Abstract
The polymerase chain reaction (PCR) is widely used for applications which require a high level of specificity and reliability, such as genetic testing, clinical diagnostics, blood screening, forensics and biodefense. Great improvements to PCR performance have been achieved by the use of Hot Start activation strategies that aim to prevent DNA polymerase extension until more stringent, higher temperatures are reached. Herein we present a novel Hot Start activation approach in PCR where primers contain one or two thermolabile, 4-oxo-1-pentyl (OXP) phosphotriester (PTE) modification groups at 3′-terminal and 3′-penultimate internucleotide linkages. Studies demonstrated that the presence of one or more OXP PTE modifications impaired DNA polymerase primer extension at the lower temperatures that exist prior to PCR amplification. Furthermore, incubation of the OXP-modified primers at elevated temperatures was found to produce the corresponding unmodified phosphodiester (PDE) primer, which was then a suitable DNA polymerase substrate. The OXP-modified primers were tested in conventional PCR with endpoint detection, in one-step reverse transcription (RT)–PCR and in real-time PCR with SYBR Green I dye and Taqman® probe detection. When OXP-modified primers were used as substitutes for unmodified PDE primers in PCR, significant improvement was observed in the specificity and efficiency of nucleic acid target amplification.
INTRODUCTION
The polymerase chain reaction (PCR) is a powerful technique used to produce multiple copies of a nucleic acid region of interest. The ‘essential reaction components’ in a PCR cocktail include the DNA polymerase, two oligonucleotide primers, deoxynucleoside 5′-triphosphates (dNTPs), magnesium ion and other buffer components (1,2). This wide-spread technique finds utility in applications that require the amplification of a target region of interest with high specificity and accuracy (3–8). Robust PCR performance is of the utmost importance for high-sensitivity analytical PCR schemes, such as detection of single-copy DNA molecules (9), blood-borne infectious agents (5,7), biohazardous microbes (3), defective or cancerous genes (4,10,11), single nucleotide polymorphisms (10,11) and forensic samples (3,8). In addition, the need for improved PCR performance is imperative for the preparation of samples for cloning and next-generation sequencing applications (12,13).
Although primer sequence and length can be carefully designed to optimize its hybridization to only the intended target sequence at the annealing temperature, PCR amplification reactions can still be plagued by off-target amplification (14). Off-target amplifications are thought to occur during the lower temperature conditions of PCR sample preparation and thermal cycler ramping to the initial denaturation temperature. Under these less stringent conditions, the primers, which are in a large molar excess over target, can bind nonspecifically to regions of the nucleic acid target with partial complementarity or to other primer molecules (14). These nonspecific primer complexes may initiate the synthesis of undesired ‘mis-priming’ and ‘primer dimer’ extension products, respectively. As has been discussed by Chou et al. (14), mis-priming can compete with amplification of the desired target sequences, thereby significantly reducing the efficiency of the amplification of the desired sequence, especially for low copy number targets. Furthermore, primer dimers may undergo amplified oligomerization during PCR to create a complex mixture of primer artifacts, the quantity of which often varies inversely with the yield of specific PCR product in low copy number amplifications.
One widely used means of improving the specificity of PCR is to employ a Hot Start activation technique. The goal of this technique is to prevent the DNA polymerase from premature extension of primer complexes with lesser degrees of complementarity during the low stringency conditions of pre-PCR sample preparation. In Hot Start activation, primer extension is blocked until the reaction mixture reaches an elevated, Hot Start temperature, where the stringency of the primer/target hybridization is optimal for specificity, and primer complexes are dissociated. As first described, a critical component of the PCR mixture, such as magnesium chloride or DNA polymerase, was added under high stringency conditions to a ‘hot’ sample containing all other components of the PCR cocktail (15). Other noteworthy approaches include the physical separation of reaction components (14,16,17), DNA polymerase inhibition at lower temperatures (18–24), the use of accessory proteins (25–27) and modified primer constructs (28–36). As PCR-based applications continue to evolve, so does the need for approaches to Hot Start activation in PCR that allow for further improvements to performance.
Approaches that employ modified primers for improved PCR specificity have been diverse and have included competitor sequences (33,37), primers with secondary structure (28,31), modifications that improve hybridization selectivity (32,35), 3′-modifications that block primer extension until 3′–5′ exonuclease removal (29,34) and nucleobase blocking modifications that are removable by UV irradiation (36) or by thermal deprotection (30). While each of these approaches has demonstrated remarkable reductions in off-target product formation, limitations have included the lack of generality in the approach as well as the requirement for additional enzymes, special activation conditions, specific nucleoside modifications or longer and structurally complicated primers. With these considerations in mind, we sought to develop a novel primer modification that can be introduced into any oligonucleotide sequence and that can allow for primer-based Hot Start activation without any additional special conditions. Phosphotriester (PTE) oligonucleotide derivatives (38–42), members of P-modified oligonucleotide class (43–45), were considered for this approach as the most ‘activated’ phosphoester bond in the PTE fragment can be preferentially cleaved to produce oligonucleotides containing a natural phosphodiester (PDE) linkage (46–67). While earlier approaches used chemical treatment for removal, several new PTE-protecting groups have recently been described that can be introduced using solid-phase oligonucleotide synthesis, with removal at elevated temperature to generate the corresponding unmodified PDE oligonucleotide (68–75). The identification of temperature-sensitive PTE derivatives, combined with literature evidence that the presence of PTE primer modifications blocks DNA polymerase primer extension (76–78), showed great promise for utility in a Hot Start PCR approach.
Herein we will describe the design, synthesis and evaluation of modified oligonucleotide primers that contain one or two internucleotide PTE linkages at the 3′-terminus of the primer. Proof of principle studies showed significant promise for the 4-oxo-1-pentyl (OXP) group, as it displayed the desired characteristics of reduced elongation at low temperatures and rapid conversion to an extendable primer at elevated temperatures, without being too unstable for routine handling (74). We therefore sought to further explore these OXP-modified primers in a number of problematic PCR-based primer/template systems. These experiments will include investigation of primers that are prone to primer dimer formation and mis-priming using endpoint analysis and real-time measurements. Additional experiments will evaluate the detection of low copy number templates and the detection of RNA templates using a one-step reverse transcriptase PCR approach.
MATERIALS AND METHODS
Materials
The chemicals used in this study were commercially available through Acros (Geel, Belgium), Fisher Scientific (Pittsburgh, PA, USA), Sigma-Aldrich (St. Louis, MO, USA), or ChemGenes (Wilmington, MA USA). All oligonucleotide synthesis reagents were obtained from Glen Research Corporation (Sterling, VA, USA). Unmodified gene-specific PCR primers, unmodified poly-dT18 primers and probes were ordered through TriLink BioTechnologies, Inc. Synthesis of modified phosphoramidites is described in the Supplementary Data section. Primers which contained either one PTE modification to the 3′-terminal internucleotide linkage or two modifications to the 3′-terminal and penultimate internucleotide linkages were also prepared by TriLink, on an ABI Expedite 8909 DNA synthesizer using standard manufacturer-suggested procedures as detailed in the Supplementary Data section. HPLC was accomplished on a Beckman, Inc. (Fullerton, CA, USA) System Gold Nouveau Model 126 with Model 168 photodiode array detector. NMR spectra were recorded on Bruker Model AX 500 spectrometer (NuMega, San Diego, CA, USA). Electrospray mass spectrometry analyses were done by HT Laboratories (San Diego, CA, USA).
Bacteriophage λ genomic DNA was purchased from Roche Applied Science (Indianapolis, IN, USA), HIV-1 genomic DNA was a component of the Gene Amplimer kit purchased from Applied Biosystems (Foster City, CA, USA), and Human genomic DNA was purchased from Promega (Fitchburg, WI, USA). Thermus aquaticus (Taq) DNA polymerase (recombinant) and Platinum® Taq DNA Polymerases were purchased from Invitrogen (Carlsbad, CA, USA). DyNAzyme™ II Hot Start DNA polymerase and the deoxynucleotide solution set (dNTPs) were purchased from New England Biolabs (Ipswich, MA, USA). AmpliTaq Gold® DNA Polymerase was purchased from Applied Biosystems and HotStart-IT® Taq DNA polymerase was purchased from USB (Cleveland, OH, USA). SYBR Green® I nucleic acid stain was purchased from Invitrogen and passive reference ROX dye (1 mM) was purchased from Stratagene (La Jolla, CA, USA). Human Liver Total RNA was purchased from Clontech (Mountain View, CA, USA), RNase Inhibitor was purchased from Ambion (Austin, TX, USA) and Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) was purchased from Invitrogen. All endpoint PCR experiments were performed on either a Perkin Elmer GeneAmp® 9600 or a Perkin Elmer GeneAmp® 2400 thermal cycler. All real-time PCR experiments were performed using a Stratagene Mx3005P® QPCR System instrument.
Study of kinetics of conversion of OXP-modified oligonucleotides to corresponding PDE oligonucleotides in PCR buffer
Twenty-eight microliters of OXP-modified oligonucleotide solution (50 μM) was mixed with 532 μl of water and 140 μl of 5× PCR buffer [250 mM KCl, 7.5 mM MgCl2, 50 mM Tris (pH 8.4 at 25°C)] and aliquots of 95 μl each were placed in thin-walled 200 μl PCR tubes. One control tube was placed on dry ice immediately while other tubes were placed in GeneAmp 2400 PCR Thermal Cycler running at 95°C. At a specified time, the tube was removed and frozen on dry ice. Each sample was diluted with 105 μl of water and analyzed by reverse-phase HPLC on Waters (Milford, MA, USA) Micro-Bondapak C18 10 μ analytical column (3.9 × 30 mm) using a gradient of acetonitrile (Buffer B) in 100 mM TEAA (pH 7.5; Buffer A). The gradient was 0–50% of Buffer B over 40 min, 1 ml/min. On the resultant chromatogram, the HPLC peaks corresponding to OXP-modified oligonucleotide and unmodified PDE oligonucleotide were integrated at 260 nm wavelength.
Estimation of the number of PDE primer molecules generated from an OXP primer during typical PCR amplification
The estimation is based on kinetic data (Figure 3). Since the PCR is not an isothermal process, the following calculations are considered to be an approximation. A typical amount of DNA target introduced into a PCR experiment is 101–103 copies. The upper theoretical limit of amplicon molecules produced during PCR would be: 104–106 molecules after 10 cycles; 107–109 molecules after 20 cycles; 1010–1012 molecules after 30 cycles; and 1013–1015 molecules after 40 cycles. A typical initial concentration of the primers in the PCR reaction mixture is 0.1–1.0 μM, and the reaction volume is 20–50 μl. That is equal to 2–50 pmol or 1.2 × 1012 to 3.0 × 1013 molecules of each primer.
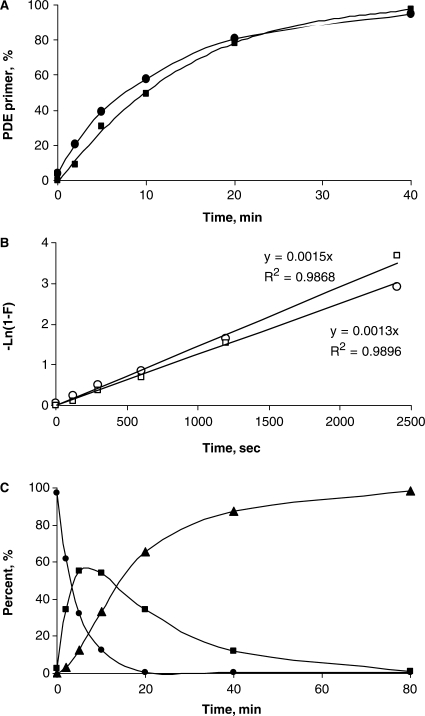
Figure 3.
