phenomenon was observed only with filaments whose distal ends were restrained, we suggest that F-pili rotate during elongation, and that the abrupt transitions reflect the formation of interwound supercoils as torsional strain was exchanged for bending strain on the flexible filaments.
Fig. 4.
Supercoiling of F-pili during extension and retraction. This figure shows three examples in which extension of a pilus continues after its distal portion has become immobilized by binding to another pilus (A and B) or to the substratum (C). As each pilus continues to elongate, it curves in an arc, then abruptly generates a supercoil (arrowhead). After this transition, further extension of the pilus leads to elongation of the supercoiled segment (evident in all three time series), whereas retraction leads to shortening (B). All parameters are as described in Fig. 1 except that row C shows an F' lac strain. Each row of images consists of frames from a time series: (A) Movie S7; (B) Movie S8; (C) Movie S9. Time is shown as seconds after the first frame. (Scale bar, 5 μm; all images are at the same magnification.)
Whereas these observations were striking and unexpected, additional evidence for rotation derived from occasional observation of a cell tethered to the coverslip by an F-pilus, leaving the cell itself free to move. An example is shown in Movie S10; the cell is tethered by a short F-pilus heavily labeled with R17. Rotation of the cell is evident (Fig. S1). Its slow and diminishing rate of rotation may reflect the decreasing rate of retraction characteristic of heavily labeled F-pili (Movie S3) and of drag forces on the tethered cell.
F-pilus rotation tightly coupled to extension and retraction would suggest a mechanism whereby the molecular motor(s) driving extension and retraction remains stationary whereas the filament rotates as successive subunits are added or removed (19).
Discussion
The present studies resolve several persistent uncertainties about F-pili. The most important with respect to F-pilus function is dynamic length variation. We have shown that F-pili constantly undergo cycles of extension and retraction, as suggested earlier by Jacobson (14) and by Ippen-Ihler and colleagues (17) based on indirect evidence. These cycles, along with the flexibility of F-pili, would allow F-pilus tips to sample the volume around each donor cell quickly and efficiently. Once contact is made with another cell, retraction generates enough force to pull the cells toward each other. By drawing donor and recipient cells together, retraction would also allow stabilization of cell–cell contacts, which is critical to efficient DNA transfer from F+ and related donors (8, 10). Retraction and stabilization would be especially important in liquid media, where bacterial motility and flow forces would contribute to instability. Thus, F+ cells function well as DNA donors both in liquid and on solid media, whereas conjugative systems lacking retractable pili function well only on solid surfaces (7, 27). DNA transfer from F+ cells also occurs on solid and even in liquid media without stabilization, albeit at a very much reduced frequency (21, 22, 28).
The dynamics of F-pili may be compared with those of type IV pili, whose function in gliding motility depends on their ability to retract (25, 26). Several contrasts suggest fundamental mechanistic differences. First, live cell imaging did not indicate that type IV pili rotate as they extend or retract (26). Second, type IV pili extend and retract at rates approaching 1 μm/sec (25, 26), much faster than F-pili. Finally, the energy sources for type IV pilus dynamics are hexameric ring ATPases, one for extension (PilF) and a different one for retraction (PilT) (29). In contrast, F encodes only a single candidate ATPase, TraC, that is required for F-pilus assembly (8). TraC ATPase function appears to be essential because a mutation in the TraC Walker A box abolished F-pilus formation (Harris and Silverman, unpublished data). An extension/retraction switch at the base of individual F-pili (30) could regulate TraC ATPase activity in such a way that the protein is functionally equivalent to both PilF and PilT. Alternatively, retraction might not require ATP hydrolysis at all (15).
In summary, our observations show that F-pili undergo cycles of extension and retraction, that extension occurs by subunit addition at the cell-proximal end; that retraction can bring two cells together; and that extension and retraction could be accompanied by a rotation about the long axis of the filament, although this remains inferential. With these basic facts in hand, we look forward to investigating the synthesis and functions of F-pili in interactions between F+ and other cells and in DNA transfer per se. In addition, although a detailed mechanical analysis of F-pili is beyond the scope of this communication, it will be of great interest to understand the structural basis for the adaptive mechanical properties of these filaments.
