Abstract
Sterol regulatory element binding protein-1c (SREBP-1c) is a basic helix–loop–helix (bHLH) homodimeric transactivator, which induces itself and several lipogenic enzymes, notably fatty acid synthase (FAS). We demonstrated that hypoxia-inducible factor (HIF) represses the SREBP-1c gene by inducing Stimulated with retinoic acid (Stra)13/Differentiated embryo chondrocyte 1(DEC1) and its isoform, DEC2. Stra13/DEC1 and DEC2 are bHLH homodimeric transcription repressors. We found that both Stra13 and DEC2 inhibit SREBP-1c-induced transcription by competing with SREBP-1c for binding to the E-box in the SREBP-1c promoter and/or by interacting with SREBP-1c protein. DEC2 is instantly and temporarily induced in acute hypoxia, while Stra13 is induced in prolonged hypoxia. This expression profile reflects the finding that Stra13 represses DEC2, thus maintains low level of DEC2 in prolonged hypoxia. DEC2-siRNA restores the hypoxic repression but Stra13-siRNA fails to do so, suggesting that DEC2 is the major initiator of hypoxic repression of SREBP-1c, whereas Stra13 substitutes for DEC2 in prolonged hypoxia. Our findings imply that Stra13 and DEC2 are the mediators to repress SREBP-1c gene in response to hypoxia. By doing so, HIF and its targets, Stra13 and DEC2 reduce the ATP consuming anabolic lipogenesis prior to the actual decrease of ATP acting as a feed-forward mechanism.
INTRODUCTION
Under hypoxic conditions, cells cannot maintain the aerobic respiration that is required for oxidative phosphorylation by mitochondria, and this leads to decreased generation of ATP. Many types of hypoxia-tolerant cells avoid the risk of energy failure not only by increasing anaerobic glycolysis, but also by decreasing O2 consumption (1). The hypoxia-inducible factor-α/β (HIF-α/β) heterodimeric transcription factor plays a central role in both processes. HIF represses the respiration and biogenesis of mitochondria by inducing pyruvate dehydrogenase kinase 1 and the c-Myc antagonist, MXI-1, respectively (2–4).
The HIF-α and β subunits belong to the basic helix–loop–helix (bHLH)-Per-Arnt-Sim (PAS) protein family. In normoxia, HIF-α is ubiquitinated and rapidly degraded. It contains a binding site for the ubiquitin E3 ligase, von Hippel-Lindau protein (pVHL), which ubiquitinates it, targeting it for degradation. pVHL recognizes and binds to hydroxylated proline residues in HIF-α. Proline hydroxylation of HIF-α is catalyzed by HIF-α-specific proline hydroxylases, using O2, α-ketoglutarate, Vitamin C and Fe2+ (5). Another HIF-α-specific asparaginyl hydroxylase named factor-inhibiting HIF-1α uses the same cofactors to inhibit the transactivation activity of HIF-1α. Therefore, in addition to hypoxia, HIF-α can be stabilized and transactivated by other factors that inhibit these hydroxylation reactions, such as divalent metals, oxidizing agents, succinate and an increased oxygen consumption rate of mitochondria (3,4).
HIF-1α was the original HIF-α isoform identified by affinity purification, while HIF-2α/EPAS-1 was identified in a homology search (6). Both HIF-1α and HIF-2α form functional heterodimers with HIF-1β, also referred as aryl hydrocarbon receptor nuclear translocator (Arnt). Although knockout mice experiments showed that HIF-1α and 2α have distinctly different functions and play nonredundant roles (7), no target genes specific for HIF-2α have been identified. HIF-1α and HIF-2α share many target genes, but HIF-1α appears to be the predominant form responsible for induction of the target genes (8).
There is some evidence that a decreased demand for ATP is also important for hypoxic adaptation. In hypoxia-adapted hepatocytes, global protein synthesis is suppressed rapidly, thereby decreasing the consumption of ATP (9). Another ATP-consuming anabolic process is lipogenesis, which encompasses the processes by which glucose is converted to triglyceride by lipogenic enzymes, and takes place in both liver and adipose tissue (10). Fatty acid synthase (FAS), the key lipogenic enzyme responsible for the endogenous synthesis of fatty acids, has been shown to be regulated by hormonal and nutritional effects at the levels of transcription and activity (11,12). Insulin and sterols also have long-term effects on the expression of FAS genes (13), probably via the transcription factor, sterol regulatory element binding protein-1c (SREBP-1c), also referred to as adipocyte determination, and differentiation-dependent factor 1 (ADD1) (14). The SREBP-1 gene encodes two almost identical proteins, SREBP-1a and SREBP-1c transcripts from two different promoters. Besides the first four unique amino acids, SREBP-1c is identical to SREBP-1a (15). In the mouse liver, the SREBP-1c is 9-fold more than SREBP-1a. The SREBP-1c protein retains a greater ability to stimulate transcription of genes involved in fatty acid synthesis while SREBP-1a for cholesterol metabolism (15). SREBP-1c promoter contains a sterol regulatory element (SRE) and can be induced by SREBP-1c itself. Therefore, the SREBP-1c promoter makes it possible to form a positive feedback loop expression of SREBP-1c (16,17).
SREBP-1c/ADD1 belongs to the bHLH leucine zipper family, and is synthesized as a 125-kDa precursor protein bound to the endoplasmic reticulum (ER). When it is cleaved during sterol deprivation, its N-terminal region (amino acids 1–480) is released from the ER membrane into the nucleus as a 68-kDa mature transcription factor. The active SREBP-1c makes homodimer, which has dual DNA-binding specificity; it binds not only to the SRE, but also to the E-box (14). Besides being regulated by proteolytic release, transcription of the SREBP-1c gene is regulated by many hormonal and nutritional signals, including fasting and re-feeding (18), and insulin (19). SREBP-1s are known to contribute the adipogenesis by promoting that synthesis of the endogenous ligands for the adipogenic transactivator PPARγ. Yun et al. (20) showed that Stra13, a hypoxia-induced transcription repressor family, represses PPARγ2 promoter and functions as a mediator of hypoxic inhibition of adipogenesis. Stra13 is also referred to as Differentiated embryo chondrocyte 1 (DEC1). Stra13/DEC1 and its isoform DEC2 are class B type bHLH proteins which make homodimer. Both Stra13 homodimer and DEC2 homodimer are able to bind the E-box sequences (21). Stra13/DEC1 and DEC2 homodimers play a key role in cell differentiation, circadian rhythms, immune regulation and carcinogenesis (22). In the current study we investigated how HIF and its targets, Stra13/DEC1 and DEC2 bring about hypoxic repression of FAS and SREBP-1c.
MATERIALS AND METHODS
Materials and plasmids
The anti-HIF-1α antibody was obtained from Novus Biochemicals. The anti-HIF-1β/Arnt antibody and anti-human-SREBP-1 antibody were purchased from BD Biosciences (Palo Alto, CA, USA) and Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-mouse-SREBP-1 antibody was also generated, as described previously (23). The following cDNAs were used: HIF-1α (human, U22431), HIF-1β (human, NM_001668), Stra13/DEC1 (mouse, AF010305), DEC2 (mouse, NM_024469) and SREBP-1c (amino acids 1–403 of rat AF286469). The plasmid pEBG-SREBP-1c encodes rat SREBP-1c (amino acid 1–403) fused to Glutathione-S-transferase (GST) under the control of the mammalian elongation factor 1 promoter. The FAS promoter-driven luciferase reporter plasmid contains the upstream regulatory region (−220 bp to +25 bp) of the rat FAS promoter (24). The SREBP-1c promoter-driven luciferase reporter plasmid contains the enhancer and promoter region (−2.7 kb to +1 bp) of the mouse SREBP-1c gene (23). All chemicals were purchased from Sigma Co.