Evaluation of the rate of unmodified primer formation after incubation of OXP-modified HIV-1 primers at elevated temperatures. (A) Kinetic curves of conversion of forward (filled circle) and reverse (filled square) single OXP-modified primers to corresponding PDE primers at 95°C in 1× PCR buffer (see, Figure 2). (B) A semi-logarithmic plot of kinetics of conversion of forward (open circle) and reverse (open square) single OXP-modified primers to a corresponding PDE primer at 95°C in 1× PCR buffer (see, Figure 2). (C) Kinetic curves of conversion of a double OXP-modified forward primer to the corresponding PDE primer in 1× PCR buffer at 95°C (see, Supplementary Scheme 3). Starting double OXP-modified primer (filled circle); intermediate isomeric single OXP-modified primers (filled square); and unmodified PDE primer (filled triangle) are indicated with the appropriate symbols.
If the initial amount of OXP primer would be 5 × 1012 molecules and pre-PCR denaturation step at 95°C is set for 60 s, 4 × 1011 PDE primer molecules would be generated from an OXP primer before the first PCR cycle. Note, that the number of generated PDE primer molecules is 4 × 108 to 1010 times more than the initial number of target DNA molecules and is sufficient to support PCR for ∼30 cycles of amplification. Assuming that each subsequent PCR cycle is equivalent to ∼60 s of an additional incubation at 95°C, then after 30 PCR cycles there will be ∼1013 PDE primer molecules produced. This amount is enough to support PCR amplification up to 35 cycles. Since a practical number of the amplicon molecules produced in typical amplification in PCR is limited to a factor 1010–1012 (1,2), the number of PDE primer molecules generated after 35 cycles (∼1013) will be sufficient for most PCR applications.
Primer extension experiments
Primer extension with Klenow fragment of DNA polymerase
Primer extension experiments using large fragment (Klenow) of DNA polymerase I (New England Biolabs) were performed at 25°C using the HIV-1 tat reverse primer (5′-AATACTATGGTCCACACAACTATTGCT-3′) that was unmodified or contained a single OXP modification. For each primer, a 30 μl mixture was prepared, which contained the primer at 5 μM in NEBuffer 2 (50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris–HCl, pH 7.9 at 25°C). Each mixture was split into two thin-walled 200 μl PCR tubes (15 μl each) and covered with a drop of mineral oil. The first tube of each pair was stored at 4°C for 45 min (‘no heat pretreatment’), while the second tube of each pair was incubated at 95°C for 40 min (‘with heat pretreatment’), followed by 4°C for 5 min using a GeneAmp 2400 PCR Thermal Cycler (Perkin Elmer, Waltham, MA, USA). From each of these four tubes, 14 μl was transferred to four fresh tubes that contained a 54.6 μl reaction mixture comprised of NEBuffer 2 (50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris–HCl, pH 7.9 at 25°C), 256 μM dNTPs and 1.9 μM of template oligonucleotide 1 (5′-AATCTTAGCAATAGTTGTGTGGACCATAGTATTTTTTTTT-3′). The resultant mixture was allowed to stay at 25°C for 5 min and a 9.8 μl aliquot was removed as a ‘zero time point’ and quenched on dry ice with 10.2 μl of Novex® TBE-Urea Sample Buffer (2×; Invitrogen). To start the reaction, 1.2 μl of a 0.5 U/μl dilution of DNA polymerase was added and resultant mixture was incubated at 25°C, removing 10 μl aliquots and quenching with 10 μl of Novex® TBE-Urea Sample Buffer (2×) on dry ice at time 2 min, 5 min, 13 min, 32 min and 80 min (PAGE analysis: Figure 4A and B).
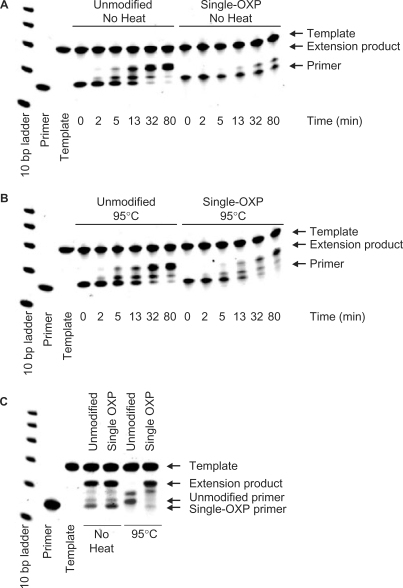
Figure 4.
PAGE analysis of primer extension experiments with single OXP-modified and PDE primers. Primer extension with Klenow fragment of DNA polymerase I of nonheated (A) and preheated (B) single OXP-modified reverse primer, respectively along template 2. The extension reactions were incubated at 25°C for the indicated times after which the reaction mixtures were quenched and analyzed. (C) Primer extension with Taq DNA polymerase of PDE and OXP forward primers (nonheated control and preheated sample) along template oligonucleotide 1. Extension reactions were incubated at 25°C for 15 min, after which the aliquots from reaction mixtures were quenched and analyzed.
Primer extension with Taq DNA polymerase
Primer extension experiments using recombinant Taq DNA polymerase (Invitrogen) were performed at 25°C using the OXP-modified forward primer and control PDE forward primer (5′-GAATTGGGTGTCAACATAGCAGAAT-3′). For each primer, a 30 μl mixture was prepared, which contained the primer at 5 μM and 1× PCR buffer [50 mM KCl, 2.5 mM MgCl2, 10 mM Tris (pH 8.4 at 25°C)]. Each mixture was split into two thin-walled 200 μl PCR tubes and covered with mineral oil. The first tube of each pair was stored at 4°C for 45 min (‘no heat pretreatment’), while the second tube was stored at 95°C for 40 min (‘with heat pretreatment’), followed by 4°C for 5 min. From each of these four tubes, 10 μl was transferred to four fresh tubes that contained a 40 μl reaction mixture comprised of 1× PCR buffer, 253 μM dNTPs, 1.9 μM template oligonucleotide (5′-TAATGCCTATTCTGCTATGTTGGCACCCAATTCTTTTTTT-3′), and 80 U of Taq polymerase. The reaction started upon addition of the pretreated primer at 25°C. At determined time intervals, 10 μl aliquots were removed and quenched with 10 μl of Novex® TBE-Urea Sample Buffer (2×) on dry ice at time 15 min, 25 min, 60 min and 125 min (PAGE analysis shown for 15 min time point only, Figure 4C).
PAGE analysis of extension products
Aliquots (5 μl) of each quenched sample were analyzed on a 10% TBE-Urea Gels (1.0 mm, 15 wells, Invitrogen), using 1× TBE running buffer which had been prewarmed to 80°C. The bromophenol blue dye was run to the bottom of the gel, and the gel was stained with SYBR® Gold Nucleic Acid Gel Stain (Invitrogen) according to the manufacturer's recommendations. An image of the stained gel was acquired using UV transillumination with an Alpha Innotech Corporation (San Leandro, CA, USA) Multi Image Light Cabinet with CCD Camera and quantified using AlphaEaseFC™ software.
Endpoint PCR experiments
Endpoint PCR protocols were set up by combining the following components in a single, thin-walled 200 μl tube. All Taq DNA polymerase reactions contained 1× PCR buffer [20 mM Tris (pH 8.4), 50 mM KCl, 2.5 mM MgCl2], 0.2 mM dNTPs and 1.25 U Taq DNA polymerase, recombinant, in a 50 μl reaction volume. Reactions that amplified a 365-bp fragment of the tat gene from HIV genomic DNA typically included 0.5 μM forward and reverse primers (5′-GAATTGGGTGTCAACATAGCAGAAT-3′ and 5′-AATACTATGGTCCACACAACTATTGCT-3′) and 0, 1, 5, 25 or 125 copies of HIV recombinant DNA (as standardized from the Gene Amplimer kit), with 10 ng of human genomic DNA as a carrier. The primers employed for these studies were either unmodified, single OXP-modified, single MAF-modified or double OXP-modified. Unless otherwise stated, thermal cycling conditions were 94°C for 10 min denaturation step, followed by 30 PCR cycles at 94°C for 30 s, 56°C for 30 s, 72°C for 1 min and final extension at 72°C for 7 min. Reactions that amplified a 653-bp region of the β-actin gene from human genomic DNA included 0.1 μM forward and reverse primers (5′-AGAGATGGCCACGGCTGCTT-3′ and 5′-ATTTGCGGTGGACGATGGAG-3′) and 0 ng, 0.1 ng, 1 ng, 10 ng or 100 ng of Human genomic DNA. The primers employed for these studies were either unmodified or double OXP-modified. Thermal cycling conditions were 94°C for 2 min denaturation step, followed by 35 PCR cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 45 s and final extension at 72°C for 7 min.
Endpoint PCR protocols that evaluated other Hot Start DNA polymerases all employed 1.25 U of DNA polymerase, five copies of HIV recombinant DNA (as standardized from the Gene Amplimer kit), 10 ng of human genomic DNA as a carrier, 0.2 mM dNTPs, in a 50 μl reaction volume. In each reaction, a 365-bp fragment of the tat gene from HIV recombinant DNA was amplified by included 0.5 μM forward and reverse primers (5′-GAATTGGGTGTCAACATAGCAGAAT-3′ and 5′-AATACTATGGTCCACACAACTATTGCT-3′) that were either unmodified, single OXP-modified or double OXP-modified. Reactions containing Platinum® Taq DNA Polymerase employed the same 1× PCR buffer as was employed for Taq DNA polymerase [20 mM Tris (pH 8.4), 50 mM KCl, 2.5 mM MgCl2]. The 1× buffer for reactions with DyNAzyme™ II Hot Start DNA polymerase included 15 mM Tris–HCl (pH 8.2), 30 mM KCl, 5 mM (NH4)2SO4, 2.5 mM MgCl2 and 0.02% BSA. Reactions employing AmpliTaq Gold® DNA Polymerase employed 1× Buffer II [50 mM KCl and 10 mM Tris–HCl (pH 8.3) and 2.5 mM MgCl2]. The buffer for HotStart-IT® Taq DNA polymerase included 10 mM Tris–HCl (pH 8.6), 50 mM KCl and 1.5 mM MgCl2. The thermal cycling protocol employed was the same as above: 94°C for 10 min denaturation step, followed by 35 PCR cycles (94°C for 40 s, 56°C for 30 s and 72°C for 60 s) and a final extension at 72°C for 7 min.
Endpoint PCR experiments were performed on either a GeneAmp 2400 or a GeneAmp 9600 PCR Thermal Cycler. After PCR, 20 μl of each sample was loaded on 4% agarose E-gel cartridge (Invitrogen) and run for 25 min according to the manufacturer's suggested protocol. An image of the gel was acquired using UV transillumination with an Alpha Innotech Corporation Multi Image Light Cabinet with CCD Camera and quantified using AlphaEaseFC™ software.
Real-time PCR experiments, with SYBR green detection
A quantitative real-time protocol was used in which the components were combined in a single, thin-walled 200 μl tube. Reaction conditions were 1× PCR buffer [20 mM Tris (pH 8.4), 50 mM KCl and 2.5 mM MgCl2], 0.5 μM gene-specific PCR primers (5′-GAATTGGGTGTCAACATAGCAGAAT-3′ and 5′-AATACTATGGTCCACACAACTATTGCT-3′), 0.2 mM dNTPs, 0.15× SYBR Green® I nucleic acid stain, 30 nM passive reference ROX dye, 0, 1, 5, 25 or 125 copies of HIV recombinant DNA (as standardized from the Gene Amplimer kit), 10 ng of human genomic DNA and 1.25 U Taq DNA polymerase (recombinant), in a 25 μl reaction volume. Thermal cycling conditions were 95°C for a 10 min denaturation step, followed by 40 PCR cycles (95°C for 40 s, 56°C for 30 s and 72°C for 1 min). The primers employed for these studies were either unmodified, single OXP-modified or double OXP-modified. Each template concentration was set up in quadruplicate, and reactions were performed in a Stratagene Mx3005P® QPCR System instrument.