Materials and Methods
Bacterial Strains and Growth.
E. coli K-12 strains HfrH and AE2386/JCFL0 were from the authors' strain collection and have been described (23). The strains expressed the cytoplasmic, IPTG-inducible fluorescent marker DsRed Express (31). Cultures were grown with aeration at 37°C in Luria-Bertani (LB) medium containing chloramphenicol (5 μg/ml) and IPTG (0.5 mM). Culture densities were measured as the optical density (OD) at 600 nm.
For microscopy, overnight cultures were diluted into fresh medium and grown to an OD of 0.15–0.2. Bacteria were collected by sedimentation from 0.5 ml of culture, washed once in PBMC buffer [10 mM Na2HPO4/10 mM KH2PO4/ 2 mM MgCl2/0.2 mM CaCl2 (pH 6.3)], and suspended in 0.5 ml of PBMC.
Imaging Methods.
Covered microscope chambers with glass coverslip bottoms were used (GWSt-5030, www.willcowells.com). Microscope chambers were prepared by incubating the glass surface for 10 min with either normal goat serum (diluted 1:100 in PBMC) or normal rabbit serum (diluted 1:10,000 in PBMC), then rinsing three times with PBMC. One hundred μL of the bacterial suspension was vortexed and added to a microscope chamber; the bacteria were allowed to settle for 10 min at room temperature. The sample was then overlaid with a thin, 1-cm2 layer of agarose (1.8% agarose in PBS) (34), and excess liquid was carefully removed. A preparation of R17 bacteriophage labeled with Alexa-488 (23) was diluted 20 μl to 500 μl in PBMC, and 20 μl was added to the surface of the agar square and allowed to soak in for 5 min before excess liquid was wicked away and the sample was viewed by microscopy. Usually samples were viewed at room temperature; occasional samples were viewed at 30°C using a heated microscope stage. Samples were imaged by using a Zeiss LSM 510 confocal microscope with a Plan-Apochromat 63 × 1.4 N.A. DIC objective. Alexa-488 was excited with the 488-nm line of an argon laser with a 505–530-nm filter for emission, and DsRed was excited with the 543-nm line of a HeNe laser, with a 560-nm long pass filter for emission. An HFT UV/488/543 beam splitter was used. Time series were collected in a single focal plane at 1.6 to 2.0-s intervals. Laser power was kept low to minimize photodamage and allow cells to be observed for up to 20 min.
Supplementary Material
Acknowledgments.
P.M.S. acknowledges informative discussions with Dr. E. H. Egelman, University of Virginia. We thank the Imaging Core Facility of the Oklahoma Medical Research Foundation for access to microscopy equipment. This work was supported by National Science Foundation Grant MCB-065583 (to P.M.S.) and Grant MCB-0344541 (to M.C.) and the Oklahoma Institute for the Advancement of Science and Technology Grant HR05–020 (to M.C.) and Grant HR04–082 (to P.M.S.). P.M.S. acknowledges support as the Marjorie Nichlos Chair in Medical Research and M.C., as the J.P. Hannigan Distinguished Research Scientist.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806786105/DCSupplemental.