Measurement of ATP
A constant-light signal luciferase assay developed by Boehringer-Mannheim (ATP Bioluminescence Assay Kit CLS II) was utilized to determine levels of ATP. Wild-type mouse Hepa1c1c7 cells were plated in triplicate at 5 × 104 cells in a 35-mm tissue culture plate and allowed to incubate overnight. After 16 h, the cells were exposed to hypoxia for the indicated times. Molar amounts of ATP were determined using ATP standards (10–4 to 10–11 M ATP) versus the relative luciferase units. Luciferase units were normalized for total protein concentration as determined by the Bradford assay using bovine serum albumin as a standard. We present the averages and standard deviations of at least three experiments.
Northern analysis and quantitative real-time reverse transcription (RT)–polymerase chain reaction (PCR) (Q-PCR)
Total RNA was isolated using an RNeasy spin column (Qiagen Inc., Valencia, CA, USA). Northern analyses were performed as described previously (25). cDNA was reverse transcribed from total RNA (1 μg) using AMV reverse transcriptase with dNTPs and random primers (Promega, Madison, WI, USA). For quantitative real time reverse transcription (RT)–polymerase chain reaction (PCR) (Q-PCR) analysis, the iQTM SYBR Green Supermix and MyiQ single color real-time PCR detection system (Bio-Rad, Hercules, CA, USA) were used. The expression level of 18S rRNA was used for normalization. All PCRs were performed in triplicate. We present the average and standard deviation of at least three experiments. Primer sequences are given in Supplementary Table S1.
Electrophoretic mobility shift assays (EMSA)
GST-SREBP-1c (amino acids 1–403) fusion protein was expressed in Escherichia coli (BL21) and purified using glutathione uniflow resin according to the instruction of manufacturer (Amersham Biosciences, Uppsala, Sweden). The oligonucleotides used for the E-box-containing FAS promoter (−74 to −51 bp); the oligonucleotides used for the SRE complex sequences of SREBP-1c promoter (−89 to −53 bp); the SRE mutant sequences and the E-box mutant sequences are shown in Figure 5B and Supplementary Figure S2C. Each pair of oligonucleotides (1.75 pmol) was annealed and labeled with α-[32P]-dATP and Klenow enzyme. Recombinant GST-SREBP-1c (amino acids 1–403) protein were preincubated with polydeoxyinosinic-deoxycytidylic acid (1 μg) in 20 μl binding reactions containing reaction buffer [10 mM Tris pH 7.5, 50 mM KCl, 2.5 mM MgCl2, 0.05 mM EDTA, 0.1% (v/v) Triton X-100, 8% (v/v) glycerol, 1 mM dithiothreitol and 0.1% (w/v) non-fat dry milk] for 30 min on ice, as described (26). The radiolabeled oligonucleotides (4 × 105 c.p.m., approximately 0.3 pmol) were incubated with recombinant GST-SREBP-1c protein (5 μg) for 45 min on ice, and reaction mixtures were then separated by 6% PAGE at 4°C and exposed to X-ray film.
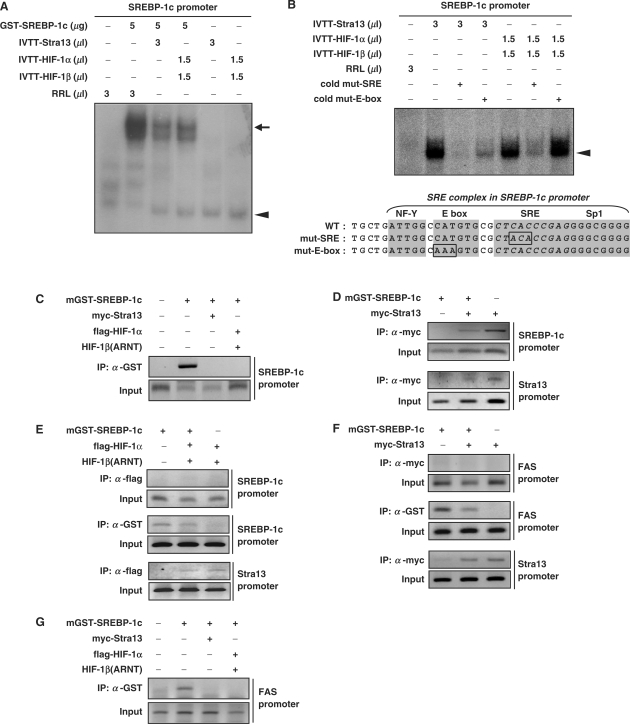
Figure 5.
DNA-binding activity of SREBP-1c on the SREBP-1c promoter. (A) EMSAs were performed using the radiolabeled oligonucleotides for the SREBP-1c promoter (−89 to −53 bp of mouse SREBP-1c gene) shown below. Recombinant GST-SREBP-1c protein was incubated with the indicated amount of either Stra13- or HIF-1α/HIF-1β-programmed rabbit reticulocyte lysate, followed by incubation with radiolabeled oligonucleotides. The upper arrow indicates the DNA–SREBP-1c complex, whereas the lower arrowhead indicates the DNA-Stra13 or DNA–HIF-1α/HIF-1β complex. (B) For competition assays, the indicated amount of either Stra13- or HIF-1α/β-programmed rabbit reticulocyte lysate was incubated with a 50-fold molar excess of either unlabeled SRE mutant oligonucleotides or E-box mutant oligonucleotides, followed by incubation with the radiolabeled oligonucleotides containing the wild-type SREBP-1c promoter for 30 min at 4°C prior to loading. (C–G) pEBG-SREBP-1c was transfected into 293 cells together with the indicated plasmids. Thereafter, ChIP assays were performed with the indicated antibodies as described in Materials and methods section.
Co-immunoprecipitation and GST pull-down
Human 293 cells were transfected with pEBG-SREBP-1c together with either pCMV-myc-Stra13, pCMV-myc-DEC2 or pCMV-3flag-HIF-1α and whole-cell extracts were prepared. For immunoprecipitation, 300 μg samples of whole-cell lysates were analyzed as described (25). The cleared extracts were mixed and precipitated with 2 μg of the indicated antibody.
[35S]-labeled SREBP-1c, HIF-1α, HIF-1β or Stra13 proteins were in vitro translated using a rabbit reticulocyte lysate (Promega), then incubated for 2 h at 4°C with immobilized GST or GST-SREBP-1c in 500 μl of NETN buffer [20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40 and 1 mM PMSF]. GST-SREBP-1c (amino acids 1–403) bound to the glutathione-uniflow resin was washed three times with 1 ml of NETN buffer at 4°C and eluted by boiling in SDS sample buffer. Boiled samples were subjected to SDS–PAGE and autoradiography.
Gene silencing using small interfering RNA (siRNA)
siRNAs specific for HIF-1α, HIF-2α, Stra13, DEC2 and green fluorescent protein (GFP) were synthesized by Samchully Pharm. Co. (Seoul, Korea). Sequence of each siRNA is shown in Supplementary Table S1. For siRNA transfection, Hepa1c1c7 cells or 3T3-L1 cells were plated at 5 × 105 cells in a 60-mm plate. Eighteen hours later, transfection was carried out using PolyMAG according to the instructions of the manufacturer (Chemicell GmBH, Germany). Forty-eight to 72 hours after transfection, total RNA or whole-cell extracts were prepared for further assays. To generate stably HIF-1α knockdown cells, we used a retroviral vector system. We ligated a short-hairpin RNA (shRNA) against HIF-1α into pSIREN-RetroQ vector (BD Biosciences) to generate pSIREN-RetroQ-shHIF-1α, according to the instructions of the manufacturer (BD Biosciences). pSIREN-RetroQ-shcontrol were provided from BD Biosciences. Sequence of each shRNA is shown in Supplementary Table S2.