Real-time PCR experiments, with TaqMan® probe detection
A quantitative real-time protocol was used in which the components were combined in a single, thin-walled 200 μl tube. Reaction conditions included 1× PCR buffer [20 mM Tris (pH 8.4), 50 mM KCl and 2.5 mM MgCl2], 0.2 mM dNTPs, and 30 nM passive reference ROX dye, and 1.25 U Taq DNA polymerase (recombinant) in a 25 μl reaction volume. The 533-bp region of the DNA packaging protein of enterobacteria phage λ was amplified with 0.5 μM gene-specific PCR primers (5′-CAGGAGCTGGACTTTACTGATGC-3′ and 5′-CGGGATATCGACATTTCTGCACC-3′), 0.1 μM TaqMan® probe (5′-6-FAM-TCTGTTCATCGTCGTGGCGGCCCA-BHQ1-3′) and 0, 10, 50, 100, 500, 1000, 5000 and 10 000 copies of λ genomic DNA. Thermal cycling conditions were 95°C for 10 min denaturation step, followed by 40 PCR cycles at 95°C for 40 s, 56°C for 30 s and 72°C for 2 min. The primers employed for these studies were either unmodified or double OXP-modified. For each primer/template target, each template concentration was set up in quadruplicate, and reactions were performed in a Stratagene Mx3005P® QPCR System instrument.
One-step reverse transcription–PCR
A one-step reverse transcription (RT)–PCR protocol was used in which the components were combined in a single tube. Reaction conditions were 1× PCR buffer [20 mM Tris (pH 8.4), 50 mM KCl and 1.5 mM MgCl2), gene-specific PCR primers (0.5 μM), poly-dT18 primer (1 μM), 0.16 mM dNTPs, 0.25 μg of Human Liver Total RNA, 10 U RNase Inhibitor, 25 U M-MLV RT and 0.3 U Taq DNA polymerase (recombinant) in a 25 μl reaction volume. Thermal cycling conditions were 42°C for 30 min (RT step), 95°C for 10 min inactivation and denaturation step, followed by 30 PCR cycles at 95°C for 30 s, 60°C for 30 s, 72°C for 30 s and final extension at 72°C for 5 min. Gene-specific PCR primers for the PBGD, ABCA1 and β-actin genes (79,80) were prepared as unmodified, single OXP-modified and double OXP-modified. In particular, the PCR primer sequences for the 205-bp PBGD amplicon (5′-GAGTGATTCGCGTGGGTACC-3′ and 5′-GGCTCCGATGGTGAAGCC-3′), the 264-bp ABCA1 amplicon (5′-GCACTGAGGAAGATGCTGAAA-3′ and 5′-AGTTCCTGGAAGGTCTTGTTCAC-3′) and the 446-bp β-actin amplicon (5′-AGAGATGGCCACGGCTGCTT-3′ and 5′-ATTTGCGGTGGACGATGGAG-3′) were prepared. One-step RT–PCR experiments were performed on either a GeneAmp 2400 or a GeneAmp 9600 PCR Thermal Cycler. After PCR, 20 μl of each sample was loaded on 4% agarose E-gel cartridge (Invitrogen) and run for 25 min according to the manufacturer's suggested protocol. An image of the gel was acquired using UV transillumination with an Alpha Innotech Corporation Multi Image Light Cabinet with CCD Camera and quantified using AlphaEaseFC™ software.
RESULTS
In these studies, thermolabile PTE primer modifications were evaluated for their ability to improve the performance of problematic PCR systems. Experiments compared the performance of unmodified PDE primers to primers containing either one or two PTE primer modifications at the 3′-terminal and/or penultimate PDE linkage(s). These modifications were evaluated for their ability to reduce off-target amplifications by providing a Hot Start activation step to PCR (Figure 1A). Herein, we will investigate whether the presence of one or more PTE modifications will significantly reduce DNA polymerase-mediated primer extension at the lower, less stringent temperatures during the reaction set-up process, while allowing for efficient ‘Hot Start’ activation at elevated temperature by conversion to the corresponding unmodified primer. Further studies on the leading PTE-modified primers will evaluate several primer/template systems prone to primer dimer formation and mis-priming, with endpoint or real-time detection.
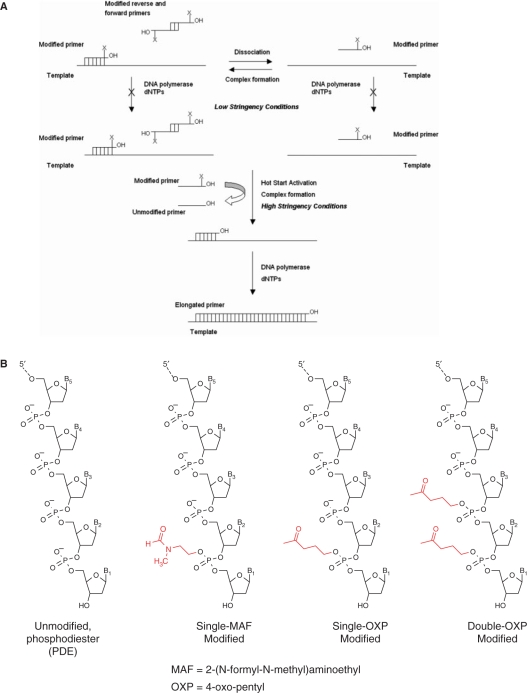
Figure 1.
(A) Possible Hot Start activation mechanism for PCR amplifications employing PTE-modified primers. X indicates the thermolabile PTE modification group. (B) PTE modifications examined in these studies.
Synthesis of OXP and MAF phosphoramidites and modified primers
To test the hypothesis that PTE-modified oligonucleotides can be converted to the corresponding unmodified PDE oligonucleotides at elevated temperatures in PCR buffer, two candidate thermolabile PTE modification groups were identified from a systematic analysis of published data: OXP and 2-(N-formyl-N-methyl)aminoethyl (MAF) (Figure 1B) (46–75). Details of synthesis of the OXP- and MAF-modified 2′-deoxynucleoside 3′-phosphoramidites of dA, dG, dC and dT are described in Supplementary Data section. The phenoxyacetyl group was used for protection of the exocyclic amino group of dA, dC and dG. The 5′-DMT-protected OXP-modified nucleoside 3′-phosphoramidites were isolated by silica gel chromatography in 50–70% overall yields. These phosphoramidites were stable for a week as 0.1 M acetonitrile solutions and were shown to be as stable as any conventional nucleoside phosphoramidite at −20°C or −70°C. A similar route was used to prepare MAF-modified 3′-phosphoramidites. Although faster deprotecting PTE groups, such as 3-(N-tert-butylcarboxamido)-1-propyl (TBCA) and 4-methylthio-1-butyl (MTB) (72,75) were examined, their low stability during preparation and isolation limited their development.
Several pairs of forward and reverse primers with OXP and MAF modifications as well as the corresponding control PDE primers were synthesized. The modified primer pairs each contained either a single OXP or a single MAF PTE group at the last 3′-internucleotide linkage of primer. Other primer pairs contained double OXP modifications, with two consecutive OXP groups at terminal and penultimate 3′-internucleotide linkages of the primer. The incorporation of PTE group in the primer results in a new chiral center and leads to the formation of diastereomers. From HPLC analysis it was evident that diastereomeric forms of single OXP-modified primers (and MAF primers, not shown) are separable at conditions used, while those for double OXP-modified primers were not (Figure 2). The isolated and desalted PTE primers were shown to be stable for at least 6 months when kept as a frozen solution at −20°C. Subsequent studies have shown that they are stable for over a year in DMSO at −20°C (data not shown).
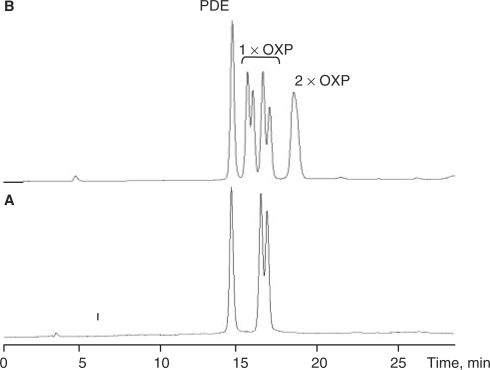
Figure 2.
Conversion of OXP-modified primers to the corresponding PDE primer. (A) Reverse phase HPLC analysis of a single OXP-modified primer after 5 min incubation at 95°C in 50 mM KCl, 1.5 mM MgCl2 and 10 mM Tris (pH 8.4 at 25°C). (B) Reverse phase HPLC analysis of the double OXP-modified primer after 8 min incubation at 95°C in 50 mM KCl, 1.5 mM MgCl2 and 10 mM Tris (pH 8.4 at 25°C). Peaks which are due to primers containing a single OXP modification are labeled 1× OXP, while peaks with two OXP modifications are labeled 2× OXP.
Kinetics of conversion of PTE primers to PDE primers
Using HPLC analysis, the kinetics of conversion of single OXP-modified primers to the corresponding unmodified PDE primers was investigated at 95°C in PCR buffer (pH 8.4 at 25°C). A 40-min incubation resulted in nearly complete conversion of OXP-modified primer to PDE primer (Figure 3A). The half-conversion time was determined to be 8.5 ± 1.5 min, with no significant difference in rate for both diastereomeric forms of the OXP primer. Detailed analysis of the kinetics for both reverse and forward HIV-1 primers revealed that OXP to PDE conversion follows a first (or pseudo-first)-order process with a rate constant K = (1.4 ± 0.1) × 10−3 s–1 (Figure 3A and B). The observed first-order kinetics is consistent with the intramolecular fragmentation mechanism proposed by Beaucage and coworkers (74).
The kinetics of conversion of the double OXP-modified primer to the corresponding unmodified PDE primer is more complicated (Supplementary Scheme 3, Figures 2B and 3C), as it requires two sequential OXP-modification removal steps. When the first OXP modification is removed from a double OXP-modified primer, two isomeric forms of a single OXP-modified primer can be formed as intermediates—one has a single OXP modification at the 3′-terminal internucleotide linkage modification, while the other has a single OXP modification at the 3′-penultimate internucleotide position. Both of these intermediates convert to unmodified PDE primer in a second OXP removal step. HPLC analysis of the mixture at an intermediate stage indicated the presence of all four compounds (unmodified, two single OXP-modified isomers and double OXP-modified), with partial separation of respective diastereomers (Figure 2B). Due to the more complicated route, the rate of formation of PDE primer from double OXP-modified primer was delayed compared to the conversion rate of single OXP-modified primer to PDE primer. Incubation at 95°C for 1–1.5 h (t½ = ∼15 min) was needed to completely convert the double OXP-modified primer to PDE primer. The forward and reverse primers with a single MAF modification at the 3′-terminal internucleotide linkage were also tested for conversion to corresponding PDE primers and found to have a half time of ∼20 min in PCR buffer at 95°C.
