Abstract
We report molecular dynamics simulations of the equilibrium folding/unfolding thermodynamics of an all-atom model of the Trp-cage miniprotein in explicit solvent. Simulations are used to sample the folding/unfolding free energy difference and its derivatives along 2 isochores. We model the ΔGu(P,T) landscape using the simulation data and propose a stablility diagram model for Trp-cage. We find the proposed diagram to exhibit features similar to globular proteins with increasing hydrostatic pressure destabilizing the native fold. The observed energy differences ΔEu are roughly linearly temperature-dependent and approach ΔEu = 0 with decreasing temperature, suggesting that the system approached the region of cold denaturation. In the low-temperature denatured state, the native helical secondary structure elements are largely preserved, whereas the protein conformation changes to an “open-clamp” configuration. A tighter packing of water around nonpolar sites, accompanied by an increasing solvent-accessible surface area of the unfolded ensemble, seems to stabilize the unfolded state at elevated pressures.
Keywords: folding, free energy, hydrostatic pressure, simulations
The stability of natively folded proteins in solution is determined by the competition of many effects that reach a balance near physiological conditions. As a consequence, the stability of a protein can be affected in many different ways (1–5). High-temperature denaturation is mostly accompanied by a dramatic loss of protein secondary structure (6). However, elevated pressures (2, 3), changing pH (2), and cosolvents such as salts (7) and osmolytes (4) also affect the stability of the native state, often destabilizing in character but under certain conditions also significantly stabilizing (4). In addition, many globular proteins are also destabilized at low (subzero) temperatures, a process known as “cold denaturation” (8). Cold denaturation is experimentally accomplished with the help of elevated pressures (2), leading to a characteristic tongue-shaped P,T-stability diagram, found for many globular proteins (9–15). Hydrophobic forces play a key role in the protein folding process (16–18), but it is the balance of hydrophilic and hydrophobic forces that determines the conformational equilibrium. The notion that proteins under native conditions are only “marginally stable” (7) seems to be an important requirement for their ability to explore different conformational substates (19) and hence for protein function. The application of high hydrostatic pressure (20) has been shown to be able shift the equilibrium of conformational states (21, 22), promoting denaturation (23) but also altering the native state (24) and modifying protein–protein interaction (20). Pressure effects on protein structure appear to be determined mostly by changing the balance between hydrophilic and hydrophobic interactions (25–27).
Model peptides and proteins have long been sought as templates for understanding protein structure and function. The Trp-cage miniprotein (28) is a relatively well-characterized system, designed to shed light on protein folding pathways and understanding stability of globular proteins. Trp-cage is a 20-residue protein (28), exhibiting a cooperatively folded tertiary structure (29). Its structure has been determined by NMR [Protein Data Bank (PDB) ID code 1L2Y], and its melting behavior has been characterized using CD (28, 30, 31), changes in chemical shifts, fluorescence quenching (30), fluorescence correlation (32), and UV-resonance Raman spectroscopy (33). The small size and the rapid kinetics of Trp-cage have made it an attractive target for computer simulation studies using implicit (34–40) and explicit solvent models (41–44). The specific virtue of an explicit water solvent is that it can model the different contributions stabilizing both the native and the unfolded state, such as hydrophobic and hydrophilic hydration effects, better than implicit solvent models. Of particular importance is the correct balance of enthalpic and entropic solvation effects and their temperature and pressure dependence. However, due to the large computational burden, the folding/unfolding equilibrium of even small biomolecules still represents a major computational challenge. Here, we present a previously undescribed unbiased computation of the P,T-stability diagram of an atomic detail model of a computer simulated protein starting from an unfolded initial structure. The P,T-stability diagram has been calculated for small peptides (45, 46) and an RNA oligomer (47). Our simulations demonstrate that the Trp-cage, although small in size, exhibits essential thermodynamic features also found in globular proteins (5, 12, 31, 48). Replica-exchange molecular dynamics simulations are performed on the Trp-cage protein. To study the effect of volume changes on protein hydration, we study the folding/unfolding thermodynamics on 2 different isochores.
Results and Discussion
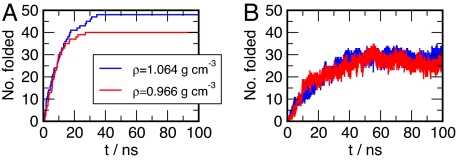
Replica exchange simulations at 2 different densities are used to compute the folding/unfolding equilibrium thermodynamics of the Trp-cage. Time series of the number of replicas that have reached the folded state at least once during the simulation [number of folded replicas (NFR)] and the total number of folded replicas (rmsd ≤0.22 nm) are shown in Fig. 1. A single exponential fit of the NFR gives folding times of 8.5 and 10.5 ns in the replica ensemble for the 0.966 and 1.064 gcm−3 isochores, respectively. The number of folded states for both isochores have reached steady states after ≈40 ns. Consequently, the final 60 ns of the simulation runs are used for analysis.
Fig. 1.
Convergence of the REMD simulations. (A) Time history of the number of replicas that have folded (rmsd ≤ 0.22 nm) at least once in the simulation (NFR). (B) Number of replicas sampling the folded state as a function of time. The total number of replicas sampling the 0.966 gcm−3 and 1.064 gcm−3 isochores is 40 and 48, respectively. After 40 ns all replicas have reached folded state at least once.