Chromatin immunoprecipitation (ChIP) assay
Human 293 cells were transfected with pEBG-SREBP-1c together with pCMV-myc-Stra13, pCMV-myc-DEC2 or pCMV-3flag-HIF-1α/pcDNA3-HIF-1β (Arnt). ChIP assays were performed according to the instructions of the manufacturer (Upstate Biotechnology, Lake Placid, NY, USA). The transfected cells were cross-linked in 1% formaldehyde at 37°C for 10 min and resuspended in 200 μl of lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1)]. Lysates were sonicated. We measured OD 260 of the sonicated lysate solution to ensure that the amount of chromatin used in each sample is similar (27). Then we diluted the sonicated lysate 10-fold with ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl (pH 8.1), 167 mM NaCl]. The diluted lysates were immunoprecipitated with 2 μg of anti-GST antibody (Upstate Biotechnology), anti-flag antibody (Sigma) or anti-myc antibody (clone 9E10, Boehringer Mannheim). The immunoprecipitates were washed with four kinds of buffers; low salt buffer, high salt buffer, LiCl wash buffer and TE buffer as described (23). The immune complexes were eluted with 300 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and reversed (23). The isolated DNAs were used for PCR and the primers are shown in Supplementary Table S3.
Transient transfection and luciferase assay
Cells were plated at 1 × 105 cells/well in 24-well plates. Eighteen hours later, transfection was carried out using Lipofectamine plus reagent (Invitrogen, Carlsbad, CA, USA). Forty-eight hours after transfection, cell extracts were prepared and analyzed with a luminometer (Turner TD-20/20, Promega) using the luciferase assay system (Promega). Luciferase activity was normalized for total protein concentration as determined by the Bradford assay using bovine serum albumin as a standard. The transfection efficiency was monitored by measuring β-galactosidase activity of the cotransfected β-galactosidase-encoding plasmid (pCHO110).
RESULTS
Hypoxia reduces the expression of FAS and SREBP-1c
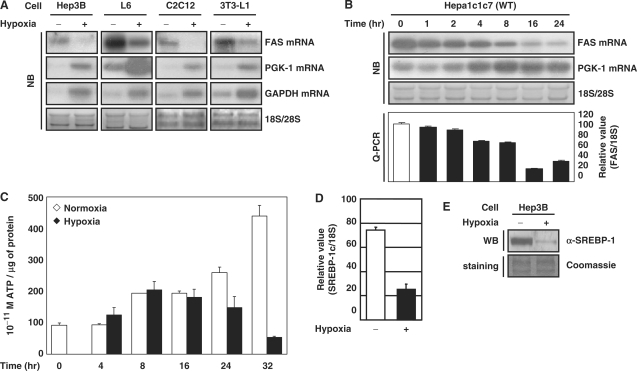
In adipocytes, the exposure of hypoxia reduces the content of triglyceride and cholesterol (Supplementary Figure S1A and B) (28–31). To determine whether hypoxia influences the expression of the lipogenic enzymes, we measured FAS mRNA. Hypoxia reduced FAS mRNA in Hep3B human hepatocytes, L6 mouse skeletal myocytes, C2C12 mouse myoblasts, 3T3-L1 mouse preadipocytes and Hepa1c1c7 mouse hepatoma cells (Figure 1A). In contrast, transcripts of hypoxia-inducible genes such as phosphoglycerate kinase-1 (PGK-1) and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) increased. After 16 h of hypoxic exposure, FAS mRNA was reduced by 20% in Hepa1c1c7 cells (Figure 1B) but the ATP level was unaffected (Figure 1C) (32,33). These findings imply that cells can shut down anabolic genes prior to an actual reduction of ATP, ultimately by reducing the levels of anabolic enzymes such as FAS. Lipogenic gene expression is promoted by potent lipogenic activators such as SREBP-1c. Quantitative real-time RT–PCR (Q-PCR) showed that hypoxic treatment reduced SREBP-1c mRNA in human hepatoma Hep3B cells (Figure 1D). The amount of SREBP-1 protein was also reduced (Figure 1E).
Figure 1.
Effect of hypoxia on the expression of FAS and SREBP-1c. (A and B) The indicated cells were incubated under hypoxic conditions (1% O2) (Model 1029 Forma Scientific, Inc.) for 16 h or for the indicated times. The levels of mRNAs were analyzed by northern blot (NB) and Q-PCR. (C) Wild-type Hepa1c1c7 cells were exposed to hypoxia for the indicated times. ATP content was measured using an ATP bioluminescence assay (Boehringer–Mannheim). (D) Hep3B cells were incubated in 1% O2 for 16 h. The level of SREBP-1c mRNA was quantified by Q-PCR using the ABI PRISM 7000 Sequence Detection System (Applied BioSystems). (E) Hep3B cells were incubated in 1% O2 for 6 h. Western blot (WB) analysis was performed using anti-SREBP-1 antibody (BD Biosciences).
HIF is involved in hypoxic repression of FAS and SREBP-1c
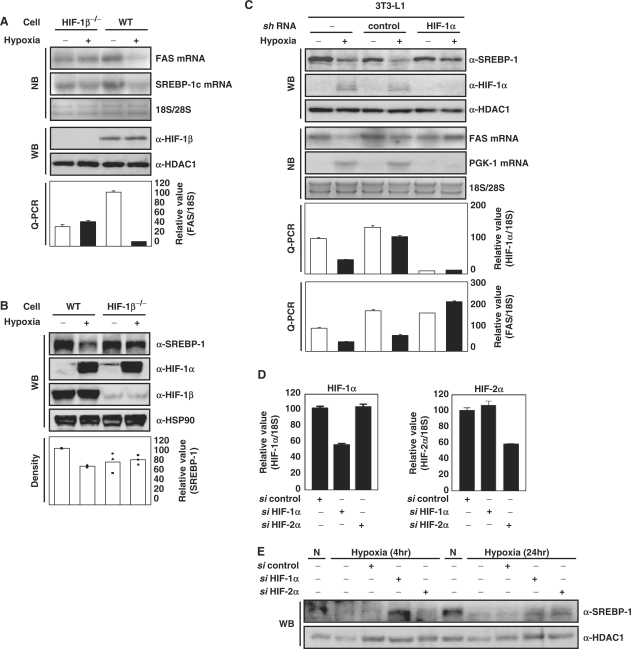
To test whether HIF is involved in this process, we measured transcript and protein levels of FAS and SREBP-1c in wild-type mouse hepatoma Hepa1c1c7 cells and HIF-1β-defective Hepa1c1c7 variant cells (Figure 2A and B) (34). Hypoxia failed to reduce mRNA of either FAS or SREBP-1c in the HIF-1β defective cells, indicating that HIF-1β is required for their repression by hypoxia. We also tested hypoxic repression of FAS mRNA and SREBP-1c protein in HIF-1α knockdown 3T3-L1 cells generated by infection with a retrovirus encoding shRNA against HIF-1α. We confirmed a specific reduction of protein and mRNA of HIF-1α by the cognate shRNA in 3T3-L1 cells (Figure 2C), and hypoxic treatment failed to repress FAS mRNA and SREBP-1c protein in the HIF-1α-knockdown 3T3-L1 cells.