Extension of PTE primers by DNA polymerases
The ability of Klenow fragment of DNA polymerase I and Taq DNA polymerase to perform template-dependent extension and elongation of the HIV-1 single OXP-modified primers was investigated. Due to the possibility of spontaneous conversion of OXP primers into the corresponding PDE primers at elevated temperatures, all elongations were performed at room temperature. Under these reaction conditions, it was found that a standard PDE primer can be elongated to a full-length extension product, while the corresponding OXP primer was poorly extendable by Klenow fragment of DNA polymerase I (Figure 4A). The appearance of a small amount of truncated extension product for the OXP primer may be attributed to a selective (although inefficient) single-nucleotide extension of one of the two diastereomeric forms of OXP primer that were present in the mixture. After partial extension to an (n + 1) product, no further primer extension was observed. When the OXP-modified primer was converted to the PDE primer by heating at 95°C for 40 min, primer extension was no longer hindered by the presence of the OXP group and its extension became equal to that of a control PDE primer (Figure 4B). As anticipated, the unheated OXP-modified primer was found to migrate on a gel more slowly than the corresponding PDE primer and heat-deprotected OXP primer.
Although the detailed extension experiments were performed with the mesophilic Klenow fragment of DNA polymerase I, similar results were obtained with the thermophilic Taq DNA polymerase suitable for PCR (Figure 4C). To accommodate for the slower polymerization efficacy of Taq DNA polymerase at room temperature, we used a ∼60-fold higher concentration of the polymerase compared to the standard PCR conditions. Nevertheless, no extension of the primer to a full-length product was observed unless the OXP primer was preheated at 95°C for 40 min.
The double OXP-modified primers were also tested in the same DNA polymerase-mediated primer extension reactions. Incubation of double modified OXP forward primer with Klenow fragment of DNA polymerase I at conditions identical to those in Figure 4 showed no detectable primer extension. As was the case with the single OXP-modified primer, when double OXP-modified primer was preincubated in PCR buffer at 95°C for 40 min, the extension reaction resulted in predominant formation of full-length extension product, as was seen for the control PDE primer (data not shown).
Performance of single PTE-modified primers in HIV-1 PCR system that is prone to primer dimer formation—comparison of OXP and MAF
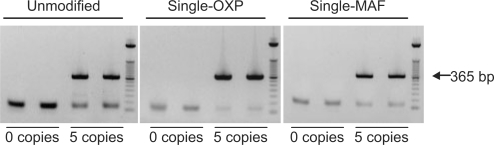
A well-characterized amplification system described by Chou et al. (14) to be prone to primer dimer formation was selected to test the utility of single PTE-modified primers. We first compared the performance of unmodified primers, single OXP-modified primers and single MAF-modified primers by performing 40 cycles of PCR in the presence or absence of five copies of HIV-1 recombinant DNA template (Figure 5). Agarose gel analysis of the resultant amplification products revealed that the presence of PTE modifications significantly improved reaction performance by reducing the amount of primer dimer formation. Furthermore, data indicated a comparable but slightly better performance for OXP primers compared to MAF primers. Under the conditions used, the ratios of amplicon to primer dimer formed were found to be 5.6 (PDE primers), 22.8 (OXP primers) and 13.5 (MAF primers) after 40 PCR cycles. Therefore, the OXP primer modification was identified as the lead modification for further investigation.
Figure 5.
Agarose gel analysis of PCR amplification of 365-bp fragment of HIV-1 DNA template using PDE, OXP and MAF primer sets at concentration of 0.5 μM, volume of 25 μl. Lanes 1–4 contain unmodified PDE primers, where lanes 1 and 2 are nontemplate control (NTC) and lanes 3 and 4 have five copies of HIV-1 recombinant DNA. Lanes 5–8 contain single OXP-modified primers, where lanes 5 and 6 are NTC and lanes 7 and 8 have five copies of HIV-1 recombinant DNA. Lanes 9–12 contain single MAF-modified primers, where lanes 9 and 10 are NTC and lanes 11 and 12 have five copies of HIV-1 recombinant DNA. The 50-bp ladder is loaded after lanes 4, 8 and 12. Thermal cycling parameters: 95°C (10 min), 40 cycles of [95°C (40 s), 56°C (30 s) and 72°C (2 min)].
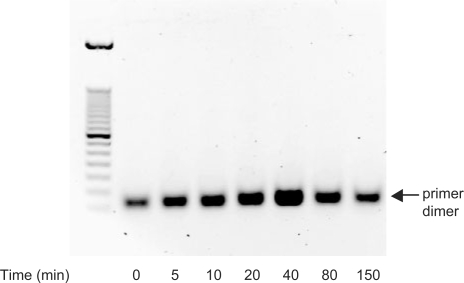
To explore the effect of the OXP group on primer dimer formation, PCR conditions that generate a high yield of primer dimer in the absence of template were evaluated. For the HIV-1 single OXP-modified primer pair, a primer concentration of 4.5 μM, approximately 5- to 20-fold higher than in a typical PCR procedure (Figure 6), was chosen, since robust primer dimer formation occurred with unmodified PDE primers. In these studies, the OXP primers were preincubated at 95°C for increasing the time intervals in order to prepare mixtures containing different proportions of PTE and PDE primers under simulated PCR conditions. The preheated mixtures were cooled and employed in the subsequent PCR experiment using Taq DNA polymerase. When the OXP primer did not undergo any preheating, only a small amount of primer dimer was observed after 30 cycles of PCR amplification (Figure 6, lane 2). This observation was consistent with a small level (<1%) of contamination of the starting OXP primers with PDE primers. As the length of preheating of OXP primer at 95°C was increased, a corresponding increase in the amount of the primer dimer accumulated in PCR was observed (Figure 6, lanes 3–6). For preheating times of 0–40 min, the yield of primer dimer was proportional to the time of the preheating, and it correlated with the kinetics of conversion of the OXP primers to the PDE primers at 95°C (Figure 3). However, when the OXP primers underwent an extensive preheating treatment (Figure 6; 80 min and 150 min, lanes 7 and 8, respectively), the yield of the primer dimer decreased. This was likely a consequence of possible primer degradation by depurination at high temperatures (81). Overall, in the absence of primer preheating, the accumulation of primer dimer in nontemplate PCR system was very low with the use of OXP primers and shows promise for use in PCR.
Figure 6.
Dependence of pre-PCR heating of PTE on primer dimer accumulation. Agarose gel analysis of primer dimer accumulation with preheated single OXP-modified primers in nontemplate system. Mixture of both primers was preheated at 95°C in 1× PCR buffer (pH 8.4 at 25°C) for increasing amounts of time. Samples were cooled on ice water, the Taq DNA polymerase was added followed by PCR amplification. PCR cycle parameters: 95°C (2 min); 30 cycles of [95°C (40 s); 56°C (30 s); and 72°C (2 min)]; 72°C (7 min). Primer dimer amplicon, indicated on the gel image, ran as a 50–80 bp DNA fragment (left lane: 50-bp ladder).
Evaluation of the effect of multiple thermolabile OXP primer modifications in a primer/template system that is prone to primer dimer formation
Since a comparison of different PTE modification groups revealed that the single OXP-modified primers showed great promise as the lead modification for improved PCR performance, further studies were conducted in which the effect of two OXP modifications on PCR performance was ascertained. Unmodified, single OXP-modified and double OXP-modified PCR primers were evaluated for their ability to amplify a 365-bp fragment of the HIV-1 tat gene (14). In all PCR experiments, reactions containing 0, 1, 5, 25 or 125 copies of HIV-1 recombinant DNA were amplified by Taq DNA polymerase, with analysis by agarose gel electrophoresis or real-time PCR.
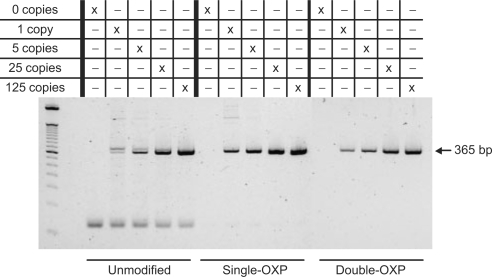
In the first experiment (Figure 7), 35 thermal cycles of amplification were performed, where the amount of product and primer dimer formed for four different amounts of input template was analyzed by agarose gel electrophoresis. When unmodified primers were employed, primer dimers were found to form efficiently over the entire range of template amounts. Furthermore, a double amplicon band was discovered when using unmodified primers. This is most likely due to the formation of the 405-bp mis-priming product, as described for this primer/template system by Chou et al. (14). When a single OXP PTE modification was introduced onto the 3′-terminal internucleotide linkage of each primer, a significant improvement in PCR performance was observed. In particular, a considerable drop in primer dimer formation was evident, with a corresponding increase in amplicon formation relative to reactions employing unmodified primers. Furthermore, formation of the double amplicon band was not observed. When two OXP PTE primer modifications were introduced, successful amplicon formation was evident, with no detectable presence of primer dimer or other off-target amplification products. Although the reaction specificity is significantly improved, a decrease in amplicon yield in comparison to the single OXP-modified primers was observed. This is likely due to the slower rate of formation of PDE primer from the double OXP-modified primers, which may result in decreased availability of viable (unmodified) primer during PCR, thereby lowering the efficiency of amplicon formation. This reduction in yield was not sustained when thermal cycling was increased to 40 cycles, as the final yield in amplicon formation was equivalent to that of the single OXP-modified primers (see Supplementary Figure S1).
Figure 7.
Agarose gel analysis of the PCR products resulting from the amplification of a 365-bp fragment from the HIV-1 tat gene using 0.5 μM unmodified, single OXP-modified and double OXP-modified primers. Reactions contained 10 ng of human genomic DNA and 0, 1, 5, 25 or 125 copies of HIV recombinant DNA as calibrated from the Gene Amplimer kit.
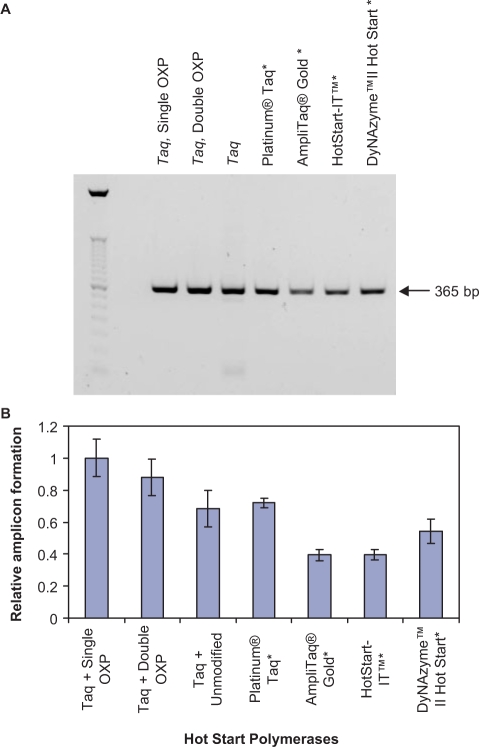
In the second set of endpoint PCR experiments, the performance of primers containing one or two OXP PTE modifications was compared to other Hot Start DNA polymerases for their ability to amplify a 365-bp fragment of the tat gene of HIV-1 recombinant DNA. Thirty-five thermal cycles of amplification were performed, where the amount of product and primer dimer formed for reactions containing five copies of input template and 1.25 U of DNA polymerase was analyzed by agarose gel electrophoresis (Figure 8A). Reactions containing single and double OXP-modified primers were amplified using Taq DNA polymerase. Reactions containing unmodified primers were amplified by Taq DNA polymerase or one of the following commercially available Hot Start DNA polymerases: Platinum® Taq DNA Polymerase, AmpliTaq Gold® DNA Polymerase, HotStart-IT™ Taq DNA Polymerase and DyNazyme ™ II Hot Start DNA polymerase. For all Hot Start technologies evaluated, each was able to suppress primer dimer formation, with the only reaction that formed detectable amounts of primer dimer was the one containing unmodified primers and Taq DNA polymerase. To determine how significant the observed variations in amplicon yield for each of the examined Hot Start technologies were, the amplicon density for three independent experiments was averaged and plotted in a bar graph, with all results normalized to single OXP-modified primers plus Taq DNA polymerase (Figure 8B). Reactions containing single OXP-modified and double OXP-modified primers formed high-amplicon yields. The reactions containing Platinum® Taq DNA Polymerase formed comparable yields to that of unmodified primers plus Taq or DyNazyme™ II Hot Start DNA polymerase, with reactions containing AmpliTaq Gold® DNA Polymerase and HotStart-IT™ Taq DNA polymerase presented the lowest yields. In summary, all Hot Start approaches provided adequate suppression of primer dimer formation, with amplicon yields varying by a factor of two. The most robust amplicon formation was evident for single and double OXP-modified primers.