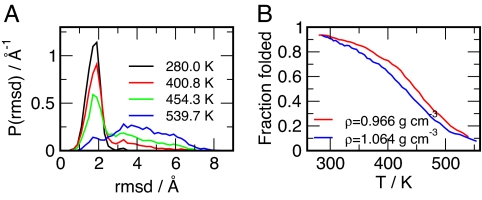
We use the rmsd from the NMR structure (backbone atoms of frame 1 of PDB ID code 1L2Y) as a measure to distinguish between folded and unfolded configurations. The rmsd distributions shown in Fig. 2A exhibit a narrow peak, representing the folded configurations, centered at ≈0.18 nm. With increasing temperatures, the narrow peak diminishes at the expense of the appearance of a broad distribution of rmsds between 0.3 and 0.8 nm, representing unfolded configurations. The system apparently shows a 2-state folding behavior, as proposed for Trp-cage from fluorescence quenching data (30), but recently has been taken into question according to an extensive fluorescence correlation spectroscopy study (32).
Fig. 2.
Folding/unfolding equilibrium of trp-cage. (A) Backbone atom rmsd distributions obtained from the 0.966 gcm−3-isochore for 4 selected temperatures. (B) Fraction of folded configurations (rmsd ≤ 0.22 nm) as a function of temperature for both studied isochores.
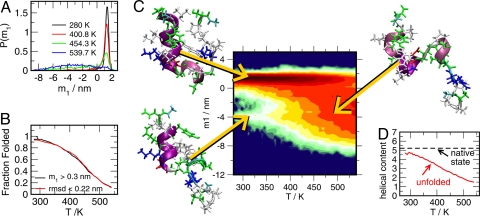
The fraction of folded states with rmsd ≤0.22 nm as a function of temperature has a quasisigmoidal shape, indicating transition temperatures of 445 and 425 K for 0.966 and 1.064 gcm−3, respectively. Alternatively, we have also determined the equilibrium of folded and unfolded states using principal component analysis (PCA) based the entire replica exchange molecular dynamics (REMD) ensemble of configurations. The largest eigenmode m1 describes a clamp-like opening of the cage, as suggested by the representatative low-temperature configurations shown in Fig. 3. A quantitative analysis based on the projection of the configurational density on the largest PCA-eigenmode m1 is given in Fig. 3 A–C. Similar to the rmsd distributions given in Fig. 2A, the projection of density of states on m1 is represented by a large narrow peak at low temperatures, which is decreasing at the expense of a broad distribution at higher temperatures. Employing a threshold of m1>0.3 nm to separate the “folded” basin from the “unfolded” configurations, we obtain a similar temperature dependence of the fraction of folded stated as due to the rmsd distributions, as shown in Fig. 3B.
Fig. 3.
Structural aspects related to the folding/unfolding of Trp-cage. (A–C) Principal component analysis (70) of the entire REMD configurational ensemble obtained for the 0.966 gcm−3 isochore: (A) Probability distributions of the largest eigenmode m1 for 4 selected temperatures. (B) Comparison of the temperature dependence of fraction of folded states as obtained from the rmsd and m1 distributions. (C) Free energy landscape of Trp-cage (in units of kT) projected on the m1 eigenmode. Representative configurations of Trp-cage for selected states are indicated. (D) Helical content (defined as in ref. 71) of unfolded Trp-cage as a function of temperature. The helical content of the native state (PDB ID code 1L2Y) is given as a reference.
We would like to emphasize that our simulations differ in certain aspects from the simulations of Trp-cage reported recently by Juraszek and Bolhuis (43). Their extensive simulation study using the OPLS-AA forcefield model with explicit simple point charge model (SPC) water suggests the existence of 2 different folding pathways: One in which the helix forms first (i), and 1 in which a contact between Trp-6 and a polyproline forms before the helix (ii). The latter pathway is found to be ≈4 times more likely in their study. It is not unlikely, however, that the formation of a Trp-6-polyproline contact could represent a kinetic bottleneck, hampering the formation of the helices and therefore reaching the fully folded state. In our simulations, pathway i is observed almost exclusively, with the cage opening and the helix melting having roughly the same transition temperature (44). The restriction to path i apparently allows the molecule to reach the folded state (and the folding/unfolding equilibrium) more quickly, also suggesting that the ff94-model has a less rugged, more funnel-like free energy landscape, which is enabling our REMD simulations to equilibrate within a 40 ns time-window.