Figure 2.
Effect of HIF on hypoxic-repression of FAS and SREBP-1c. (A and B) Wild-type mouse Hepa1c1c7 cells and HIF-1β-defective Hepa1c1c7 cells were incubated in 1% O2 for 16 h. The levels of FAS and SREBP-1c mRNA were analyzed by NB and Q-PCR. WB analysis was performed using anti-SREBP-1 antibody. The level of SREBP-1 protein was estimated by measuring band intensities (LAS 3000, Fuji) and numbers represent averages and standard deviations of three independent experiments. WB with anti-HDAC1 antibody or anti-Hsp90 antibody were used as loading controls. (C) HIF-1α-knockdown 3T3-L1 cells and control 3T3-L1 cells were generated using the retroviral system as described in Materials and methods section. The cells were incubated in 1% O2 for 16 h. WB analyses, NB analyses and Q-PCR were performed. (D and E) Hepa1c1c7 cells were transfected with the indicated siRNAs as described. Before harvest, the transfected cells were exposed to hypoxia (1% O2, 4 h or 24 h). The mRNA levels were quantified by Q-PCR. Values represent means and standard deviations of three experiments.
HIF-1α and 2α induce HRE-dependent expression of same target genes with different temporal patterns (35). Small inhibitory RNAs (siRNAs) against HIF-1α and HIF-2α were transfected into Hepa1c1c7 cells and we confirmed the specific reduction of HIF mRNA by the cognate siRNAs (Figure 2D). We tested whether HIF-2α is also involved in hypoxic repression of SREBP-1c protein. Western analysis showed that HIF-1α siRNA but not HIF-2α siRNA restored the expression of SREBP-1c protein which was repressed by acute hypoxic exposure (4 h). In contrast, in prolonged hypoxia, both HIF-1α siRNA and HIF-2α siRNA partially restored the expression of SREBP-1c protein (Figure 2E) and FAS (Supplementary Figure S1C). Taken together, our findings demonstrate that HIF-1α and HIF-2α act on SREBP-1c repression but in different temporal windows, with HIF-1α acting in the acute hypoxic phase and both HIF-1α and-2α in the prolonged phage.
Stra13/DEC1 is involved in hypoxic repression of SREBP-1c
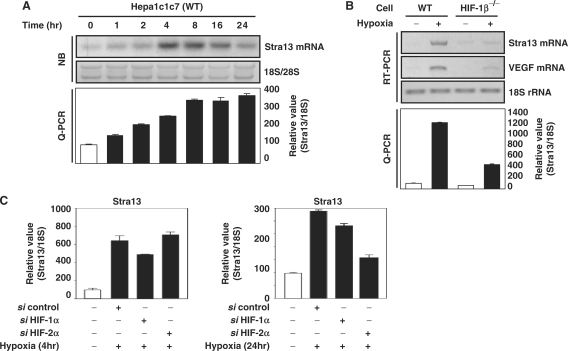
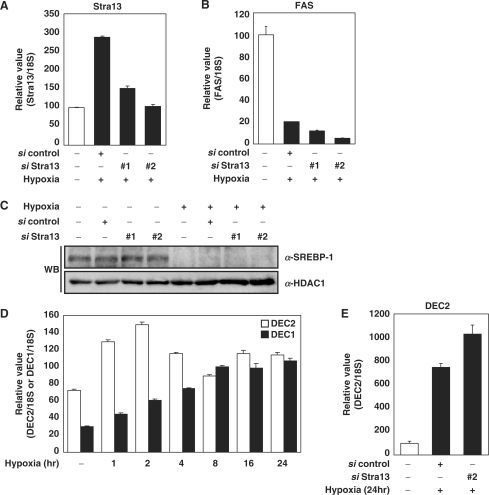
We investigated whether a transcription repressor, Stra13/DEC1 is involved in HIF-dependent repression of SREBP-1c. Northern analyses confirmed that hypoxia increases mRNA level of Stra13/DEC1 prior to maximum decrease of FAS expression (Figures 3A and 1B). By using HIF-1β defective cells, we confirmed that Stra13 induction was HIF-1-dependent (Figure 3B). Consistent with the case of SREBP-1c, in acute (4 h) hypoxic exposure, siRNA against HIF-1α reduced the hypoxic induction of Stra13, whereas in prolonged hypoxic exposure (24 h) HIF-2α siRNA was more effective than HIF-1α siRNA (Figure 3C) (35).
Figure 3.
Involvement of Stra13 on the expression of SREBP-1c. (A) Hepa1c1c7 cells were incubated in hypoxic conditions (1% O2) for the indicated times. The levels of Stra13 mRNA were analyzed by northern analysis and Q-PCR. (B) Wild-type Hepa1c1c7 cells and HIF-1β-defective Hepa1c1c7 cells were exposed to hypoxia for 16 h. The levels of mRNAs were detected by RT–PCR analysis and Q-PCR. (C) Hepa1c1c7 cells were transfected with the indicated siRNAs as described. Before harvest, the transfected cells were exposed to hypoxia (1% O2, 4 h or 24 h). The level of Stra13 mRNA was quantified by Q-PCR. Values represent means and standard deviations of three experiments.
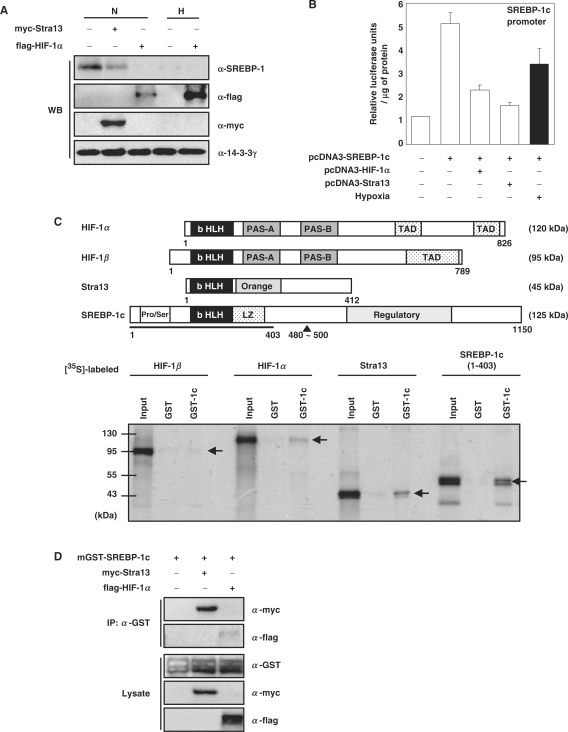
We measured SREBP-1c protein in human 293 cells transfected with either HIF-1α or Stra13. The results, shown in Figure 4A indicate that even in normoxic cells, forced expression of either HIF-1α or Stra13 reduces the amount of the endogenous SREBP-1c protein. These results indicate that either HIF-1α or Stra13 is sufficient to mediate hypoxic repression of SREBP-1c. The promoter of the mouse SREBP-1c gene contains two binding sites for Liver X Receptor (LXR), a sterol regulatory element complex (−84 to −53 in the mouse SREBP-1c gene) which consists of an E-box and SRE, and recognition sites for nuclear factor-Y (NF-Y) and Sp1 (Supplementary Figure S2A) (16,36). We transiently cotransfected a plasmid encoding SREBP-1c cDNA together with a reporter plasmid driven by the upstream regulatory region (−2700 to +1) of the mouse SREBP-1c gene. Since SREBP-1c transactivates its own promoter, overexpression of SREBP-1c increased the activity of SREBP-1c promoter (Figure 4B) (14,16). Even in the presence of excess SREBP-1c, hypoxia reduced SREBP-1c promoter activity. Cotransfection of either HIF-1α or Stra13 reduced SREBP-1c promoter activity even in normoxic cells (Figure 4B). Again, these results indicated that HIF-1α and Stra13 repress the activity of SREBP-1c promoter not only by reducing the amount of SREBP-1c, but also by reducing its activity. We also found that HIF-1α and Stra13 repress the activity of FAS promoter (Supplementary Figure S2B). In order to test whether HIF-1α and Stra13 interact with SREBP-1c, thereby preventing its activity, we used bacteria-expressed GST-SREBP-1c (amino acids 1–403) fusion protein and [35S]-labeled HIF-1α, HIF-1β, and Stra13 in GST pull-down assays. The results, shown in Figure 4C, indicate that SREBP-1c interacts with itself as a homodimer (23), that it interacts with Stra13 as strongly as it does with itself, that it also interacts with HIF-1α, though to a lesser degree, and that HIF-1β fails to interact with SREBP-1c. ChIP assays confirmed that SREBP-1c interacts with Stra13 in vivo, but scarcely with HIF-1α (Figure 4D). Our results suggest that HIF-1α induces Stra13, and then Stra13 interacts with SREBP-1c.
