Abstract
The biological role of manganese (Mn2+) has been a long-standing puzzle, since at low concentrations it activates several polymerases whilst at higher concentrations it inhibits. Viral RNA polymerases possess a common architecture, reminiscent of a closed right hand. The RNA-dependent RNA polymerase (RdRp) of bacteriophage ϕ6 is one of the best understood examples of this important class of polymerases. We have probed the role of Mn2+ by biochemical, biophysical and structural analyses of the wild-type enzyme and of a mutant form with an altered Mn2+-binding site (E491 to Q). The E491Q mutant has much reduced affinity for Mn2+, reduced RNA binding and a compromised elongation rate. Loss of Mn2+ binding structurally stabilizes the enzyme. These data and a re-examination of the structures of other viral RNA polymerases clarify the role of manganese in the activation of polymerization: Mn2+ coordination of a catalytic aspartate is necessary to allow the active site to properly engage with the triphosphates of the incoming NTPs. The structural flexibility caused by Mn2+ is also important for the enzyme dynamics, explaining the requirement for manganese throughout RNA polymerization.
INTRODUCTION
Virus encoded RdRps are essential to the life cycle of RNA viruses, being responsible for genome replication and mRNA transcription when associated with other proteins in the replication complex (1). In recent years, the structures of a number of viral RdRps have been determined, in some cases in complex with the template, NTP and/or metal ions (2–5). They exhibit structural conservation of a particular variant of the canonical right hand motif which not only possesses palm, fingers and thumb sub-domains, but is also rather spherical due to the linking of fingers and thumb sub-domains, using the so-called ‘fingertips’. This gives viral RdRps a ‘closed hand’ appearance and creates two positively charged tunnels: the template and NTP-binding channels (5,9,14).
RNA synthesis by an RdRp involves template binding, initiation complex formation, transition from initiation to elongation and finally termination. Correct initiation and adequate processivity of the RNA synthesis are essential for maintaining the integrity of the viral genome. The first insights into the molecular details of the initiation mechanism were provided by structural studies of ϕ6 RdRp (6) in complex with a 5-nt long DNA template and NTP substrates. These studies provide an atomic model for the de novo (primer-independent) initiation of RNA polymerization (6,7). In brief, in the transcription catalysed by ϕ6 RdRp, the template RNA is bound in the template tunnel, with the 3′ base (T1) being recognized by a specificity pocket (site S) within the C-terminal domain (abbreviations are defined in ref. 14). An interrogation site (I) is defined by a set of arginine residues at the entrance of the NTP tunnel which lock the phosphate moieties of incoming NTPs in position, allowing the base component to interrogate the template sequence. Incoming GTP is stabilized as it base pairs to the penultimate cytidine (T2) by packing against Y630 (defined as site P, the initiation stabilizing platform). The template then moves backwards, releasing the 3′-cytidine from pocket S. This allows a second GTP to enter and base pair to the T1 position of the template (at site P), stabilized by Y630, locking the initiation complex into its final conformation. Catalysis can then occur, with release of PPi. The next NTP slips in, setting the ratchet, and chain elongation is underway. In ϕ6 RdRp, the double-stranded RNA (dsRNA) exit path is blocked by the C-terminal sub-domain. Therefore, displacement of this domain is necessary for elongation to occur. When the nascent dsRNA product reaches a critical length, the C-terminal domain must be displaced to allow the extending product to exit the contained active site of the polymerase (5,8).
The addition of manganese ions is known to stimulate RNA polymerization in a number of viruses including ϕ6 (9,10), related phages ϕ8, ϕ12 and ϕ13 (11,12), Bovine viral diarrhea virus (BVDV) (13), classical swine fever virus (CSFV) (14), GB virus (15), and hepatitis C virus (HCV) (16–19). Furthermore, the same phenomenon has also been observed with ϕ6 polymerase complex particles (20,21), which are active assemblies of four viral proteins forming the innermost capsid of the ϕ6 virion (22,23), as well as with bluetongue virus (24). Previous structural studies identified a high affinity manganese-binding site in ϕ6 RdRp palm domain (6), coordinated by D454 (one of the invariant catalytic aspartates), E491 and A495. This binding site is conserved across a number of viral RNA polymerases, such as West Nile virus (WNV: pdb ID 2HCN), foot-and-mouth disease virus (FMDV: pdb ID 1WNE), Norwalk virus (NV: pdb ID 1SH3), Dengue virus (DV: pdb ID 2J7U), poliovirus (PV: pdb ID 1RDR) and reovirus (RV: pdb ID 1MWH), although the bound ion appears to vary (25–30).