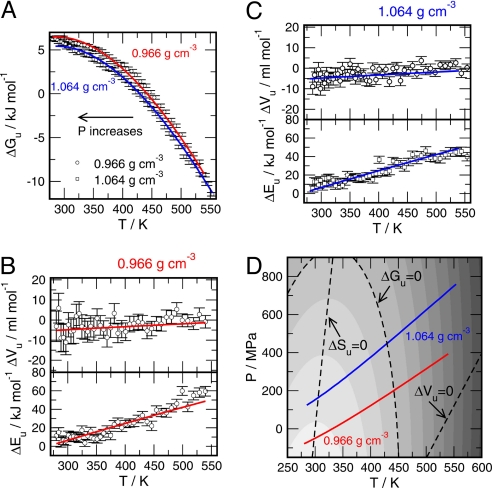
Because we can distinguish folded and unfolded ensembles, we can calculate the free energy of unfolding according to ΔGu(T,P) = Gunfolded − Gfolded = −RT ln [(1−xfolded)/xfolded], where xfolded is the fraction of folded states obtained from the REMD at each T, and P is the average pressure, <P>V. In a similar fashion we can determine the energy and volume changes associated with unfolding according to ΔEu = Eunfolded − Efolded and ΔVu = Vunfolded − Vfolded. All 3 properties (ΔGu, ΔEu, and ΔVu) were sampled for the states along the 2 isochores and are shown in Fig. 4 A–C. ΔEu is directly available via the average potential energies obtained for the folded and unfolded ensembles sampled by the REMD simulation. ΔVu is not directly available due to the application of constant volume conditions during the REMD simulation. However, the free energy change associated with volume changes becomes evident by observing slightly differing average pressures for the folded and unfolded ensembles following VΔP≈ PΔV. To determine the associated volume changes directly and not rely on the observed pressure differences only, we performed a posteriori additional short (80 ps) constant-pressure simulation runs at the average pressures obtained for each of the statepoints. The simulations were performed under isobar isothermal conditions using a Nosé–Hoover (49, 50) thermostat and a Rahman–Parrinello barostat (51, 52) with coupling times of τT = 0.5 ps and τP = 1.0 ps, using a molecular dynamics timestep of Δt = 2.0 fs. Exactly 200 NPT simulations were conducted for each statepoint: 100 simulations representing the folded and 100, the unfolded ensemble. The starting configurations for these simulations (in total: 17,600 simulation runs) were sampled randomly from the set of REMD configurations obtained from the final 60 ns. The length of the simulations was chosen such that a volume relaxation was feasible, whereas the configuration of the protein stayed in sufficiently close proximity to the initial configuration. The calculated ΔEu obtained from the NPT simulations are within the errorbars of ΔEu obtained from REMD, and the observed volume changes were found to be consistent with the calculated pressure difference for the subensembles. The data shown Fig. 4 depict the data according the NPT simulations.
Fig. 4.
Averages of the difference upon unfolding of the (A) free energy, (B and C) total energy, and specific volume, calculated for the 2 indicated isochores. (D) Free energy surface ΔGu(P,T) obtained by fitting to a Hawley-type model the free energy and its derivatives calculated for the 2 isochores. Different grayscale colors indicate changes of ΔGu of 2 kJ mol−1.
Having traced the free energy and 2 of its derivatives along the 2 isochores, both of them effectively crossing a large portion of the experimentally accessible P- and T-ranges, we are able to construct the stability diagram of the Trp-cage in terms of ΔGu as a function of temperature and pressure by fitting the calculated data to a Hawley-type (9, 12, 13) free energy surface ΔGu (P,T) = Δβ/2 (P−P0)2 + Δα(P−P0)(T−T0) − ΔCP [T(ln T/T0 −1) + T0] + ΔV0 (P−P0) − ΔS0 (T−T0) + ΔG0. Here, Δβ, Δα, and ΔCP are the changes in compressibility, expansivity, and heat capacity, and ΔV0, ΔS0, and ΔG0 are the unfolding volume, entropy, and free energy at the reference state (T0 = 331 K, P0 = 0 MPa), correspondingly. All coefficients in this expansion are assumed to be constant with temperature and pressure. The fitted values are given in Table 1. First, we would like to emphasize that the independently calculated isochores shown in Fig. 4A directly demonstrate the destabilizing effect of an increasing hydrostatic pressure. In addition, the fitted free energy landscape ΔGu(P,T), shown in Fig. 4D, indicates that simulated Trp-cage is very likely to exhibit a ellipse shaped stability diagram, similar to globular proteins. The condition for an elliptical phase diagram, Δα2 > ΔCP Δβ/T0, is satisfied here with a positive ΔCp and a negative Δβ (12). The Trp-cage is found to be quite stable with respect to changes in temperature and pressure, as the high unfolding temperatures (≈450 K at normal pressure) and pressures (≈>800 MPa at ambient temperatures) suggest. Moreover, the transition ranges are found to be very broad, which is apparently due to the small size of the molecule and associated with small energy and volume changes. Fig. 4A strongly suggests a parabolic shape of the unfolding free energy vs. temperature curve for Trp-cage, which is further supported by temperature dependence of the corresponding ΔEu-isochores, both increasing almost linearly and qualitatively resembling very much the curves reported by Makhatadze and Privalov for myoglobin (5). In addition, the calculated heat capacity of 0.2 ± 0.05 kJ K−1mol−1 is found to be close to the experimental value of 0.3± 0.1 kJ K−1·mol−1 reported recently by Streicher and Makhatadze (31). From the Hawley model we can extrapolate the free energy to 25°C and 1 atm (0.1 MPa) and get ΔGu = 6.2 ± 0.5 kJ mol−1, ΔHu = 36.0 ± 5 kJ mol−1 (at Tm = 448 K). These values can be compared with the measured ΔGu = 3.2 ± 5 kJ mol−1, and ΔHu = 56.0 ± 2 kJ mol−1 (at Tm = 317 K).
Table 1.
Thermodynamics parameters describing the Hawley-type (9, 12) free energy surface ΔGu(P,T) for simulated Trp-cage shown in Fig. 4
| ΔG0 | 5.9 ± 0.5 kJ mol−1 |
| ΔV0 | −4.3 ± 1.0 ml·mol−1 |
| ΔS0 | 1.9 ± 0.5 × 10−2 kJ mol−1·K−1 |
| ΔCp | 2.0 ± 0.5 × 10−1 kJ mol−1 K−1 |
| Δα | 2.2 ± 2.0 × 10−2 ml· mol−1 K−1 |
| Δβ | −4.0 ± 6.0 × 10−6 kJ mol−1 MPa−2 |
The chosen reference state is at T0 = 331 K, and P0 = 0 MPa.