Figure 8.
Comparison of the performance of OXP-modified primers to other Hot Start DNA polymerases. (A) Agarose gel analysis of the PCR products resulting from the 35 thermal cycles of amplification of five copies of a 365-bp fragment from the HIV-1 tat gene using 0.5 μM unmodified, single OXP-modified and double OXP-modified primers. Reactions containing unmodified primers were amplified by Taq DNA polymerase, Platinum® Taq DNA Polymerase, AmpliTaq Gold® DNA Polymerase, HotStart-IT™ Taq DNA Polymerase and DyNazyme™ II Hot Start DNA Polymerase. Reactions containing single and double OXP-modified primers were amplified by Taq DNA polymerase. (B) Graphical representation of PCR amplicon yield. The results from triplicate experiments were averaged and are normalized to the yield of reactions containing single OXP-modified primers plus Taq DNA polymerase. Error bars represent the SD. (*), indicates Hot Start DNA polymerases.
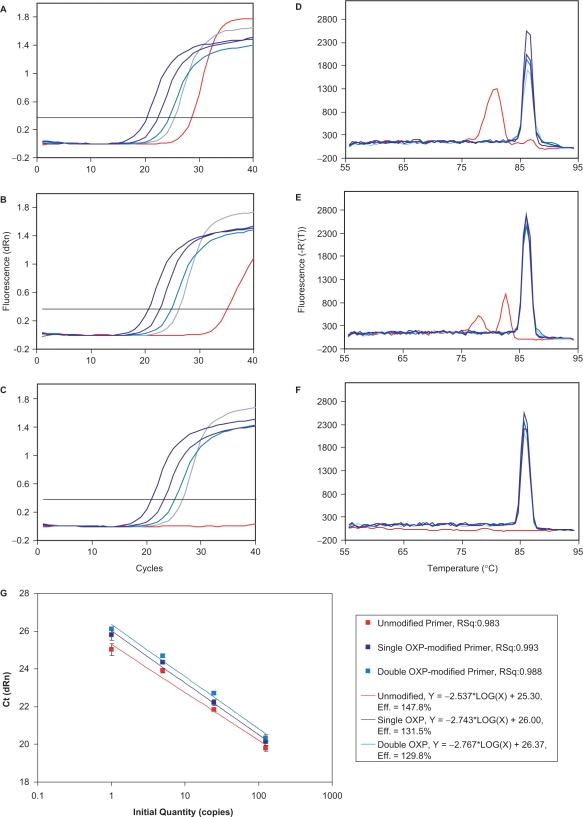
In the third set of experiments, unmodified, single OXP-modified and double OXP-modified primers were evaluated in real-time PCR, with detection by the commonly employed intercalating dye, SYBR Green®. The nonspecific binding of SYBR Green® to double-stranded DNA allowed for detection of both amplicon and off-target products during the thermal cycling process (82,83). These experiments were set-up in quadruplicate, using the same set-up as the endpoint experiments, with the exception that 40 thermal cycles were performed. When unmodified primers were employed (Figure 9A and D), all four input template concentrations were detected in real-time, with Cts (threshold cycles) ranging from 20 to 23 calculated from the sigmoidal amplification plots. However, reactions containing no template (NTC) had a significant Ct of 28.3. The corresponding dissociation curves demonstrated amplicon formation for reactions containing template (87°C) and primer dimer formation for reactions performed in the absence of template (81°C). When single OXP-modified primers were employed (Figure 9B and E), all four templates were detected with similar efficiency to the unmodified primers. The NTC reactions had a marked reduction in primer dimer formation, which was evidenced by a much delayed Ct of 36.7. The dissociation curves revealed clean amplicon formation for all plus-template reactions (87°C) and much reduced primer dimer formation. When double OXP-modified primers were evaluated (Figure 9C and F), all four plus-template reactions displayed robust amplification plots, with a slight delay in Ct relative to the unmodified and single OXP-modified primers. One notable occurrence was that the double OXP-modified primers had no measurable Ct for the NTC reactions and gave very slight indication of primer dimer formation (melting temperature of 79°C in the dissociation curve).
Figure 9.
Real-time PCR analysis of the formation of a 365-bp amplicon of the HIV-1 tat gene using detection by SYBR Green® I nucleic acid stain. Reactions, which contained 10 ng of human genomic DNA and 0, 1, 5, 25 or 125 copies of HIV recombinant DNA as calibrated from the Gene Amplimer kit, were performed in quadruplicate and employed 0.5 μM unmodified, single OXP-modified and double OXP-modified primers. This figure displays amplification plots: (A) unmodified primers; (B) single OXP-modified primers; and (C) double OXP-modified primers; dissociation curves: (D) unmodified primers; (E) single OXP-modified primers; and (F) double OXP-modified and a standard curve (G).
When the performance of the three different types of primer was evaluated by plotting the Ct values versus a known amount of input template in a standard curve (Figure 9G), each displayed good linearity over the high end of the template concentration range (5–125 copies). On the lower end of the template concentration range, the Ct values deviated from linearity, with earlier Ct values than those expected from extrapolation of the trend line. Presumably, the earlier Ct for these reactions is due to the detection of both primer dimer and amplicon formation, as primer dimers form most efficiently when the DNA template is limiting (as seen for Figure 7). For each of the OXP-modified primers, the lowest copy number amplifications were closer to linearity than the corresponding unmodified primers as determined by the R2-value of the curve fit. The linearity and lower efficiency values are indicative of the improved specificity of amplicon formation. Although the Cts were delayed for both the single and double OXP-modified primers, they were all within one Ct of one another and will result in no significant delay in data acquisition. More importantly, the improved quality of the data generated with the modified primers suggests that implementation of OXP-modified primers will allow for better estimation of copy numbers in unknown samples by significantly reducing false positives.
Evaluation of thermolabile primer modifications in a primer/template system that is prone to mis-priming
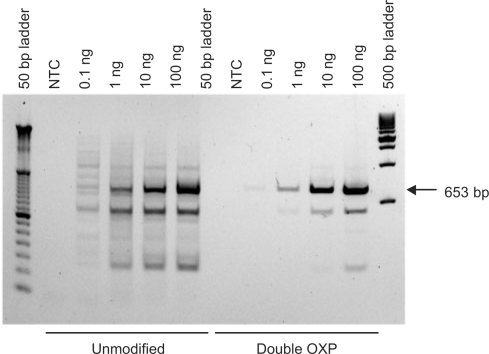
Mis-priming is the result of unintended primer binding to one or more off-target regions of a DNA template resulting in spurious amplification (14). These off-target amplification products are problematic, as they reduce the efficiency of formation of the amplicon of interest by competing for key components within the reaction [namely, primer, DNA polymerase and deoxyribonucleotide 5′-triphosphates (dNTPs)], and may give rise to false positives, depending on the method of detection. In these studies, a problematic primer/template system, which is targeted to a 653-bp fragment of the β-actin gene from human genomic DNA, was selected for evaluation. When this target is amplified using unmodified primers, multiple bands were formed, with sizes ranging from 150 bp to >1000 bp (80). An initial evaluation of unmodified, single OXP-modified and double OXP-modified primers revealed that the presence of two OXP PTE primer modifications provided the greatest benefit in improving the specificity of amplification (Supplementary Figure S2). Further investigations were performed by comparing the performance of unmodified and double OXP-modified primers in PCR, using endpoint analysis as a measure of reaction performance. In these studies, input amounts of genomic DNA ranging from 0 to 100 ng were evaluated (Figure 10). When unmodified primers were utilized, reactions employing 0.1 ng of template displayed low efficiency and the highest degree of nonspecificity, with at least 12 additional off-target amplification products evident by agarose gel analysis. As the amount of input template was increased, so did the formation of the desired amplicon. However, high levels of off-target amplification products still formed, and the amplification products ran as smears on the gel. In contrast, when the double OXP-modified primers were tested, the 653-bp amplicon formed with much higher specificity for all template concentrations, without the smearing that was observed for reactions containing the unmodified primers. Although off-target amplicon was not entirely suppressed, this significant improvement will be beneficial for use in downstream applications, such as cloning and sequencing.
Figure 10.
Amplification of a 653-bp fragment of the β-actin gene of human genomic DNA using unmodified and double OXP-modified primers. Reactions contained 0 ng, 0.1 ng, 1 ng, 10 ng or 100 ng of human genomic DNA.
Evaluation of thermolabile primer modifications for improved detection of a low copy number target
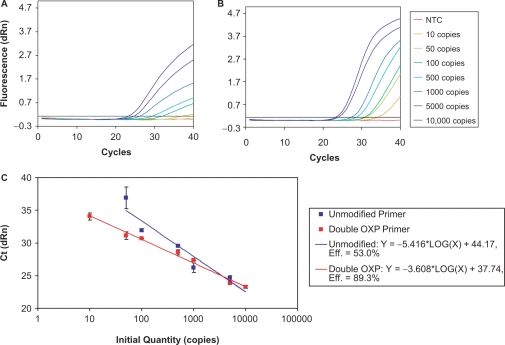
PCR-based detection of low copy number samples can be compromised by competing amplification of nontarget sequences, which can have a significant effect on the specificity and sensitivity of detection. A 533-bp fragment of the DNA packaging protein of enterobacteria phage λ was selected for evaluation as it is prone to off-target amplifications, especially for low amounts of input genomic DNA. Using real-time TaqMan® probe detection, unmodified and double OXP-modified primers were compared for their ability to amplify 0–10 000 copies of input genomic DNA (see Supplementary Figure S3 for performance of single OXP-modified primers). When unmodified primers were employed (Figure 11A), reactions containing between 100 and 10 000 copies of λ genomic DNA showed robust detection of amplicon formation. However, amplicon formation for reactions containing 50 copies was barely detectable in 40 cycles. Furthermore, the amplification curve was low in amplitude and indicative of low amplification efficiency (84). When primers containing two OXP modifications were employed (Figure 11B), amplification products were detected for all seven amounts of input template, including 10 copies. In addition, the amplification plots displayed much higher fluorescence amplitudes and were indicative of a much more efficient reaction. When the results from these studies were plotted in a standard curve (Figure 11C), the data from the double OXP-modified primers followed a linear trend for all template concentrations examined. In contrast, the unmodified primers displayed a linear trend between 100 and 10 000 copies, with the 50 copy sample significantly deviating from the line and the 10 copy sample being undetectable over the course of the experiment. At the higher template concentrations, both unmodified and double OXP-modified primers detected the amplicon with similar efficiency (as determined by Ct values). However, the poor detection of amplicon for the lower copy number samples with the unmodified primers is likely due to predominating amplification of off-target regions of the template. These findings indicate the promise of primers with thermolabile PTE modifications for improving the lower limit of detection of difficult primer/template targets.
Figure 11.
Real-time PCR detection of the amplification of a 533-bp fragment of the DNA packaging protein of enterobacteria phage λ using TaqMan® probe detection. Reactions were performed in quadruplicate and compared the performance of unmodified and double OXP-modified primers for their ability to detect 10, 50, 100, 500, 1000, 5000 and 10 000 copies of λ genomic DNA. This figure displays the amplification plots for unmodified (A) and double OXP-modified (B) primers as well as the corresponding standard curve (C).