Both de novo and primer-dependent RNA synthesis of CSFV and GB virus RdRps are stimulated by Mg2+ and Mn2+ ions. However, for BVDV, CSFV and C-terminally truncated HCV RdRps, de novo initiation in vitro is more dependent on Mn2+ than on Mg2+ ions (14,31,32). Similar results were obtained with mutant forms of ϕ6 polymerase where deletions were introduced at the initiation platform loop (10,33). Mn2+ ions also modulate the substrate specificity and nucleotide misincorporation of several polymerases (32,34–38). However, because of the low physiological concentration of manganese [∼10 µM compared to >10 mM Mg2+; (39)], it remains uncertain if it has a general physiological role in polymerization.
In this study, we address the role of Mn2+ using a ϕ6 RdRp mutant with decreased Mn2+-binding affinity. One of the residues coordinated by Mn2+, E491, was mutated to glutamine and the structure and activity of the mutant RdRp (denoted E491Q) analysed. The isolated mutant polymerase is less effective at template binding, initiation and elongation, and we were able to capture an inhibited state of the enzyme. Overall, our structural and biochemical results shed light on the functions of Mn2+ in de novo RNA-dependent RNA synthesis. Manganese binding destabilizes the enzyme, suggesting that a degree of structural flexibility is required for the efficient binding of template RNA and for the dynamic processes involved in complementary strand synthesis.
MATERIALS AND METHODS
Plasmids
Plasmid pEM15 contains the ϕ6 S segment with an internal deletion (9) and was used to prepare sΔ+13 ssRNA template. Plasmids pLM659 (40) and pLM687 (41) were templates for the production of the 5′-terminal fragment of s+ and full-length l+, respectively [where s+ and l+ are single stranded, message sense RNAs for the small (S) and large (L) genome segments]. Plasmid pEM2 [containing wild type (wt) ϕ6 gene 2 encoding the RdRp (9)] was cut with AvrII and NotI, treated with Klenow polymerase and ligated. The resultant plasmid pEM33 encodes recombinant RdRp with a C-terminal hexahistidine tag tethered to the protein with an AlaAlaLeuGlu linker. A point mutation resulting in substitution E491Q was introduced to pEM33 using QuickChange mutagenesis kit (Stratagene), together with the upstream 5′-CACCAAGGAAGGCGCCACCGTGCTGGTAGGAGATCTTCATGTAAGG-3′ and 5′-CCTTACATGAAGATCTCCTACCAGCACGGTGGCGCCTTCCTTGGTG-3′ downstream primers, resulting in plasmid pNL18. To construct a plasmid for the expression of E491Q polymerase without tag, pNL18 was cut with NdeI and NsiI and ligated with similarly cut pEMG2 expression vector for wt ϕ6 RdRp, (42). The resultant plasmid, pSVe4 was partially sequenced to verify the mutation. Wild-type ϕ6 RdRp was expressed from pEMG2 plasmid (42).
Preparation of RNA substrates
Synthetic ssRNAs were produced by run-off in vitro transcription with T7 polymerase (43). Templates for the T7 transcription were prepared from plasmid DNA either by cutting with suitable restriction endonucleases or by PCR amplification. The spac fragment was amplified from pLM659 with the upstream oligonucleotide primer 5′-GCGCTGCAGTAATACGACTCACTATAGGAAAAAAACTTTATATAACTCTTATATAAGTGC-3′ and the downstream oligonucleotide primer 5′-GCGGAATTCAATTAACCCTCACTAAAGGTATTCCTTTATTATTAGTGGGAACAACACC-3′. Templates for sΔ+13 and l+ RNAs were produced as previously described (9,44). All ssRNAs were successively extracted with TRIzol (Invitrogen, Carlsbad, California, USA)/chloroform (5:1), precipitated with 4 M LiCl, and with 0.75 M NH4Ac and dissolved in sterile water. The ssRNA concentrations were measured (A260) and the quality of each preparation was checked by electrophoresis in 1% agarose gels. For gel retardation assays, the spac RNA was purified in denaturing polyacrylamide gel as described by Frilander and Turunen (43).