Figure 4.
Effect of hypoxia on the SREBP-1c promoter activity and protein–protein interactions between bHLH proteins. (A) Human 293 cells were transfected with either pCMV-myc-Stra13 or pCMV-3flag-HIF-1α. The transfected cells were incubated in hypoxia (1% O2, 6 h) before harvesting. WB analysis was performed using the indicated antibodies. WB with anti-14-3-3γ was used as loading controls. (B) The mouse SREBP-1c promoter-driven reporter plasmid (250 ng) was transfected into 5 × 104 NIH 3T3 cells together with 250 ng of the indicated plasmids and 50 ng of pCHO110 which encodes β-galactosidase. The transfected cells were incubated in hypoxia (1% O2, 16 h) before harvesting, and luciferase assays were performed as described previously (26). Numbers represent averages and standard deviations of three independent experiments. (C) Immobilized GST-SREBP-1c (amino acids 1–403, GST-1c) was incubated with [35S]-labeled in vitro transcribed and translated (IVTT) proteins for 2 h at 4°C and washed as described (25). Protein bound to the glutathione-uniflow resin with unincubated IVTT proteins (input, 10%) was subjected to SDS–PAGE and visualized by exposure to X-ray film. In the diagram above, the structure of each protein is shown schematically. bHLH: basic helix–loop–helix, TAD: transactivation domain, PAS, Per-Arnt-Sim homology; LZ, leucine zipper; Pro/Ser, proline and serine rich region. (D) pEBG-SREBP-1c which encodes GST-SREBP-1c was transfected into 293 cells together with the indicated plasmids. The transfected cell lysates (300 μg) were immunoprecipitated (IP) with resin-bound anti-GST antibody. Then the resulting immunocomplexes or total lysates (30 μg, 10% input) were analyzed by western blotting.
Interactions between bHLH proteins and SREBP-1c promoter
Next, we investigated whether either HIF-1α or Stra13 inhibits DNA binding by SREBP-1c, since each has a bHLH domain required for dimerization-dependent binding to the E-box motif (-CANNTG-). The SREBP-1c proximal promoter contains the SRE complex, which contains SRE (-ATCACCCCAC-), E-box and cis-acting elements for NF-Y and Sp1 (Figure 5B and Supplementary Figure S2A) (16). SREBP-1c is a bHLH-leucine zipper protein that has dual DNA-binding specificity; it binds not only to the E-box, but also to the SRE (14). EMSAs showed that GST-SREBP-1c binds to SRE complex-containing oligonucleotides. Addition of either Stra13 homodimer or HIF-1α/β heterodimer prevented this binding (indicated by the higher arrow in Figure 5A). Interestingly, both Stra13 homodimer and HIF-1α/β heterodimer are able to interact with the SRE complex (indicated by the lower arrowhead in Figure 5A). In order to test whether the interactions between the SREBP-1c promoter and Stra13 and HIF-1α/β are specific for E-box sequences or SRE sequences, we added an excess of unlabeled mutant oligonucleotides (Figure 5B). Addition of unlabeled SRE mutant oligonucleotides more effectively diminished both Stra13 and HIF-1α/β binding than addition of unlabeled E-box mutant oligonucleotides, indicating that the Stra13 homodimer and HIF-1α/β heterodimer interact with the SREBP-1c promoter, E-box specifically (Figure 5B). Using the ChIP technique, we confirmed these findings in vivo. The transfected GST-SREBP-1c was recruited to the endogenous chromosomal SREBP-1c promoter, and cotransfection with either myc-Stra13 or flag-HIF-1α/HIF-1β prevented SREBP-1c from binding to SREBP-1c promoter (Figure 5C). ChIP analyses showed that both Stra13 homodimer and HIF-1α/β heterodimer bound to the chromosomal promoter of their target gene, Stra13 (the lowest panel in Figure 5D–F) (21). Interestingly, the Stra13 homodimer is recruited to the endogenous SREBP-1c promoter (the upper panel of Figure 5D), whereas HIF-1α/β fails to do so (the upper panel of Figure 5E). These findings suggest that HIF-1 induces Stra13, and that then Stra13 interacts with the E-box sequence in the SREBP-1c promoter and/or with the SREBP-1c protein itself, preventing SREBP-1c from binding to its recognition site. We tested whether Stra13 directly interacts with E-box sequences in FAS promoter by using EMSA (Supplementary Figure S2C) and ChIP analyses (Figure 5F). We found that Stra13 fails to do so (the upper panel of Figure 5F). Instead, the presence of both Stra13 and HIF-1α/β prevents SREBP-1c from binding to the FAS promoter (Figure 5G and the middle panel of Figure 5F). Our findings suggest that HIF-induced Stra13 prevents SREBP-1c from binding to the FAS promoter, not by competing for binding to E-box in FAS promoter, but presumably by interacting with SREBP-1c protein.
Effects of Stra13 siRNA on hypoxic repression of SREBP-1c
In order to test the contribution of Stra13 to the hypoxic repression of SREBP-1c and FAS, siRNA against Stra13/DEC1 was transfected into Hepa1c1c7 cells. We noted a reduction of about 50% in Stra13/DEC1 mRNA and protein by two different siRNAs (Figure 6A and Supplementary Figure S3A). The results in Figure 6B and C demonstrate that Stra13 siRNA failed to recover hypoxic repression of FAS and SREBP-1c, suggesting that Stra13 is not unique repressor that mediates hypoxic repression of SREBP-1c and FAS. DEC2, an isoform of Stra13/DEC1 is identified in each mammalian species. Both Stra13/DEC1 and DEC2 are induced by hypoxia. The mRNA expression of Stra13/DEC1 gradually increased and reached the maximum after 8-h exposure to hypoxia, while that of DEC2 instantly and temporarily increased during acute hypoxic exposure (1–2 h) (Figure 6D). Li et al. (37) had showed that Stra13/DEC1 represses the expression of DEC2 through binding to E-box in DEC2 promoter. Consistently, we found that the induction of DEC2 is decreased as Stra13/DEC1 is gradually increased (Figure 6D), and that the siRNA against Stra13 increases the expression of DEC2 (Figure 6E). We also confirmed that hypoxic induction of DEC2 also depends on the HIF-1 (Supplementary Figure S3B and C) (21).
Figure 6.