The following aspects of the calculated diagram seem to be noteworthy. First, the observed variation of the unfolding energy ΔEu over the entire temperature interval is much larger (a factor of ≈4) than the variation of the free energy ΔGu, leading to a similar kind of enthalpy/entropy compensation behavior as observed in globular proteins (12). In addition, for the lowest temperatures, ΔEu approaches 0, indicating that at ambient temperature conditions the Trp-cage protein can unfold with only little expense of energy, which is another significant thermodynamic signature of globular proteins in the native state. Moreover, the almost constant heat capacity, as indicated by the linear T-behavior of ΔEu, and ΔEu≈ 0 at low T, strongly suggests that the lowest temperatures of the 2 isochores are in close proximity to the ΔSu = 0-line (also drawn in Fig. 4D for the fitted data), where the Trp-cage enters the cold denaturation regime (8). As suggested by Fig. 3D, the observed small energy differences ΔEu at low temperatures go along with the preservation of the secondary structure elements of the native fold. With decreasing temperatures, the helical content of the unfolded Trp-cage approaches the value for native state.
The details of the P-T stability diagram presented here depend on the chosen forcefield. The ff94 force field is known to exaggerate α- helix stability. The thermodynamic stability diagram for Trp-cage has been calculated only for the ff94 forcefield (44) and showed that the fraction of the states occupying the folded state is correct for low T but failed to describe the stability for higher temperatures. The high propensity to form α-helices may be partly resposible for the enhanced stability of the protein at high temperature. Our calculations show the same effect and we expected that the stability with pressure will also be exaggerated and that the Trp-cage will unfold (at low temperatures) at pressures below the 800 MPa predicted here. However, the general features of the stability diagram (like cold denaturation) are similar to the features found for globular proteins. Further calculations of the P-T stability diagram for other force fields and water models may be needed to better understand the forcefield dependence of the folding free energy, energy and volume changes.
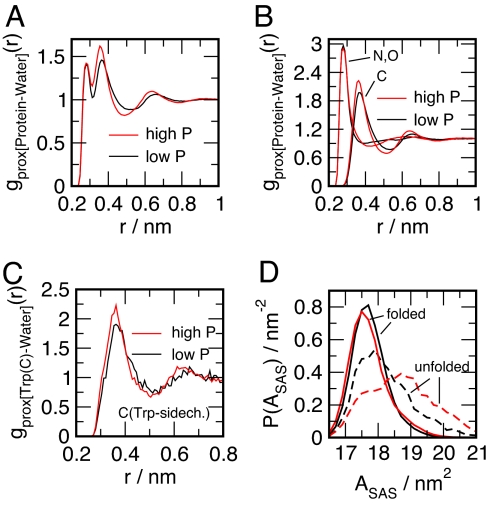
To elucidate the effect of pressure on the stability of the native fold, we inspect the water density in close proximity to the protein. We use a procedure for calculating protein-water “proximal pair correlation functions” gprox(r), similar to the 1 used by Ashbaugh and Paulaitis (53, 54), suggested earlier by others (55, 56). As reference sites we use heavy atoms of the protein (C, N, O) divided in “polar” (O, N) and “nonpolar” (C) groups. The normalization volume sα(r)dr is defined by volume elements with a shortest distance to any atom belonging to 1 of those groups of atoms, taken out of the set of atoms containing all protein heavy atoms. Additivity of the partial surface area sα(r) leads to the relation [sα(r)+sβ(r)] gtotalprox(r) = sα(r) gαprox(r)+sβ(r)gβprox(r), where sα represents the surface area of a subgroup of heavy atoms as a function of distance to the protein r. Fig. 5A shows the proximal radial distribution function of all heavy atoms of the entire protein at low and high pressures. The given gprox(r) exhibit a typical 2-peak feature, which has been reported to be according to the hydration of polar and nonpolar atoms (56, 57), which is also clearly demonstrated for the Trp-cage in Fig. 5A. Note that the proportion between the 2 peaks is markedly different from for proteins reported hitherto (56, 57), with the nonpolar peak being significantly more dominant in the Trp-cage case. This suggests that the surface of larger size proteins is on average significantly more polar, which, however, might be also a consequence of the small size of the molecule. When comparing the low and high density isochores, it is observed that the water packing (density) around the nonpolar sites changes significantly more than the packing around the polar sites, as revealed by the increase of the second peak, and by the individual component proximal radial distribution functions shown in Fig. 5 A and B. A similar trend is observed for the packing of water around the C atoms of the Trp-sidechain in the unfolded state given in Fig. 5C, where the Trp-sidechain has been previously shown to be significantly exposed to the water solvent (44). A tighter packing of water around nonpolar, or in particular hydrophobic atoms, might explain the increasing surface area observed for the unfolded configurations at elevated pressures shown in Fig. 5D. A tighter water packing efficiency around unpolar atoms might hence also be responsible for shifting the configurational equilibrium toward the unfolded state. Our results are in agreement with the description of hydrophobic hydration as “soft” and polar hydration as “hard” in response to high pressures (58).