Evaluation of thermolabile primer modifications in one-step RT–PCR
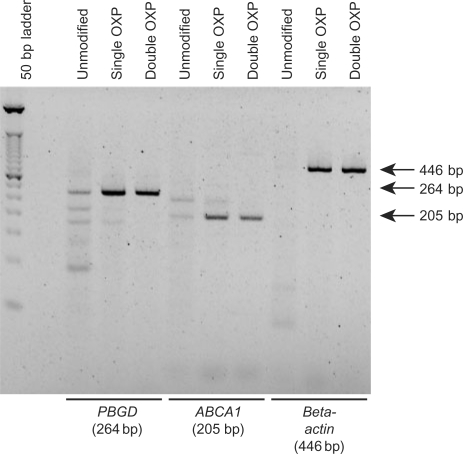
Modified PCR primers were evaluated as a potential improvement to a one-step RT–PCR approach as they are not expected to be extendable during the cDNA synthesis step, thereby reducing the probability of off-target amplicon formation. Human ABCA1, PBGD and β-actin genes were arbitrarily selected as representative RNA targets to investigate the potential benefit of PCR primers with thermolabile PTE modifications in one-step RT–PCR (79,80). In this approach, the unmodified poly-dT18 primer, a pair of PCR primers, MMLV RT and Taq DNA polymerase were included in a single reaction tube. The relative performance of unmodified, single OXP-modified and double OXP modified PCR primers was evaluated in one-step RT–PCR using a poly-dT18 primer for cDNA synthesis (Figure 12). For PBGD, use of unmodified PCR primers resulted in formation of several nonspecific amplicons, with the desired amplicon (264 bp) being formed at low efficiency. For ABCA1, two amplicons of equal efficiency were formed when unmodified PCR primers were employed, where one was the desired 205-bp amplicon. For both targets, amplicon formation was enriched when single OXP-modified PCR primers were employed, with further improvement using double OXP-modified primers. For β-actin, no detectable amplicon was formed with unmodified PCR primers. However, the desired 446-bp amplicon was formed with high specificity using either single OXP-modified or double OXP-modified primers. Overall, substitution of OXP-modified primers for corresponding unmodified primers was found to significantly improve the specificity of one-step RT–PCR. The double OXP-modified primers, which underwent slower conversion to the corresponding unmodified primers, enhanced highly specific amplicon formation in all cases.
Figure 12.
Evaluation of thermolabile primers in one-step RT–PCR. For each gene of interest (PBGD, ABCA-1 and β-actin), the PCR primers were unmodified, single OXP-modified or double OXP-modified primers. RT utilized a polydT18 primer. Reactions contained Taq DNA polymerase and M-MLV reverse transcriptase.
DISCUSSION
Hot Start PCR has proven an invaluable tool to amplify DNA targets by decreasing nonspecific target amplification. Commercially available Hot Start methodologies rely on specialized DNA polymerase compositions, such as chemical modifications, antibodies or other accessory proteins which block DNA polymerase activity at lower temperatures (18–27). The described approach investigates the introduction of modification groups onto one of the other key PCR components, the PCR primer. The described OXP and MAF PTE modifications were easily introduced via a modified phosphoramidite reagent using conventional solid phase oligonucleotide synthesis.
In proof of principle experiments, the presence of one to two internucleotide PTE modifications at the 3′-end of a primer was found to block DNA polymerase primer extension at lower, less stringent temperatures, while allowing for facile conversion to the corresponding unmodified primer using a Hot Start activation step. PTE-modified primers were not extendable by DNA polymerase prior to Hot Start activation. This property ensures that primer extension will not occur during the nonstringent pre-PCR conditions of sample preparation and manipulation. Furthermore, the PTE group is efficiently removed at stringent hybridization conditions by a thermal treatment step. Hot Start activation promotes release of the corresponding unmodified PDE oligonucleotide, which is extendable by the DNA polymerase, thus allowing for utilization of PTE-modified primers in PCR. Notably, no other reagents or special PCR conditions are necessary to activate the PTE primers.
Detailed kinetic investigation of the conversion of single OXP-modified primers agrees with the intramolecular fragmentation mechanism postulated by Beaucage and coworkers (74). Although water is not directly involved in the proposed mechanism of OXP group deprotection, our preliminary data demonstrated that anhydrous storage conditions further improved stability of the OXP primers, and may reflect the influence of solvent polarity (i.e. dielectric constant) on the rate of deprotection. While our initial expectation for optimal Hot Start activation of PTE-modified primer was t½ ∼2 min, the actual experiments have shown that a single OXP modification with a longer t½ conversion of 8.5 min provided improved PCR performance and is comparable with typical Hot Start activation conditions for Hot Start enzymes [95°C, 2–15 min (24)]. The success of PCR with longer t½ for activation is supported by our calculations (see Materials and Methods section), which predict that incubation of a single OXP-modified primer pair at 95°C for 1 min should generate enough unmodified PDE primer molecules to support PCR amplification for ∼30 cycles. After 30 PCR thermal cycles, it was estimated that the concentration of PDE primer molecules would still be adequate to support PCR amplification up to cycle 35. We have also investigated faster PTE deprotecting oligonucleotides with groups, such as TBCA and MTB (72,75). They were examined but failed to maintain stability during preparation and isolation of the PTE oligonucleotides (see Supplementary Data). Primers containing the slower deprotecting MAF group showed improvement over PDE primers. However, MAF-modified primers were less efficient in PCR amplification and showed more primer dimer formation as compared to OXP-modified primers.
Overall, the studies herein reveal a significant benefit to the substitution of OXP-modified primers for the corresponding unmodified PDE primers. In problematic primer/template systems that are prone to primer dimer formation, a marked improvement in PCR performance was evident by employing single OXP-modified primers, which significantly reduced off-target amplifications. Further benefit was evident with double OXP-modified primers, as off-target amplification products became virtually undetectable. These findings were evident in endpoint PCR analysis, as a gel-image after 35 thermal cycles revealed enrichment of the desired amplicon, with little to no primer dimer formation. Further endpoint experiments demonstrated that the presence of OXP primer modifications provided comparable if not improved PCR performance relative to other Hot Start technologies.
When PCR amplifications were monitored using real-time detection, the presence of OXP primer modifications greatly improved reaction performance by significantly delaying the Ct for off-target products and by allowing for improved linearity for all template concentrations employed. In the presence of template, the subtle delays in Ct that were observed for the modified primers may be indicative of the limiting rate of unmodified primer formation during PCR. Furthermore, these slight delays in Ct may be indicative of the presence of unextendable OXP-modified primer complexes with the DNA polymerase that dominate the early stages of PCR. However, the rate of release of the OXP modification did not significantly affect PCR performance. Although both OXP-modified PCR primers were found to benefit a number of problematic primer/template systems, the rate of unmodified PDE primer formation may complicate more advanced PCR applications, such as those that employ fast thermal cycling conditions. However, recent fast cycling work indicates that higher primer concentrations can compensate for the slower release of the OXP modification to the corresponding unmodified primers.
When the aforementioned PTE-modified primers were evaluated for their ability to support the amplification of targets that were prone to mis-priming or were limited by the sensitivity of detection, the slower converting double OXP-modified primers provided the greatest benefit. These findings are indicative of three plausible mechanisms, which may contribute to the observed improvement in PCR specificity and efficiency. First, should off-target amplicons form by primer hybridization and extension during sample set-up and consequently predominate the amplification reaction, then the presence of two modifications should block DNA polymerase extension by maintaining their protecting groups until the Hot Start, elevated temperatures are reached. Second, since PTE-modified oligonucleotides form duplexes of weaker stability than the corresponding PDE oligonucleotides (85–87), it is anticipated that the presence of PTE modifications may further disrupt the formation of extendable duplexes at the less discriminating temperatures of reaction set-up. Furthermore, the presence of diastereomeric mixtures at each PTE modification will further disrupt complex formation by further weakening duplex stability relative to PDE oligonucleotides (85–87). Third, should the off-target amplicons form during the thermal cycling process, then the limiting, temperature-dependent release of unmodified primer into the reaction should increase the probability of the primers annealing to regions of high complementarity. Regardless of the mechanism of the action of OXP-modified primers, the combination of poor extension and hybridization at low temperatures and the delayed release of unmodified primer at elevated temperatures were found to provide a significant improvement in PCR performance.
Primers with thermolabile PTE-modification groups were also found to allow selective primer usage in sequential enzymatic reactions, such as one-step RT–PCR. Although one-step RT–PCR protocols provide a more streamlined, high-throughput technique that reduces the probability of contamination by minimizing the number of handling steps (88), the technique has inherent problems and has often been found to be less sensitive than two-step (89,90). One likely cause for this lack of sensitivity is competing extension of PCR primers by reverse transcriptase (91) or DNA polymerase (14). At lower, less discriminatory temperatures of RT (42°C), PCR primer extension may result in off-target amplification products such as primer dimer and mis-priming products. PCR primers containing PTE modifications displayed diminished ability to extend during pre-PCR steps, allowing for predominant elongation of the unmodified RT primer during RT, with concomitant reduction in lower temperature, nonspecific amplicon formation. These properties allowed for improved specificity in amplicon formation, with the double OXP-modified primers revealing the greatest overall benefit.
While the current studies have described the benefit of PTE-modified primers in more traditional PCR-based applications, it is anticipated that this technology may provide benefit to more advanced applications. The promise of modified primers in one-step RT–PCR is an important finding which may benefit in a number of downstream applications where a low-temperature enzymatic step precedes an amplification step. Further advancements are envisioned for low copy number amplification reactions in which competing primer dimer formation may complicate analysis (9,92,93). Another promising area of exploration for this modified primer technology is in multiplex PCR (5,27,94). In this technology, multiple primer pairs targeted to different locations along the DNA template are introduced into a single reaction. The diversity of the primer sequences and the overall higher concentration of primer molecules in the mixture lend the reactions to a high potential for off-target amplicon formation, such as primer dimer. By employing primers with thermolabile modification groups, primer extension should be significantly suppressed during the less stringent sample preparation steps, thereby providing abundant primer for extension during the thermal cycling steps.
CONCLUSIONS
In the years since the introduction of PCR, a variety of alternate Hot Start technologies have been developed to mitigate the problems of PCR. The primary objective of our research was to develop a general strategy to address PCR specificity problems by the use of modified primers. Our approach was based on a chemical modification of the primer where the OXP modification group is incorporated into the primer during the oligonucleotide synthesis. The extension of OXP-modified primers is suppressed until the OXP group is released under desired high stringency conditions. This technology works with many thermostable DNA polymerases including commonly used Taq and does not require the use of Hot Start enzymes. However, while this approach has been shown to be a completely independent alternative Hot Start method, it potentially may provide synergistic advantages when used in conjunction with alternative Hot Start technologies. Overall, the described modified primer technology (95) represents an orthogonal approach to Hot Start activation in PCR, which may provide alterative benefits to PCR that have not been possible with current Hot Start DNA polymerase-based approaches.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (Awarding numbers 1R43GM072177-01A1, 2R44GM072177-02 and 5R44GM072177-03). Funding for open access charge: National Institute of General Medical Sciences.
Conflict of interest statement. A.V.L., N.P., V.T., J.S., K.M. and R.I.H. are employed by TriLink BioTechnologies, Inc. G.Z., J.K. and J.Y. were formerly employed by TriLink BioTechnologies, Inc. and performed this work while employed there.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Hidalgo-Mendez, L. Olivier, T. Beck, I. Koukhareva, O. Adelfinskaya and D. Combs for helpful suggestions and for critical reading of the article; P. Imperial and K. Murphy for help in the synthesis of oligonucleotides.