Effect of siRNA against Stra13/DEC1. (A–C) Hepa1c1c7 cells were transfected with the siRNAs against Stra13/DEC1 as described. Before harvest, the transfected cells were exposed to hypoxia (1% O2, 24 h). The levels of mRNA in each sample were quantified by Q-PCR. Values represent means and standard deviations of three experiments. WB analysis was performed using anti-SREBP-1 antibody (Santa Cruz Biotechnology) and anti-HDAC1 antibody. (D) Hepa1c1c7 cells were incubated in hypoxic conditions (1% O2) for the indicated times. The levels of Stra13/DEC1 and DEC2 mRNA were analyzed by Q-PCR. The expression level of 18S rRNA was used for normalization. (E) 3T3-L1 cells were transfected with the siRNAs against Stra13/DEC1 as described. Before harvest, the transfected cells were exposed to hypoxia (1% O2, 24 h). The level of DEC2 mRNA in each sample was quantified by Q-PCR. Values represent means and standard deviations of three experiments.
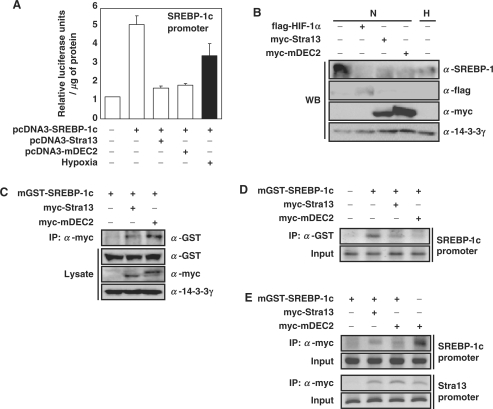
Effect of DEC2 on hypoxic repression of SREBP-1c and FAS
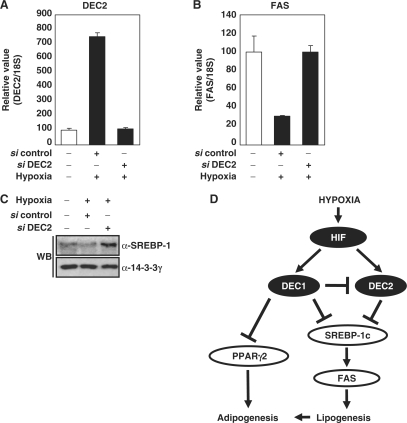
We investigated whether DEC2 also mediates the HIF-dependent repression of FAS and SREBP-1c. We confirmed that overexpression of DEC2 reduced SREBP-1c promoter activity, FAS promoter activity (Figure 7A and Supplementary Figure S3D), and the expression level of the endogenous SREBP-1c protein (Figure 7B). Similar to Stra13, ChIP assay showed that DEC2 also interacts with SREBP-1c (Figure 7C). ChIP analyses showed that DEC2 prevents GST-SREBP-1c from binding to the endogenous SREBP-1c promoter (Figure 7D), and that DEC2 homodimer is able to bind SREBP-1c promoter (upper panel) and Stra13 promoter (lower panel) through E-box (Figure 7E) (38). Our results suggest that not only Stra13 but also DEC2 prevents SREBP-1c protein from binding to its promoter by competing for binding to the E-box and/or by interacting with SREBP-1c protein. We tested whether siRNA against DEC2 can restore the hypoxic repression of FAS and SREBP-1c. We noted a reduction in DEC2 mRNA and protein by a siRNA against DEC2 (Figure 8A and Supplementary Figure S3E). The result in Figure 8B and C showed that treatment of DEC2 siRNA recovers the hypoxic repression of both FAS and SREBP-1c. Accordingly, hypoxic repression of FAS and SREBP-1c is regulated by both Stra13 and DEC2; however, DEC2 plays more pivotal role in hypoxic repression of SREBP-1c and FAS (Figure 8D).
Figure 7.
Effect of DEC2 on the repression of SREBP-1c. (A) The mouse SREBP-1c promoter-driven reporter plasmid (250 ng) and pCHO110 (50 ng) was transfected into 5 × 104 NIH 3T3 cells together with 250 ng of the indicated plasmid. The transfected cells were incubated in hypoxia (1% O2, 16 h) before harvesting, and luciferase assays were performed (26). Numbers represent averages and standard deviations of three independent experiments. (B) NIH 3T3 cells were transfected with either pCMV-myc-Stra13 or pCMV-myc-DEC2. The transfected cells were incubated in hypoxia (1% O2, 6 h) before harvesting. Immunoblot analysis was performed using the indicated antibodies. (C) pEBG-SREBP-1c which encodes GST-SREBP-1c was transfected into NIH 3T3 cells together with the indicated plasmids. The transfected cell lysates (300 μg) were immunoprecipitated (IP) with resin-bound anti-myc antibody. And the resulting immunocomplexes or total lysates (30 μg, 10% input) were analyzed by western blotting. (D and E) pEBG-SREBP-1c was transfected into 293 cells together with the indicated plasmids. Thereafter, ChIP assays were performed with the indicated antibodies as described in Materials and Methods section.
Figure 8.
Effect of siRNA against DEC2. (A–C) 3T3-L1 cells were transfected with the siRNA against DEC2 as described. Before harvest, the transfected cells were exposed to hypoxia (1% O2, 24 h). The levels of DEC2 mRNA and FAS mRNA in each sample were quantified by Q-PCR. Values represent means and standard deviations of three experiments. Immunoblot analysis was performed using anti-SREBP-1 antibody (Santa Cruz Biotechnology). WB with anti-14-3-3γ antibody was used as loading controls. (D) Schematic diagram: hypoxic repression of SREBP-1c and PPARγ2 (20) through HIF-induced Stra13/DEC1 and DEC2.
DISCUSSION
We have demonstrated that hypoxia represses the FAS and SREBP-1c genes. SREBP-1c is a major transactivator for several lipogenic enzymes, notably FAS. Since SREBP-1c transactivates its own promoter, the initial inhibition of SREBP-1c activity can trigger a positive feedback loop of SREBP-1c repression. HIF represses SREBP-1c by inducing Stra13/DEC1 and DEC2, bHLH homodimeric transcription repressors. Both Stra13 and DEC2 are also able to interact with other type of bHLH protein, including SREBP-1c. We showed that both Stra13 and DEC2 inhibit SREBP-1c-induced transcription by competing for binding to the E-box in the SREBP-1c promoter. In contrast to SREBP-1c promoter, Stra13 fails to bind to E box in the FAS promoter. Nevertheless, Stra13 prevents SREBP-1c from binding to FAS promoter (Figure 5G) (39,40). This result implies that protein–protein interaction between SREBP-1c and Stra13 also prevent SREBP-1c from binding to its target promoter. mRNA expression of DEC2 rapidly and temporarily increased in acute hypoxia, while Stra13 increased in prolonged hypoxia (Figure 6D). These expression profiles reflect the finding that Stra13 transcriptionally represses DEC2 through binding to the E-box in the DEC2 promoter, thus maintained low level of DEC2 mRNA in prolonged hypoxia (37,41). Transfection of siRNA against Stra13 failed to reverse the hypoxic repression of SREBP-1 and FAS, since knockdown of Stra13 increased the expression of DEC2 even in prolonged hypoxia, then DEC2 replaces Stra13 (Figure 6). In contrast, knockdown of DEC2 by siRNA restored the hypoxic repression of FAS and SREBP-1c, suggesting that DEC2 could be the initiator of hypoxic repression of SREBP-1c whereas Stra13 might maintain the event in prolonged hypoxia. Therefore, without initial DEC2, late-started Stra13 fails to effectively repress the SREBP-1c and FAS genes in response to hypoxia.