Fig. 5.
Water packing around Trp-cage. (A–C) Proximal radial distribution functions gprox(r) between water's center of mass and the heavy atoms of the Trp-cage molecule. All data shown refers to “low temperature configurations” with T<320 K (A) gprox(r) including both, “polar” (O and N) and nonpolar “C” atoms. (B) separate gprox(r) for “polar” (O and N) and nonpolar “C” atoms. (C) gprox(r) for nonpolar “C” atoms of the Trp-sidechain, calculated for the “unfolded” configurations. (D) Solvent accessible surface area distributions (72) calculated for folded and unfolded configurations of Trp-cage for T<320 K.
In addition to the observed differences in water packing around polar and nonpolar atoms, there might be additional factors affecting the stability of the folded state. The formation of an Asp-9/Arg-16 salt-bridge has been proposed to significantly stabilize the folded state (28, 41). Indeed, the folded conformation of ff94-model Trp-cage has been shown to largely exhibit an Asp-9/Arg-16 contact (44). Comparing low- and high- pressure configurations, we observe that the tendency to form a Asp-9/Arg-16 contact in the folded ensemble (averaging over states with T ≤ 320 K) drops from 86% to 73% with increasing pressure, possibly destabilizing the folded state. The enhanced stability of open salt bridge conformations is very likely related to the increasing ionic solvation strength of water depending on the almost linearly increasing dielectric constant due to an increasing dipole density (59). However, on a less coarse grained level, the importance of a changing local hydration of side chains and backbone cannot be ruled out, and should be further investigated.
Conclusion
We have used the REMD enhanced sampling approach to study the stability diagram of the Trp-cage miniprotein by calculating 2 isochores, scanning effectively a large portion of the experimentally accessible P,T-range.
With the help of an approximate description of the free energy of unfolding ΔGu(P,T) in terms of the Hawley theory, employing sampled free energy, energy and volume differences, we are able to generate a stability diagram for Trp-cage. With increasing pressure, as with increasing temperature the native state is found to be destabilized, leading to a typical ellipsoidal (tongue-shaped) protein stability diagram, as found for many globular proteins. The energy difference between folded and unfolded conformations is found to approach zero energy difference at the lowest temperatures, which is accompanied by a conservation of helical secondary structure in the unfolded ensemble. With increasing pressure we observe a tighter packing of water around nonpolar atoms, accompanied by an increase of the solvent accessible surface area of the unfolded ensemble, and destabilized Asp-9/Arg-16 salt-bridge in the folded ensemble.
We would like to emphasize that with a melting temperature being approximately 140 K too high on a quantitative level, the presented computer models are still far from being completely satisfactory. In addition, the used ff94-forcefield has the known tendency to overemphasize the stability of helices, which is in our case apparently leading to an overall dominance of a folding pathway with an initial α-helical segment being formed first. However, this results in a less rugged free energy surface compared with that of the OPLS-AA model, finally allowing our REMD simulations to converge in the presented 100 ns time window.
We have demonstrated that, with present existing computer simulation techniques, it is possible to obtain reasonable rigorous thermodynamic data for the temperature- and pressure-induced folding/unfolding of proteins based on atomic detail models. Together with the constantly increasing computing power, experimental P,T data might thus be used to significantly improve current forcefield models in the future.
Simulation Methods.
We use REMD simulations (60) to study the unbiased equilibrium folding of the Trp-cage miniprotein, using the Amber (ff94) (61) forcefield and TIP3P water (62). We perform calculations of 2 isochores at densities of 0.966 gcm−3 and 1.064 gcm−3, effectively scanning the (P,T)-plane in a range between −100 and 800 MPa and 280 K and 280 K 550 K using 40 and 48 replicas. The densities were chosen to obtain average pressures of 0.1 MPa (1 bar) and 200 MPa at 330 K for the low- and high-density simulations, respectively. Simulations extending over 100 ns per replica provide a total of 8.8 μs worth of trajectory data. The entire simulation of the 2 isochores represents ≈105 CPU hours (2.2 GHz AMD Opteron) on the Linux clusters at Rensselaer Polytechnic Institute and Technische Universität Dortmund.
The Trp-cage sequence (Ac-NLYIQWLKDGGPSSGRPPPS-NME) is generated in an initially all-PPII conformation by the LEAP program distributed with AMBER 6.0. The N and C termini were capped with methyl groups, and the constructed model peptide consists of 313 atoms. The LEAP-generated structure is slightly compacted during a short 25-ps simulation in the gas phase at 300 K. This structure is solvated in a cubic box 2,637 by TIP3P (62) water molecules, and the system is equilibrated during a 100-ps constant pressure simulation at 330 K and 1 bar. The final structure obtained from this simulation is placed in cubic boxes of lengths 4.40562 nm and 4.20562 nm, and is used as a starting configuration for the constant volume REMD simulations for the 0.966, and 1.064 gcm−3 isochores. The peptide is found to be completely unfolded with a (C, N, O) backbone rmsd of from the first NMR structure of 6.0 Å. Moreover it lacks any regular secondary structure elements; in particular, it has none of the helical structure elements that are present in the native state. REMD (60) has been used to study the thermodynamics of the Trp-cage protein starting from this initial structure. REMD is an enhanced sampling technique based on the parallel tempering Monte Carlo method (63–65), where multiple copies (or replicas) of identical systems are simulated in parallel at different temperatures. Periodically, state-exchange moves are attempted, where 2 neighboring replicas exchange their thermodynamic state (their temperature). The acceptance rule for each state-exchange moves between 2 neighboring states i and j is chosen to be
where β = 1/kBT and U(r⃗iN) represents the configurational energy of the system in state i. The state-exchange acceptance probability Pacc has been shown to obey the detailed balance condition for an extended ensemble of canonical states (65).