REFERENCES
- 1.Mullis KB. 1987. Process for amplifying nucleic acid sequences. U.S. Patent No. US4683202 (28 July 1987) [Google Scholar]
- 2.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 3.Budowle B, Schutzer SE, Einseln A, Kelley LC, Walsh AC, Smith JA, Marrone BL, Robertson J, Campos J. Public health. Building microbial forensics as a response to bioterrorism. Science. 2003;301:1852–1853. doi: 10.1126/science.1090083. [DOI] [PubMed] [Google Scholar]
- 4.Dahiya R, Deng G, Selph C, Carroll P, Presti J., Jr. A novel p53 mutation hotspot at codon 132 (AAG–>AGG) in human renal cancer. Biochem. Mol. Biol. Int. 1998;44:407–415. doi: 10.1080/15216549800201422. [DOI] [PubMed] [Google Scholar]
- 5.Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolmodin LA, Williams JF. PCR. Basic principles and routine practice. Methods Mol. Biol. 1997;67:3–15. doi: 10.1385/0-89603-483-6:3. [DOI] [PubMed] [Google Scholar]
- 7.Saldanha J, Minor P. A sensitive PCR method for detecting HCV RNA in plasma pools, blood products, and single donations. J. Med. Virol. 1994;43:72–76. doi: 10.1002/jmv.1890430114. [DOI] [PubMed] [Google Scholar]
- 8.Sato Y, Hayakawa M, Nakajima T, Motani H, Kiuchi M. HLA typing of aortic tissues from unidentified bodies using hot start polymerase chain reaction-sequence specific primers. Leg. Med. 2003;5(Suppl. 1):S191–S193. doi: 10.1016/s1344-6223(02)00108-6. [DOI] [PubMed] [Google Scholar]
- 9.Wabuyele MB, Soper SA. PCR amplification and sequencing of single copy DNA molecules. Single Mol. 2001;2:13–21. [Google Scholar]
- 10.Chen X, Sullivan PF. Single nucleotide polymorphism genotyping: biochemistry, protocol, cost and throughput. Pharmacogenomics J. 2003;3:77–96. doi: 10.1038/sj.tpj.6500167. [DOI] [PubMed] [Google Scholar]
- 11.Tsuchihashi Z, Dracopoli NC. Progress in high throughput SNP genotyping methods. Pharmacogenomics J. 2002;2:103–110. doi: 10.1038/sj.tpj.6500094. [DOI] [PubMed] [Google Scholar]
- 12.Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 13.Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou Q, Russell M, Birch DE, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Aquila RT, Bechtel LJ, Videler JA, Eron JJ, Gorczyca P, Kaplan JC. Maximizing sensitivity and specificity of PCR by pre-amplification heating. Nucleic Acids Res. 1991;19:3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaijalainen S, Karhunen PJ, Lalu K, Lindstrom K. An alternative hot start technique for PCR in small volumes using beads of wax-embedded reaction components dried in trehalose. Nucleic Acids Res. 1993;21:2959–2960. doi: 10.1093/nar/21.12.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanzer LR, Hu Y, Cripe L, Moore RE. A hot-start reverse transcription-polymerase chain reaction protocol that initiates multiple analyses simultaneously. Anal. Biochem. 1999;273:307–310. doi: 10.1006/abio.1999.4222. [DOI] [PubMed] [Google Scholar]
- 18.Birch DE, Laird WJ, Zoccoli A. 1998. Nucleic acid amplification using a reversibly inactivated thermostable enzyme. U.S. Patent No. US5773258 (30 June 1998) [Google Scholar]
- 19.Dang C, Jayasena SD. Oligonucleotide inhibitors of Taq DNA polymerase facilitate detection of low copy number targets by PCR. J. Mol. Biol. 1996;264:268–278. doi: 10.1006/jmbi.1996.0640. [DOI] [PubMed] [Google Scholar]
- 20.Eastlund E, Mueller E. Hot Start RT-PCR results in improved performance of the enhanced avian RT-PCR system. LifeSci. Q. 2001;2:2–5. [Google Scholar]
- 21.Kellogg DE, Rybalkin I, Chen S, Mukhamedova N, Vlasik T, Siebert PD, Chemchik A. TaqStart antibodyTM: ‘hot start’ PCR Facilitated by a neutralizing monoclonal antibody directed against Taq DNA polymerase. Biotechniques. 1994;16:1134–1137. [PubMed] [Google Scholar]
- 22.Kermekchiev MB, Tzekov A, Barnes WM. Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR. Nucleic Acids Res. 2003;31:6139–6147. doi: 10.1093/nar/gkg813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuguchi H, Nakatsuji M, Fujiwara S, Takagi M, Imanaka T. Characterization and application to hot start PCR of neutralizing monoclonal antibodies against KOD DNA polymerase. J. Biochem. 1999;126:762–768. doi: 10.1093/oxfordjournals.jbchem.a022514. [DOI] [PubMed] [Google Scholar]
- 24.Moretti T, Koons B, Budowle B. Enhancement of PCR amplification yield and specificity using AmpliTaq Gold DNA polymerase. Biotechniques. 1998;25:716–722. [PubMed] [Google Scholar]
- 25.Clark DR, Vincent SP. 2006. Amplification process. U.S. Patent No. US2006057617 (16 March 2006) [Google Scholar]
- 26.Olszewskia M, Rębałab K, Szczerkowskab Z, Kur J. Application of SSB-like protein from Thermus aquaticus in multiplex PCR of human Y-STR markers identification. Mol. Cell Probes. 2005;19:203–205. doi: 10.1016/j.mcp.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Shigemori Y, Mikawa T, Shibata T, Oishi M. Multiplex PCR: use of heat-stable Thermus thermophilus RecA protein to minimize non-specific PCR products. Nucleic Acids Res. 2005;33:e126. doi: 10.1093/nar/gni111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ailenberg M, Silverman M. Controlled hot start and improved specificity in carrying out PCR utilizing touch-up and loop incorporated primers (TULIPS) Biotechniques. 2000;29:1018–1020. doi: 10.2144/00295st03. 1022–1014. [DOI] [PubMed] [Google Scholar]
- 29.Ankenbauer W, Heindl D, Laue F, Huber N. 2003. Composition and method for Hot Start nucleic acid amplification. U.S. Patent No. US2003119150 (26 June 2003) [Google Scholar]
- 30.Bonner AG. 2003. Reversible chemical modification of nucleic acids and improved method for nucleic acid hybridization. U.S. Patent No. US2003162199 (28 August 2003) [Google Scholar]
- 31.Kaboev OK, Luchkina LA, Tret’iakov AN, Bahrmand AR. PCR hot start using primers with the structure of molecular beacons (hairpin-like structure) Nucleic Acids Res. 2000;28:E94. doi: 10.1093/nar/28.21.e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laird WJ, Niemiec JT. 2004. Amplification using modified primers. U.S. Patent No. US2003044817 (6 March 2003) [Google Scholar]
- 33.Puskas LG, Bottka S. Reduction of mispriming in amplification reactions with restricted PCR. Genome Res. 1995;5:309–311. doi: 10.1101/gr.5.3.309. [DOI] [PubMed] [Google Scholar]
- 34.Ullman EF, Lishanski A, Kurn N. 2002. Method for polynucleotide amplification. U.S. Patent No. US6482590 (19 November 2002) [Google Scholar]
- 35.Will SG. 1999. Modified nucleic acid amplification primers. U.S. Patent No. US6001611 (14 December 1999). [Google Scholar]
- 36.Young DD, Edwards WF, Lusic H, Lively MO, Deiters A. Light-triggered polymerase chain reaction. Chem. Commun. 2008:462–464.. doi: 10.1039/b715152g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vestheim H, Jarman SN. Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs. Front Zool. 2008;5:12. doi: 10.1186/1742-9994-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallo KA, Shao KL, Phillips LR, Regan JB, Koziolkiewicz M, Uznanski B, Stec WJ, Zon G. Alkyl phosphotriester modified oligodeoxyribonucleotides. V. Synthesis and absolute configuration of Rp and Sp diastereomers of an ethyl phosphotriester (Et) modified EcoRI recognition sequence, d[GGAA(Et)TTCC]. A synthetic approach to regio- and stereospecific ethylation-interference studies. Nucleic Acids Res. 1986;14:7405–7420. doi: 10.1093/nar/14.18.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaPlanche LA, James TL, Powell C, Wilson WD, Uznanski B, Stec WJ, Summers MF, Zon G. Phosphorothioate-modified oligodeoxyribonucleotides. III. NMR and UV spectroscopic studies of the Rp-Rp, Sp-Sp, and Rp-Sp duplexes, [d(GGSAATTCC)]2, derived from diastereomeric O-ethyl phosphorothioates. Nucleic Acids Res. 1986;14:9081–9093. doi: 10.1093/nar/14.22.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letsinger RL, Ogilvie KK. A convenient method to stepwise synthesis of oligothymidylate derivatives in large scale quantities. J. Am. Chem. Soc. 1967;89:4801–4808. [Google Scholar]
- 41.Michelson AM, Todd AR. Nucleotides part XXXII. Synthesis of a dithymidine dinucleotide containing a 3: 5-internucleotidic linkage. J. Chem. Soc. 1955:2632–2638.. [Google Scholar]
- 42.Stec WJ, Zon G, Gallo KA, Byrd RA, UznanskiI B, Guga P. Synthesis and absolute configuration of P-chiral O-isopropyl oligonucleotide triesters. Tetrahedron Lett. 1985;26:2191–2194. [Google Scholar]
- 43.Koziolkiewicz M, Uznanski B, Stec WJ, Zon G. P-Chiral analogues of oligodeoxyribonucleotides: synthesis, stereochemistry and enzyme studies. Chem. Scr. 1986;26:251–260. [Google Scholar]
- 44.Lebedev AL, Wickstrom E. Chirality problem in P-substituted oligonucleotides. Perspect. Drug Discov. 1996;4:17–36. [Google Scholar]
- 45.Stec WJ, Wilk A. Stereocontrolled synthesis of oligo (nucleoside phosphorothioate) s. Angew. Chem. Int. Ed Engl. 1994;33:709–722. [Google Scholar]
- 46.Adamiak RW, Barciszewska MZ, Biala E, Grzeskowiak K, Kierzek R, Kraszewski A, Markiewicz WT, Wiewiorowski M. Nucleoside-3′-phosphotriesters as key intermediates for the oligoribonucleotide synthesis. III. An improved preparation of nucleoside 3′-phosphotriesters, their 1H NMR characterization and new conditions for removal of 2-cyanoethyl group. Nucleic Acids Res. 1976;3:3397–3408. doi: 10.1093/nar/3.12.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alul RH, Singman CN, Zhang GR, Letsinger RL. Oxalyl-CPG: a labile support for synthesis of sensitive oligonucleotide derivatives. Nucleic Acids Res. 1991;19:1527–1532. doi: 10.1093/nar/19.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown JM, Christodoulou C, Jones SS, Modak AS, Reese CB, Sibanda S, Ubasawa A. Synthesis of the 3-terminal half of yeast alanine transfer ribonucleic acid (tRNAAla) by the phosphotriester approach in solution. Part 1. Preparation of the nucleoside building blocks. J. Chem. Soc., Perkin Trans. 1989;1:1735–1750. [Google Scholar]
- 49.Brown JM, Christodoulou C, Modak AS, Reese CB, Serafinowska HT. Synthesis of the 3-terminal half of yeast alanine transfer ribonucleic acid (tRNAAla) by the phosphotriester approach in solution. Part 2. J. Chem. Soc., Perkin Trans. 1989;1:1751–1767. [Google Scholar]