Supporting to this notion, this type of repressing action of Stra13/DEC1 and DEC2 is also involved in the hypoxic repression of DNA mismatch repair gene, MLH1. Forced expression of both Stra13/DEC1 and DEC2 repressed MLH1 expression. Knockdown of DEC2 by siRNA recovered the hypoxic repression of the MHL1 but knockdown of Stra13 by siRNA failed to do so, suggesting that DEC2 repress MLH1 stronger than DEC1 does (41). The functional differences between DEC1 and DEC2 are not clear. DEC2, but not DEC1, represses cholesterol 7α-hydroxylase (CYP7A), and sterol 12α-hydroxylase (CYP8B), presumably by binding to the E-boxes in their promoters. Thereby DEC2, but not DEC1 controls the circadian signals for bile acid synthesis (42).
Stra13/DEC1 and DEC2 are also involved in well-known feed-forward regulation of circadian rhythm. In mammals, the circadian clock is based on a cyclic feedback loop that includes Period (Per) and Cryptochrome (Cry) proteins. Expression of Per and Cry oscillates in phase with the day/night cycle. The Clock/Bmal1 bHLH-PAS heterodimeric transcription factor activates expression of Per and Cry genes by direct interaction with the E-boxes in their promoters. Expression of the Stra13/DEC1 and DEC2 genes is also induced by light (43). In turn, Stra13/DEC1 and DEC2 repress Clock/Bmal1-induced Per and Cry transcription through competition for the E-box and/or interaction with Bmal1 (44). Therefore, Stra13/DEC1 and DEC2 are involved in resetting the circadian clock in response to light (45). Expression of Stra13 and DEC2 in liver and fat tissues showed a strong oscillatory trend, with a peak in the light phase (46,47). In contrast both FAS and SREBP-1c increase during the dark phase and fall during the light phase (45,48). We showed above that Stra13 and DEC2 inhibit SREBP-1c in a similar manner to that in which Stra13 and DEC2 inhibits Bmal1. Based on our findings we can infer that Stra13 and DEC2 can be mediators that control the oscillation of SREBP-1c and its target in response to both light and hypoxia as a feed-forward mechanism.
Regulation of SREBP-1c activity involves interactions between bHLH proteins, and also between the E-box and these proteins. Similarly, inhibitor of DNA binding (Id), a dominant negative HLH protein, interacts with SREBP-1c and prevents it from binding to the FAS promoter (49). Upstream stimulatory factors (USF1/USF2 heterodimer), bHLH-leucine zipper transactivators, bind to the E-box in the FAS promoter and mediate insulin activation. Griffin et al. (50) showed that USF1 and SREBP-1c interact in vivo and in vitro, and synergistically activate the FAS promoter. Like USFs, the Stra13 and DEC2 homodimer can interact with the E-box, and also with SREBP-1c protein. However, in contrast to USF, it prevents SREBP-1c from binding and activating the target promoter. Interestingly, USF also interacts with Stra13, so that they inhibit each other's activity (51).
The findings that HIF is a master transcription factor of several genes involved in glycolysis, angiogenesis and metastasis, elucidate that HIF plays a pivotal role in tumor progression. Beside of glycolysis, the cancer cells also increase de novo synthesis of DNA, protein and fatty acids which are required for the cell proliferation (52,53). Treatment of tumor cells with FAS inhibitors leads to cell cycle arrest and apoptosis, suggesting that lipogenesis is essential for tumor progression (54). Tumor-associated FAS and SREBP-1c are mainly induced by a growth factor activated PI3/Akt signaling cascades which are amplified through mutations in signaling molecules such as PTEN, BCR-ABL, EGFR and HER2/neu (53,55). Our finding that HIF rather inhibits the expression of FAS contradicts with the oncological implications of HIF-α expression. However, the other important findings provided clues that neither HIF nor hypoxia promotes biosynthesis at the cellular level: (i) HIF-1 induces pyruvate dehydrogenase kinase 1 which phosphorylates and inhibits the pyruvate dehydrogenase, thereby limiting entry of pyruvate into the TCA cycle and increasing the conversion of pyruvate to lactate. This would prevent biosynthesis which relies on the availability of TCA cycle intermediates (2,4) and (ii) Lum et al. (56) showed that in hematopoietic cells hypoxia increases glycolysis but decreases lipid synthesis, and that reduction of HIF-1α expression with RNA interference rather increases lipid synthesis, cell size and rate of proliferation. In solid tumor, a growth factor-activated PI3/Akt signaling cascades trigger large increase in glycolysis, entry of carbon into TCA cycle and lipogenesis, whereas HIF-1α increases glycolysis but limits both the entry of pyruvate into TCA cycle and lipogenesis, thus prevents oxidative stress and ATP depletion (53,56). Although PI3/Akt/mTOR pathway increases the translation of HIF-1α, the HIF-1α does not gain the full transactivation activity, presumably due to the hydroxylated asparagine residue in transactivation domain (57). Our results imply that malignant cancer cells increase lipogenesis even in the presence of HIF-1α not because HIF-α itself increases FAS expression, but because the augmented PI3/Akt signaling cascades exceed HIF signaling (58). Here, our results explain a molecular mechanism by which the hypoxia-induced HIF represses lipogenesis by repressing SREBP-1c and FAS gene. By doing so, HIF reduces the ATP-consuming anabolic process prior to the actual decrease of ATP acting as feed-forward regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Basic Research Program of the Korean Science and Engineering Foundation, Korea (R200706192003 to H.P.); Brain Korea 21 Research Fellowship and a Seoul Science Fellowship (to S.M.C. and H.-J.C.). Open Access charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Pierre Chambon and Dr. Mitsuhide Noshiro for providing cDNA of Stra13 and DEC2, respectively.