Our simulations employ 40 and 48 replicas for distribution over a temperature range from 280.0 K to 539.7 K. The temperature spacing between each of the replicas was chosen such that the energy distributions overlap sufficiently, and state exchange attempts are (on average) accepted with a 20% probability. To initially set up the temperature spacings, energy distributions were obtained from a preceding series of noncoupled short (0.5 ns) constant volume MD simulations at similar density. Simulations are run at ρ = 0.966 gcm−3: 280.0, 284.1, 288.2, 292.4, 296.7, 301.1, 305.6, 310.2, 314.9, 319.7, 324.6, 329.6, 334.7, 340.0, 345.4, 351.0, 356.6, 362.5, 368.4, 374.6, 380.9, 387.3, 394.0, 400.8, 407.8, 415.1, 422.5, 430.1, 438.0, 446.0, 454.3, 462.8, 471.6, 480.6, 489.8, 499.3, 509.0, 519.0, 529.2, and 539.7 K, and at ρ = 1.064 gcm−3: 285.0, 288.3, 291.7, 295.1, 298.5, 302.1, 305.7, 309.3, 313.0, 316.8, 320.7, 324.6, 328.7, 332.8, 337.0, 341.2, 345.6, 350.1, 354.6, 359.3, 364.1, 368.9, 373.9, 379.1, 384.3, 389.7, 395.2, 400.8, 406.6, 412.6, 418.7, 425.0, 431.4, 438.0, 444.8, 451.8, 459.0, 466.5, 474.1, 481.9, 490.0, 498.3, 506.9, 515.8, 524.9, 534.3, 544.0, and 553.9 K.
State exchange attempts were undertaken with a probability of 0.05, leading to an average time of ≈1.6 ps for each replica between 2 state exchanges. The time step used in the MD steps is 2 fs, and a Nosé–Hoover (49, 50) thermostat is used with a time coupling of τT = 0.5 ps. Solvent constraints were solved using the SETTLE procedure (66), whereas the SHAKE method was used to constrain the solute bond lengths (67). The simulations were carried out with the GROMACS (68) simulation program, modified by us to allow for state-swapping moves. The electrostatic interactions were treated by smooth-particle mesh Ewald summation (69) using a 36 × 36 × 36 grid with fourth-order charge interpolation and a real-space cutoff of 0.9 nm. The Ewald convergence factor α was set to 3.38 nm−1 (corresponding to a relative accuracy of the Ewald sum of 10−5). Appropriate Lennard–Jones long-range correction for energy and pressure were taken into account.
Acknowledgments.
D.P. gratefully acknowledges financial support from the Deutsche Forschungsgemeinschaft (FOR 436) and from Technische Universität Dortmund (DOMUS). Part of the calculations were performed on the LiDO compute cluster at Technische Universität Dortmund. This work has been supported by the National Science Foundation (Grant MCB-0543769).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 2.Zipp A, Kauzmann W. Pressure denaturation of metmyoglobin. Biochemistry. 1973;12:4217–4228. doi: 10.1021/bi00745a028. [DOI] [PubMed] [Google Scholar]
- 3.Heremans K. High-pressure effects on proteins and other biomolecules. Annu Rev Biophys Biol. 1982;11:1–21. doi: 10.1146/annurev.bb.11.060182.000245. [DOI] [PubMed] [Google Scholar]
- 4.Timasheff SN. The control of protein stability and association by weak interactions with water: How do solvents affect these processes. Annu Rev Biophys Biomol Struct. 1993;22:67–97. doi: 10.1146/annurev.bb.22.060193.000435. [DOI] [PubMed] [Google Scholar]
- 5.Makhatadze GI, Privalov PL. Energetics of protein structure. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 6.Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 7.Jaenicke R. Protein stability and molecular adaptation to extreme conditions. Eur J Biochem. 1991;202:715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- 8.Privalov PL. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 9.Hawley SA. Reversible pressure-temperature denaturation of chymotrypsinogen. Biochemistry. 1971;10:2436–2442. doi: 10.1021/bi00789a002. [DOI] [PubMed] [Google Scholar]
- 10.Herberhold H, Winter R. Temperature- and pressure-induced unfolding and refolding of ubiquitin: A static and kinetic Fourier transform infrared spectroscopy study. Biochemistry. 2002;41:2396–2401. doi: 10.1021/bi012023b. [DOI] [PubMed] [Google Scholar]
- 11.Panick G, et al. Exploring the temperature-pressure phase diagram of staphylococcal nuclease. Biochemistry. 1999;38:4157–4164. doi: 10.1021/bi982608e. [DOI] [PubMed] [Google Scholar]
- 12.Smeller L. Pressure-temperature phase diagrams of biomolecules. Biochim Biophys Acta. 2002;1595:11–29. doi: 10.1016/s0167-4838(01)00332-6. [DOI] [PubMed] [Google Scholar]
- 13.Ravindra R, Winter R. On the temperature-pressure free-energy landscape of proteins. ChemPhysChem. 2003;4:359–365. doi: 10.1002/cphc.200390062. [DOI] [PubMed] [Google Scholar]
- 14.Ravindra R, Royer C, Winter R. Pressure perturbation calorimetic studies of the solvation properties and the thermal unfolding of staphylococcal nuclease. Phys Chem Chem Phys. 2003;6:1952–1961. doi: 10.1039/b516608j. [DOI] [PubMed] [Google Scholar]