References
- 1.Lawley TD, et al. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 2.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teuber M. Spread of antibiotic resistance with food-borne pathogens. Cell Mol Life Sci. 1999;56:755–763. doi: 10.1007/s000180050022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 5.Reisner A, et al. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 6.Christie PJ, Vogel JP. Bacterial type IV secretion: Conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paranchych W, Frost LS. The physiology and biochemistry of pili. Adv Microb Physiol. 1988;29:53–114. doi: 10.1016/s0065-2911(08)60346-x. [DOI] [PubMed] [Google Scholar]
- 8.Firth N, Ippen-Ihler K, Skurray RA. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB Jr, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington D.C.: American Society for Microbiology; 1996. pp. 2377–2401. [Google Scholar]
- 9.Helmuth R, Achtman M. Cell-cell interactions in conjugating Escherichia coli: Purification of F pili with biological activity. Proc Natl Acad Sci USA. 1978;75:1237–1241. doi: 10.1073/pnas.75.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Achtman M, Skurray R. In: Receptors and Recognition: Microbial Interactions. Reissig JL, editor. London: Chapman & Hall; 1977. pp. 234–279. [Google Scholar]
- 11.Valentine R, et al. The F-pilus of Escherichia coli. Adv. Microb. Physiol. 1969;3:1–52. [Google Scholar]
- 12.Marvin DA, Hohn B. Filamentous bacterial viruses. Bacteriol Rev. 1969;33:172–209. doi: 10.1128/br.33.2.172-209.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtiss R., 3rd Bacterial conjugation. Annu Rev Microbiol. 1969;23:69–136. doi: 10.1146/annurev.mi.23.100169.000441. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson A. Role of F pili in the penetration of bacteriophage fl. J Virol. 1972;10:835–843. doi: 10.1128/jvi.10.4.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novotny CP, Fives-Taylor P. Retraction of F pili. J Bacteriol. 1974;117:1306–1311. doi: 10.1128/jb.117.3.1306-1311.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panicker MM, Minkley EG., Jr DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J Bacteriol. 1985;162:584–590. doi: 10.1128/jb.162.2.584-590.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sowa BA, Moore D, Ippen-Ihler K. Physiology of F-pilin synthesis and utilization. J Bacteriol. 1983;153:962–968. doi: 10.1128/jb.153.2.962-968.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawley TD, et al. Bacterial conjugative transfer: Visualization of successful mating pairs and plasmid establishment in live Escherichia coli. Mol Microbiol. 2002;44:947–956. doi: 10.1046/j.1365-2958.2002.02938.x. [DOI] [PubMed] [Google Scholar]
- 19.Silverman PM. In: Bacterial Outer Membranes as Model Systems. Inouye M, editor. New York: Wiley Interscience; 1987. pp. 277–310. [Google Scholar]
- 20.Brinton CC., Jr The properties of sex pili, the viral nature of “conjugal” genetic transfer systems, and some possible approaches to the control of bacterial drug resistance. CRC Crit Rev Microbiol. 1971;1:105–160. doi: 10.3109/10408417109104479. [DOI] [PubMed] [Google Scholar]
- 21.Babic A, et al. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 22.Harrington LC, Rogerson AC. The F pilus of Escherichia coli appears to support stable DNA transfer in the absence of wall-to-wall contact between cells. J Bacteriol. 1990;172:7263–7264. doi: 10.1128/jb.172.12.7263-7264.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daehnel K, et al. Fluorescence assays for F-pili and their application. Microbiology. 2005;151:3541–3548. doi: 10.1099/mic.0.28159-0. [DOI] [PubMed] [Google Scholar]
- 24.Maher D, Sherburne R, Taylor DE. H-pilus assembly kinetics determined by electron microscopy. J Bacteriol. 1993;175:2175–2183. doi: 10.1128/jb.175.8.2175-2183.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 26.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christie PJ. Type IV secretion: The Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning PA, Morelli G, Achtman M. traG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc Natl Acad Sci USA. 1981;78:7487–7491. doi: 10.1073/pnas.78.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang P, et al. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology. 2008;154:114–126. doi: 10.1099/mic.0.2007/011320-0. [DOI] [PubMed] [Google Scholar]
- 30.Harris RL, Silverman PM. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol. 2004;186:5480–5485. doi: 10.1128/JB.186.16.5480-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otto GP, et al. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol Microbiol. 2003;51:63–72. doi: 10.1046/j.1365-2958.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- 32.Yumura S, Mori H, Fukui Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J Cell Biol. 1984;99:894–899. doi: 10.1083/jcb.99.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.