REFERENCES
- 1.Hochachka PW. Defense strategies against hypoxia and hypothermia. Science. 1986;231:234–241. doi: 10.1126/science.2417316. [DOI] [PubMed] [Google Scholar]
- 2.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesener MS, Jurgensen JS, Rosenberger C, Scholze CK, Horstrup JH, Warnecke C, Mandriota S, Bechmann I, Frei UA, Pugh CW, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 8.Löfstedt T, Fredlund E, Holmquist-Mengelbier L, Pietras A, Ovenberger M, Poellinger L, Påhlman S. Hypoxia inducible factor-2alpha in cancer. Cell Cycle. 2007;6:919–926. doi: 10.4161/cc.6.8.4133. [DOI] [PubMed] [Google Scholar]
- 9.Hochachka PW, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc. Natl Acad. Sci. USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001;2:282–286. doi: 10.1093/embo-reports/kve071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Jones Voy B, Urs S, Kim S, Soltani-Bejnood M, Quigley N, Heo YR, Standridge M, Andersen B, Dhar M, et al. The human fatty acid synthase gene and de novo lipogenesis are coordinately regulated in human adipose tissue. J. Nutr. 2004;134:1032–1038. doi: 10.1093/jn/134.5.1032. [DOI] [PubMed] [Google Scholar]
- 12.Bennett MK, Lopez JM, Sanchez HB, Osborne TF. Sterol regulation of fatty acid synthase promoter. Coordinate feedback regulation of two major lipid pathways. J. Biol. Chem. 1995;270:25578–25583. doi: 10.1074/jbc.270.43.25578. [DOI] [PubMed] [Google Scholar]
- 13.Assimacopoulos-Jeannet F, Brichard S, Rencurel F, Cusin I, Jeanrenaud B. In vivo effects of hyperinsulinemia on lipogenic enzymes and glucose transporter expression in rat liver and adipose tissues. Metabolism. 1995;44:228–233. doi: 10.1016/0026-0495(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 14.Kim JB, Spotts GD, Halvorsen YD, Shih HM, Ellenberger T, Towle HC, Spiegelman BM. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 17.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosmain Y, Dif N, Berbe V, Loizon E, Rieusset J, Vidal H, Lefai E. Regulation of SREBP-1 expression and transcriptional action on HKII and FAS genes during fasting and refeeding in rat tissues. J. Lipid. Res. 2005;46:697–705. doi: 10.1194/jlr.M400261-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl Acad. Sci. USA. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 21.Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 2002;277:47014–47021. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- 22.Yamada K, Miyamoto K. Basic helix-loop-helix transcription factors, BHLHB2 and BHLHB3; their gene expressions are regulated by multiple extracellular stimuli. Front Biosci. 2005;10:3151–3171. doi: 10.2741/1772. [DOI] [PubMed] [Google Scholar]
- 23.Lee YS, Lee HH, Park J, Yoo EJ, Glackin CA, Choi YI, Jeon SH, Seong RH, Park SD, Kim JB. Twist2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c. Nucleic Acids Res. 2003;31:7165–7174. doi: 10.1093/nar/gkg934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SM, Choi KO, Park YK, Cho H, Yang EG, Park H. Clioquinol, a Cu(II)/Zn(II) chelator, inhibits both ubiquitination and asparagine hydroxylation of hypoxia-inducible factor-1alpha, leading to expression of vascular endothelial growth factor and erythropoietin in normoxic cells. J. Biol. Chem. 2006;281:34056–34063. doi: 10.1074/jbc.M603913200. [DOI] [PubMed] [Google Scholar]
- 26.Yim S, Choi SM, Choi Y, Lee N, Chung J, Park H. Insulin and hypoxia share common target genes but not the hypoxia-inducible factor-1alpha. J. Biol. Chem. 2003;278:38260–38268. doi: 10.1074/jbc.M306016200. [DOI] [PubMed] [Google Scholar]
- 27.Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat. Protoc. 2008;3:398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
- 28.McGowan MW, Artiss JD, Strandbergh DR, Zak B. A peroxidase-coupled method for the colorimetric determination of serum triglycerides. Clin. Chem. 1983;29:538–542. [PubMed] [Google Scholar]
- 29.Rangaswamy S, Penn MS, Saidel GM, Chisolm GM. Exogenous oxidized low-density lipoprotein injures and alters the barrier function of endothelium in rats in vivo. Circ. Res. 1997;80:37–44. doi: 10.1161/01.res.80.1.37. [DOI] [PubMed] [Google Scholar]
- 30.Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park MG, Steffensen KR, et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor gamma expression. Mol. Cell. Biol. 2004;24:3430–3444. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savransky V, Nanayakkara A, Vinero A, Li J, Bevans S, Smith PL, Torbenson MS, Polotsky VY. Chronic intermittent hypoxia predisposes to liver injury. Hepatology. 2007;45:1007–1013. doi: 10.1002/hep.21593. [DOI] [PubMed] [Google Scholar]
- 32.Choi SM, Park H. Aryl hydrocarbon receptor nuclear translocator is involved in ATP homeostasis in both normoxic and hypoxic monolayer mouse hepatoma cells. J. Appl. Pharmacol. 2006;14:132–136. [Google Scholar]
- 33.Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol. Cell. Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko HP, Okino ST, Ma Q, Whitlock J.P., Jr. Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol. Cell. Biol. 1996;16:430–436. doi: 10.1128/mcb.16.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmquist-Mengelbier L, Fredlund E, Löfstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 36.Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB. Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem. J. 2005;385:207–216. doi: 10.1042/BJ20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Xie M, Song X, Gragen S, Sachdeva K, Wan Y, Yan B. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J. Biol. Chem. 2003;278:16899–16907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 38.Kawamoto T, Noshiro M, Sato F, Maemura K, Takeda N, Nagai R, Iwata T, Fujimoto K, Furukawa M, Miyazaki K, et al. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem. Biophys. Res. Commun. 2004;313:117–124. doi: 10.1016/j.bbrc.2003.11.099. [DOI] [PubMed] [Google Scholar]
- 39.Magana MM, Osborne TF. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J. Biol. Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 40.Magana MM, Lin SS, Dooley KA, Osborne TF. Sterol regulation of acetyl coenzyme A carboxylase promoter requires two interdependent binding sites for sterol regulatory element binding proteins. J. Lipid Res. 1997;38:1630–1638. [PubMed] [Google Scholar]
- 41.Nakamura H, Tanimoto K, Hiyama K, Yunokawa M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E, Nishiyama M. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene. 2008;27:4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- 42.Noshiro M, Usui E, Kawamoto T, Kubo H, Fujimoto K, Furukawa M, Honma S, Makishima M, Honma K, Kato Y. Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J. Biol. Rhythms. 2007;22:299–311. doi: 10.1177/0748730407302461. [DOI] [PubMed] [Google Scholar]
- 43.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Song X, Ma Y, Liu J, Yang D, Yan B. DNA binding, but not interaction with Bmal1, is responsible for DEC1-mediated transcription regulation of the circadian gene mPer1. Biochem. J. 2004;382:895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 46.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 47.Noshiro M, Furukawa M, Honma S, Kawamoto T, Hamada T, Honma K, Kato Y. Tissue-specific disruption of rhythmic expression of Dec1 and Dec2 in clock mutant mice. J. Biol. Rhythms. 2005;20:404–418. doi: 10.1177/0748730405280195. [DOI] [PubMed] [Google Scholar]
- 48.Patel DD, Knight BL, Wiggins D, Humphreys SM, Gibbons GF. Disturbances in the normal regulation of SREBP-sensitive genes in PPAR alpha-deficient mice. J. Lipid Res. 2001;42:328–337. [PubMed] [Google Scholar]
- 49.Moldes M, Boizard M, Liepvre XL, Feve B, Dugail I, Pairault J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. Biochem. J. 1999;344:873–880. [PMC free article] [PubMed] [Google Scholar]
- 50.Griffin MJ, Wong RH, Pandya N, Sul HS. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty acid synthase promoter. J. Biol. Chem. 2006;282:5453–5467. doi: 10.1074/jbc.M610566200. [DOI] [PubMed] [Google Scholar]
- 51.Dhar M, Taneja R. Cross-regulatory interaction between Stra13 and USF results in functional antagonism. Oncogene. 2001;20:4750–4756. doi: 10.1038/sj.onc.1204637. [DOI] [PubMed] [Google Scholar]
- 52.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cnacer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 53.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Kuhajda FP, Pizer ES, Li JN, Mani NS, Frehywot GL, Townsend CA. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc. Natl Acad. Sci. USA. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, Riley JL, Thompson CB. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 2004;18:1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, Thompson CB. The transcription factor HIF-1α plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arsham AM, Plas DR, Thompson CB, Simon MC. Phosphatidylinositol 3-kinase/Akt signaling is neither required for hypoxic stabilization of HIF-1α nor sufficient for HIF-1-dependent target gene transcription. J. Biol. Chem. 2002;277:15162–15170. doi: 10.1074/jbc.M111162200. [DOI] [PubMed] [Google Scholar]
- 58.Furuta E, Pai SK, Zhan R, Bandyopadhyay S, Watabe M, Mo YY, Hirota S, Hosobe S, Tsukada T, Miura K, et al. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 2008;68:1003–1011. doi: 10.1158/0008-5472.CAN-07-2489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.