- 15.Wiedersich J, Kohler S, Skerra A, Friedrich J. Temperature and pressure dependence of protein stability: The engineered fluorescein-binding lipocalin FluA shows an elliptic phase diagram. Proc Natl Acad Sci USA. 2008;105:5756–5761. doi: 10.1073/pnas.0710409105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 17.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 18.Athawale MV, Goel G, Ghosh T, Truskett TM, Garde S. Effects of lengthscales and attractions on the collapse of hydrophobic polymers in water. Proc Natl Acad Sci USA. 2007;104:733–738. doi: 10.1073/pnas.0605139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 20.Silva JL, Foguel D, Royer CA. Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem Sci. 2001;26:612–618. doi: 10.1016/s0968-0004(01)01949-1. [DOI] [PubMed] [Google Scholar]
- 21.Frauenfelder H, et al. Proteins and pressure. J Phys Chem. 1990;94:1024–1037. [Google Scholar]
- 22.Akasaka K. Highly fluctuating protein structures revealed by variable pressure nuclear magnetic resonance. Biochemistry. 2003;42:10875–10885. doi: 10.1021/bi034722p. [DOI] [PubMed] [Google Scholar]
- 23.Kitahara R, Akasaka K. Close identity of a pressure stabilized intermediate with a kinetic intermediate in protein folding. Proc Acad Natl Sci USA. 2003;100:3167–3172. doi: 10.1073/pnas.0630309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitahara R, Yamura H, Akasaka K. Two folded conformers of Ubiquitin revealed by high-pressure NMR. Biochemistry. 2001;40:13556–13563. doi: 10.1021/bi010922u. [DOI] [PubMed] [Google Scholar]
- 25.Hummer G, Garde S, García AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95:1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh T, García AE, Garde S. Molecular dynamics simulations of pressure effects on hydrophobic interactions. J Am Chem Soc. 2001;123:10997–11003. doi: 10.1021/ja010446v. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh T, García AE, Garde S. Enthalpy and entropy contributions to the pressure dependence of hydrophobic interactions. J Chem Phys. 2002;116:2480–2486. [Google Scholar]
- 28.Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-residue protein. Nat Struct Biol. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 29.Gellmann SH, Woolfson DN. Mini-protein TRP: The light fantastic. Nat Struct Biol. 2002;9:408–410. doi: 10.1038/nsb0602-408. [DOI] [PubMed] [Google Scholar]
- 30.Qiu L, Pabic SA, Roitberg AE, Hagen SJ. Smaller and faster: The 20-residue trp-cage folds within 4 μs. J Am Chem Soc. 2002;124:12952–12953. doi: 10.1021/ja0279141. [DOI] [PubMed] [Google Scholar]
- 31.Streicher WW, Makhatadze GI. Unfolding thermodynamics of trp-cage, a 20 residue miniprotein, studied by differential scanning calorimetry and circular dichroism spectroscopy. Biochemistry. 2007;46:2876–2880. doi: 10.1021/bi602424x. [DOI] [PubMed] [Google Scholar]
- 32.Neuweiler H, Doose S, Sauer M. A microscopic view of miniprotein folding: Enhanced folding efficiency through formation of an intermediate. Proc Natl Acad Sci USA. 2005;102:16650–16655. doi: 10.1073/pnas.0507351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed Z, Beta IA, Mikhonin AV, Asher SA. Uv-resonance raman thermal unfolding study of trp-cage shows that it is not a simple two-state miniprotein. J Am Chem Soc. 2005;127:10943–10950. doi: 10.1021/ja050664e. [DOI] [PubMed] [Google Scholar]
- 34.Simmerling C, Stockbine B, Roitberg A. All-atom structure prediction and folding simulations of a stable protein. J Am Chem Soc. 2002;122:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 35.Snow CD, Zagrovich B, Pande VS. The Trp-cage: Folding kinetics and unfolded state topology via molecular dynamics simulations. J Am Chem Soc. 2002;124:14548–14549. doi: 10.1021/ja028604l. [DOI] [PubMed] [Google Scholar]
- 36.Pitera JW, Swope W. Understanding folding and design: Replica exchange simulations of the “Trp-cage” mini-protein. Proc Natl Acad Sci USA. 2003;100:7587–7592. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhury S, Lee MC, Xiong G, Duan Y. Ab initio folding simulations of the trp-cage miniprotein approaches nmr resolution. J Mol Biol. 2003;327:711–717. doi: 10.1016/s0022-2836(03)00177-3. [DOI] [PubMed] [Google Scholar]
- 38.Chowdhury S, Lee MC, Duan Y. Characterizing the rate-limiting step of trp-cage folding by all-atom molecular dynamics simulations. J Phys Chem B. 2004;108:13855–13865. [Google Scholar]
- 39.Schug A, Herges T, Wenzel W. Reproducible protein folding with the stochastic tunneling method. Phys Rev Lett. 2003;91:158102. doi: 10.1103/PhysRevLett.91.158102. [DOI] [PubMed] [Google Scholar]
- 40.Schug A, Herges T, Verma A, Lee KH, Wenzel W. Comparison of stochastic optimization methods for all-atom folding of the trp-cage protein. ChemPhysChem. 2005;6:2640–2646. doi: 10.1002/cphc.200500213. [DOI] [PubMed] [Google Scholar]