- 50.Daub GW, Tamelen EE. Synthesis of oligoribonucleotides based on the facile cleavage of methyl phosphotriester intermediates. J. Am. Chem. Soc. 1977;99:3526–3528. doi: 10.1021/ja00452a069. [DOI] [PubMed] [Google Scholar]
- 51.Grzeskowiak K, Adamiak RW, Wiewiorowski M. Chromatography on Sephadex LH20 as an efficient purification step after removal of internucleotide 2,2,2-trichloroethyl protective groups from oligoribonucleotide phosphotriesters. Nucleic Acids Res. 1980;8:1097–1105. doi: 10.1093/nar/8.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katagiri N, Itakura K, Narang SA. Use of arylsulfonyltriazoles for the synthesis of oligonucleotides by the triester approach. J. Am. Chem. Soc. 1975;97:7332–7337. doi: 10.1021/ja00858a021. [DOI] [PubMed] [Google Scholar]
- 53.Kuijpers WH, Huskens J, Koole LH, van Boeckel CA. Synthesis of well-defined phosphate-methylated DNA fragments: the application of potassium carbonate in methanol as deprotecting reagent. Nucleic Acids Res. 1990;18:5197–5205. doi: 10.1093/nar/18.17.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letsinger RL, Ogilvie KK. Synthesis of oligothymidylates via phosphotriester intermediates. J. Am. Chem. Soc. 1969;91:3350–3355. [Google Scholar]
- 55.Letsinger RL, Ogilvie KK, Miller PS. Developments in synthesis of oligodeoxyribonucleotides and their organic derivatives. J. Am. Chem. Soc. 1969;91:3360–3365. [Google Scholar]
- 56.Matsuzaki J, Kohno K, Tahara S, Sekine M, Hata T. Solid phase synthesis of oligodeoxyribonucleotides utilizing the phenylthio group as a phosphate protecting group. Bull. Chem. Soc. Jpn. 1987;60:1407–1413. [Google Scholar]
- 57.Moody HM, van Genderen MH, Koole LH, Kocken HJ, Meijer EM, Buck HM. Regiospecific inhibition of DNA duplication by antisense phosphate-methylated oligodeoxynucleotides. Nucleic Acids Res. 1989;17:4769–4782. doi: 10.1093/nar/17.12.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohtsuka E, Yamane A, Doi T. Chemical synthesis of the 5′-half molecule of E. coli tRNA2GLY. Tetrahedron. 1984;40:47–57. [Google Scholar]
- 59.Reese CB, Yau L. Reaction between 4-nitrobenaldoximate ion and phosphotriesters. Tetrahedron Lett. 1978;19:4443–4446. [Google Scholar]
- 60.Reese CB, Zard L. Some observations relating to the oximate ion promoted unblocking of oligonucleotide aryl esters. Nucleic Acids Res. 1981;9:4611–4626. doi: 10.1093/nar/9.18.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sekine M, Matsuzaki J, Hata T. Oligodeoxyribonucleotide synthesis by the use of S,S-diphenyl deoxyribonucleoside 3′-phosphorodithioates and bifunctional condensing reagents in the phosphotriester approach. Tetrahedron. 1985;41:5279–5288. [Google Scholar]
- 62.Sekine M, Tanimura H, Hata T. An effective method for removal of the internucleotide phenylthio group from fully protected oligonucleotides by use of bis(tributyltin) oxide. Tetrahedron Lett. 1985;26:4621–4624. [Google Scholar]
- 63.Smrt J. Oligonucleotidic compounds. XLIV. Protection of the internucleotidic bond after its synthesis as approach to the synthesis of an oligonucleotidic chain. Collect. Czech. Chem. Commun. 1973;38:3189–3197. [Google Scholar]
- 64.Sood AK, Narang SA. A Rapid and convenient synthesis of poly-thymidylic acid by the modified triester approach. Nucl. Acids Res. 1976;4:2757–2765. doi: 10.1093/nar/4.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takaku H, Watanabe T, Hamamoto S. 1,1,1,3,3,3-Hexafluoro-2-propyl protecting group in the synthesis of DNA fragments via phosphoramidite intermediates. Tetrahedron Lett. 1988;29:81–84. [Google Scholar]
- 66.Takaku H, Yamaguchi R, Nomoto T. 5-Chloro-8-quinolyl group as high efficient phosphate protecting group for the synthesis of oligoribonucleotides. Tetrahedron Lett. 1979;20:3857–3860. [Google Scholar]
- 67.Yamakage S, Fujii M, Takaku H, Uemura M. 1,1,1,3,3,3-Hexafluoro-2-propyl group as a new phosphate protecting group for oligoribonucleotide synthesis in the phosphotriester approach. Tetrahedron. 1989;45:5459–5468. [Google Scholar]
- 68.Beaucage SL, Wilk A, Grajkowski A. 2001. Thermolabile phosphorus protecting groups, associated intermediates and methods of use. U.S. Patent No. US2001044529 (22 November 2001) [Google Scholar]
- 69.Cieslak J, Beaucage SL. Thermolytic properties of 3-(2-Pyridyl)-1-Propyl and 2-[N-Methyl-N-(2-Pyridyl)]Aminoethyl Phosphate/Thiophosphate protecting groups in solid-phase synthesis of oligonucleotides. J. Org. Chem. 2003;68:10123–10129. doi: 10.1021/jo0354490. [DOI] [PubMed] [Google Scholar]
- 70.Cieslak J, Grajkowski A, Livengood V, Beaucage SL. Thermolytic 4-methylthio-1-butyl group for phosphate/thiophosphate protection in solid-phase synthesis of DNA oligonucleotides. J. Org. Chem. 2004;69:2509–2515. doi: 10.1021/jo035861f. [DOI] [PubMed] [Google Scholar]
- 71.Grajkowski A, Pedras-Vasconcelos J, Wang V, Ausin C, Hess S, Verthelyi D, Beaucage SL. Thermolytic CpG-containing DNA oligonucleotides as potential immunotherapeutic prodrugs. Nucleic Acids Res. 2005;33:3550–3560. doi: 10.1093/nar/gki657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grajkowski A, Wilk A, Chmielewski MK, Phillips LR, Beaucage SL. The 2-(N-formyl-N-methyl)aminoethyl group as a potential phosphate/thiophosphate protecting group in solid-phase oligodeoxyribonucleotides synthesis. Org. Lett. 2001;3:1287–1290. doi: 10.1021/ol0156852. [DOI] [PubMed] [Google Scholar]
- 73.Guzaev AP, Manoharan M. 2-Benzamidoethyl group – a novel type of phosphate protecting group for oligonucleotide synthesis. J. Am. Chem. Soc. 2001;123:783–793. doi: 10.1021/ja0016396. [DOI] [PubMed] [Google Scholar]
- 74.Wilk A, Chmielewski MK, Grajkowski A, Phillips LR, Beaucage SL. The 4-oxopentyl group as a labile phosphate/thiophosphate protecting group for synthetic oligodeoxyribonucleotides. Tetrahedron Lett. 2001;42:5635–5639. [Google Scholar]
- 75.Wilk A, Chmielewski MK, Grajkowski A, Phillips LR, Beaucage SL. The 3-(N-tert-butylcarboxamido)-1-propyl group as an attractive phosphate/thiophosphate protecting group for solid-phase oligodeoxyribonucleotide synthesis. J. Org. Chem. 2002;67:6430–6438. doi: 10.1021/jo0258608. [DOI] [PubMed] [Google Scholar]
- 76.Levina AS, Nevinskii GA, Lavrik OI. E. coli DNA polymerase. A study of the mechanism of primer binding using oligothymidylate analogs with ethylated internucleotide phosphate groups. Bioorg. Khim. 1985;11:358–369. [PubMed] [Google Scholar]
- 77.Nevinskii GA, Frolova EI, Levina AS, Podust VN, Lebedev AV. Prokaryotic and eukaryotic DNA-polymerase. I. The role of internucleotide phosphate groups in the binding of a primer with the enzyme. Bioorg. Khim. 1987;13:45–57. [PubMed] [Google Scholar]
- 78.Weinfeld M, Livingston DC. Synthesis and properties of oligodeoxyribonucleotides containing an ethylated internucleotide phosphate. Biochemistry. 1986;25:5083–5091. doi: 10.1021/bi00366a016. [DOI] [PubMed] [Google Scholar]
- 79.Kielar D, Dietmaier W, Langmann T, Aslanidis C, Probst M, Naruszewicz M, Schmitz G. Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription-PCR. Clin. Chem. 2001;47:2089–2097. [PubMed] [Google Scholar]
- 80.Louwrier A, van der Valk A. Thermally reversible inactivation of Taq polymerase in an organic solvent for application in hot start PCR. Enzyme Microb. Technol. 2005;36:947–952. [Google Scholar]
- 81.Douglas A, Atchison B. Degradation of DNA during the denaturation step of PCR. PCR Methods Appl. 1993;3:133–134. doi: 10.1101/gr.3.2.133. [DOI] [PubMed] [Google Scholar]
- 82.Ririea KM, Rasmussenb RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 83.Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–131. doi: 10.2144/97221bi01. 134–138. [DOI] [PubMed] [Google Scholar]
- 84.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 85.Miller PS, Annan ND, McParland KB, Pulford SM. Oligothymidylate analogues having stereoregular, alternating methylphosphonate/phosphodiester backbones as primers for DNA polymerase. Biochemistry. 1982;21:2507–2512. doi: 10.1021/bi00539a033. [DOI] [PubMed] [Google Scholar]
- 86.Pless RC, Ts’o PO. Duplex formation of a nonionic oligo(deoxythymidylate) analogue (heptadeoxythymidylyl-(3′-5′)-deoxythymidine heptaethyl ester (d-(Tp(Et))7T)) with poly(deoxyadenylate). Evaluation of the electrostatic interaction. Biochemistry. 1977;16:1239–1250. doi: 10.1021/bi00625a033. [DOI] [PubMed] [Google Scholar]
- 87.Summers MF, Powell C, Egan W, Byrd RA, Wilson WD, Zon G. Alkyl phosphotriester modified oligodeoxyribonucleotides. VI. NMR and UV spectroscopic studies of ethyl phosphotriester (Et) modified Rp-Rp and Sp-Sp duplexes, (d[GGAA(Et)TTCC])2. Nucleic Acids Res. 1986;25:7421–7436. doi: 10.1093/nar/14.18.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wacker MJ, Godard MP. Analysis of one-step and two-step real-time RT-PCR using Superscript III. J. Biomol. Tech. 2005;16:266–271. [PMC free article] [PubMed] [Google Scholar]
- 89.Lekanne Deprez RH, Fijnvandraat AC, Ruijter JM, Moorman AFM. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal. Biochem. 2002;307:63–69. doi: 10.1016/s0003-2697(02)00021-0. [DOI] [PubMed] [Google Scholar]
- 90.Suslov O, Steindler DA. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;33:e181. doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peters IR, Helps CR, Hall EJ, Day MJ. Real-time RT-PCR: considerations for efficient and sensitive assay design. J. Immunol. Methods. 2004;286:203–217. doi: 10.1016/j.jim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 92.Griffiths AD, Tawfik DS. Miniaturising the laboratory in emulsion droplets. Trends Biotechnol. 2006;24:395–402. doi: 10.1016/j.tibtech.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 93.Musundi B, Soper SA. PCR amplification and sequencing of single copy DNA molecules. Single Mol. 2001;2:13–21. [Google Scholar]
- 94.Cheng YW, Shawber C, Notterman D, Paty P, Barany F. Multiplexed profiling of candidate genes for CpG island methylation status using a flexible PCR/LDR/Universal Array assay. Genome Res. 2006;16:282–289. doi: 10.1101/gr.4181406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lebedev AV, Paul N, Yee J, Kellum J, Timoshchuk VA, Hogrefe RI, Zon G. Chemically modified primers for improved hot start PCR. Collection Symposium Series. 2008;10:398–399. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.