Protein purification
Both wt and E491Q RdRps were expressed in Escherichia coli BL21 (45) containing the appropriate expression plasmid at 20°C for 15 h and purified to near homogeneity as described previously (9). For the gel retardation assay, pooled enzyme containing fractions were diluted 10-fold with 20 mM Tris–HCl pH 8.0, filtered and applied onto MonoQ column (Amersham Biosciences, Piscataway, New Jersey, USA) and eluted with a linear gradient of 0–1 M NaCl (buffered with) in 50 mM Tris–HCl pH 8.0, 0.1 mM EDTA.
ϕ6 polymerase assay
The replication activity of wt and E491Q polymerases was typically assayed in 10 μl reaction mixtures containing 50 mM HEPES–KOH, pH 7.5, 20 mM ammonium acetate, 6% (w/v) polyethylene glycol 4000, 5 mM MgCl2, 2 mM MnCl2, 0.1 mM EDTA, 0.1% Triton X-100, 0.2 mM UTP and CTP, 1 mM ATP and GTP, 0.8 U/μl RNasin (Promega) and 0.1 mCi/ml of [α-32P]UTP (Amersham Biosciences, 3000 Ci/mmol). The mixtures were incubated at 30°C, and stopped with 2 × U buffer (46). Reaction products were separated by standard agarose gel electrophoresis, dried on Whatman 3 filter paper followed by autoradiography and/or phosphoimaging (Fuji BAS1500 or FLA 5000) analysis of the product bands. Digital image analysis was performed using AIDA Image Analyzer v. 3.44 software (Raytest Isotopenmeβgeräte GmbH, Germany).
Heparin trap assay
The previously described protocol (12,47) was used with the following modifications. Wild type and E491Q polymerases were preincubated with l+ ssRNA template in the mixtures lacking labelled UTP and two of the unlabelled NTPs. The other components of the preincubation mixtures were as described above for the ϕ6 polymerase assay. After 20 min incubation at 30°C, heparin was added to a final concentration of 9.1 mg/ml and incubated for another 5 min at 30°C. Then, the reactions were supplemented with the rest of the NTPs and 0.1 mCi/ml [α-32P]UTP, followed by incubation at 30°C. Aliquots sampled at different time points were separated by standard agarose gel electrophoresis and analysed as described above for the ϕ6 polymerase assay.
Polymerization and elongation rate
An estimation of a lag phase prior to the appearance of relevant full-length products was obtained from linear extrapolation of the data from linear parts of the plots (9) and was used to approximate the elongation rates for wt and E491Q polymerases.
Gel retardation assay
A 10 μl reaction mixture containing 50 mM HEPES–KOH, pH 7.5, 20 mM NH4Ac, 6% (w/v) polyethylene glycol 4000, 0.1 mM EDTA, 0.1% Triton X-100, 270 nM spac ssRNA with a trace amount of γ-32P-labelled spac ssRNA (T4 polynucleotide kinase, MBI Fermentas, Burlington, Ontario, Canada, [γ-32P]ATP, Amersham Biosciences, 3000 Ci/mmol) supplemented with 5 mM MgCl2 or 2 mM MnCl2, as indicated, was incubated with 0.02–0.2 mg/ml (270–2700 nM) of wt or E491Q polymerase on ice for 10 min. In the control reactions, a corresponding volume of H2O was used instead of the polymerase. Before loading, sucrose loading dye (12) was added to the reaction mixtures and the polymerase-RNA complexes were resolved in 8% native polyacrylamide gel in 1 × Tris–glycine buffer (48). The electrophoresis was carried out at 150 V and 20 mA, 4°C for ∼17 h, the gels dried on Whatman 3 filter paper followed by autoradiography. Dissociation constant for the wt·RNA complex was obtained from equation [RNAfree]/[complex] = Kd × 1/[Proteintotal] after quantification of the RNA patterns.