- 41.Zhou R. Trp-cage: Folding free energy in explicit water. Proc Natl Acad Sci USA. 2003;100:13280–13285. doi: 10.1073/pnas.2233312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ota M, Ikeguchi M, Kidera A. Phylogeny of protein-folding trajectories reveals a unique pathway to native structure. Proc Natl Acad Sci USA. 2004;101:17658–17663. doi: 10.1073/pnas.0407015102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juraszek J, Bolhuis PG. Sampling the multiple folding mechanisms of Trp-cage in explicit solvent. Proc Natl Acad Sci USA. 2006;103:15859–15864. doi: 10.1073/pnas.0606692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paschek D, Nymeyer H, García AE. Replica exchange simulation of reversible folding/unfolding of the trp-cage miniprotein in explicit solvent: On the structure and possible role of internal water. J Struct Biol. 2007;157:524–533. doi: 10.1016/j.jsb.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 45.Paschek D, García AE. Reversible temperature and pressure denaturation of a protein fragment: A replica exchange molecular dynamics simulation study. Phys Rev Lett. 2004;93:238105. doi: 10.1103/PhysRevLett.93.238105. [DOI] [PubMed] [Google Scholar]
- 46.Paschek D, Gnanakaran S, García AE. Simulations of the pressure and temperature unfolding of an alpha-helical peptide. Proc Natl Acad Sci USA. 2005;102:6765–6770. doi: 10.1073/pnas.0408527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García AE, Paschek D. Simulation of the pressure and temperature folding/unfolding equilibrium of a small RNA hairpin. J Am Chem Soc. 2008;130:815–817. doi: 10.1021/ja074191i. [DOI] [PubMed] [Google Scholar]
- 48.Heremans K, Smeller L. Protein structure and dynamics at high pressure. Biochim Biophys Acta. 1998;1368:353–370. doi: 10.1016/s0167-4838(98)00102-2. [DOI] [PubMed] [Google Scholar]
- 49.Nosé S. A molecular dynamics method for simulating in the canonical ensemble. Mol Phys. 1984;52:255–268. [Google Scholar]
- 50.Hoover WG. Canonical dynamics: Equilibrium phase space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 51.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7180. [Google Scholar]
- 52.Nosé S, Klein ML. Constant pressure molecular dynamics for molecular systems. Mol Phys. 19893;50:1055–1076. [Google Scholar]
- 53.Ashbaugh HS, Paulaitis ME. Entropy of hydrophobic hydration: Extension to hydrophobic chains. J Phys Chem. 1996;100:1900–1913. [Google Scholar]
- 54.Ashbaugh HS, Paulaitis ME. Effect of solute size and solute-water attractive interactions on hydration water structure around hydrophobic solutes. J Am Chem Soc. 2001;123:10721–10728. doi: 10.1021/ja016324k. [DOI] [PubMed] [Google Scholar]
- 55.Mehrotra PK, Beveridge DL. Structural-analysis of molecular solutions based on quasi-component distribution-functions — Application to [H2CO]aq at 25-degrees-C. J Am Chem Soc. 1980;102:4287–4294. [Google Scholar]
- 56.Levitt M, Sharon R. Accurate simulation of protein dynamics in solution. Proc Natl Acad Sci USA. 1988;85:7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smolin N, Winter R. Molecular dynamics simulations of staphylococcal nuclease: Properties at the protein surface. J Phys Chem B. 2004;108:15928–15937. [Google Scholar]
- 58.Giovambattista N, Lopes CF, Rossky PJ, Debenedetti PG. Hydrophobicity of protein surfaces: Separating geometry from chemistry. Proc Natl Acad Sci USA. 2008;105:2274–2279. doi: 10.1073/pnas.0708088105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neumann M. Dipole moment fluctuations formulas in computer simulations of polar systems. Mol Phys. 1983;50:841–858. [Google Scholar]
- 60.Sugita Y, Okamoto Y. Replica exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 61.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 62.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 63.Hansmann UHE. Parallel tempering algorithm for conformational studies of biological molecules. Chem Phys Lett. 1997;281:140–150. [Google Scholar]
- 64.Hansmann UHE, Okamoto Y. New Monte Carlo algorithms for protein folding. Curr Opin Struct Biol. 1999;9:177–183. doi: 10.1016/S0959-440X(99)80025-6. [DOI] [PubMed] [Google Scholar]
- 65.Frenkel D, Smit B. Understanding Mol Simul — From Algorithms to Applications. 2nd Ed. San Diego: Academic; 2002. [Google Scholar]
- 66.Miyamoto S, Kollman PA. SETTLE: An analytical version of the SHAKE and RATTLE algorithms for rigid water models. J Comp Chem. 1992;13:952–962. [Google Scholar]
- 67.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motions of a system with constraints: Molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 68.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 69.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 70.García AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 71.García AE. Characterization of non-alpha helical conformations in Ala-peptides. Polymer. 2004;45:669–676. [Google Scholar]
- 72.Kabsch W, Sander C , Dictionary of protein secondary structure - pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]