Thermal shift assay
The thermal shift assay (ThermoFluor) was carried out in a real-time PCR machine (BioRad DNA Engine Opticon 2) where buffered solutions of protein and fluorophore (SYPRO Orange; Molecular Probes, Invitrogen), with and without additives, were heated in a stepwise fashion from 4°C to 99°C. Five microlitres of protein (1 mg/ml) and 5 μl SYPRO Orange (Molecular Probes, Invitrogen) were made up to a total assay volume of 50 μl with starting buffer (50 mM Tris–HCl, pH 7.5, 50 mM NaCl) in white low profile thin-wall PCR plates (Abgene) sealed with microseal ‘B’ films (BioRad). The fluorophore was excited in the range of 470–505 nm and fluorescence emission was measured in the range of 540–700 nm every 0.5°C after a 10 s hold. The melting temperature (Tm) was calculated using Opticon Monitor software. The effect of manganese ions was assayed by comparing melting temperatures of protein in starting buffer with those of protein in starting buffer containing either 5 mM EDTA, 10 mM MnCl2 or both 5 mM EDTA and 10 mM MnCl2.
Protein crystallization and soaking experiments
To determine the optimal crystallization conditions for the E491Q mutant protein, a screen of 480 conditions was carried out using the sitting drop vapour diffusion method with a 200 nl drop size (1:1 protein/precipitant ratio) in a Cartesian Robot available at the Oxford Protein Production Facility (49–51). Scaling up optimized conditions in 2–3 μl drops (1:1, 1:2 or 2:1 protein/precipitant ratio) resulted in crystals of 200 × 110 × 80 µm3 in 15–20% polyethylene glycol 4000, 8.5% isopropanol, 15% glycerol and 100 mM Hepes, pH 7.5. The protein was typically concentrated to 6–8 mg/ml in 10 mM Tris–HCl, pH8.0, 100 mM NaCl.
ϕ6 E491Q mutant polymerase was co-crystallised with a 6-nt long RNA oligonucleotide 5′-UUUUCC-3′ and its DNA equivalent 5′-TTTTCC-3′ (Eurogentec, EGT Group), chosen to mimic the 3′ end (-)ssRNA sequence of s− and m− segments [5′-…UUUUCC-3′; (52,53)], the preferred templates for transcription (11,54). Six microlitre of a protein solution at a concentration of 6–8 mg/ml was incubated with 2–8 μM of the oligonucleotides for 1 h on ice, before setting up sitting drop crystallization trays in the conditions pre-established for the mutant protein crystals. For the RNA co-crystallization experiments, 2 mM MnCl2 were added to the protein buffer, whilst in all successful DNA co-crystallization experiments, no manganese ion was present in the protein buffer or crystallization solution (see Table 1).
Table 1.
Crystallization, data collection and refinement statistics
| E491Q | E491Q-RNA-Mn2+ | E491Q-DNA-GTP-Mg2+-Mn2+ | |
|---|---|---|---|
| Crystallization conditions | |||
| Divalent cations | – | 2 mM MnCl2 | 5 mM MgCl2,, 5 mM MnCl2 (soak) |
| Nucleic acid | – | 5′-UUUUCC-3′ | 5′-TTTTCC-3′ |
| NTPs | – | – | 25 mM GTP (soak) |
| Data collection details | |||
| Wavelength (Å) | 0.98 | 0.98 | 0.98 |
| Space group | P21 | P32 | P32 |
| Unit cell a,b,c (Å); α = γ = 90°; β (°) | 106.5, 91.6, 142.7; 101.6 | 110.0, 110.0, 159.1 | 109.0, 109.0, 158.8 |
| Resolution range (Å) | 20.0–3.2 (3.3–3.2) | 18.0–3.2 (3.3–3.2) | 20.0–2.8 (2.9–2.8) |
| Observations | 458 511 | 668 591 | 610 471 |
| Unique reflections | 40 323 | 52 552 | 72 626 |
| Completeness (%) | 90.5 (80.8) | 100.0 (100.0) | 99.8 (99.9) |
| I/δ(I) (%) | 7.2 (1.6) | 6.7 (1.6) | 6.2 (1.4) |
| Rmergea (%) | 17.9 | 28.9 | 22.3 |
| Refinement statistics: | |||
| Resolution range (Å) | 20.0–3.2 | 20.0–3.2 | 20.0–2.8 |
| No. of reflections (working/test) | 38 111/2038 | 33 578/1771 | 49 085/2641 |
| Rfactorb/Rfreec | 21.5/ 27.2 | 22.8/ 28.6 | 21.6/ 29.4 |
| No. of atoms: protein/ligands | 15 629 | 15 608/2 | 15 705/394 |
| RMSD bond length (Å) | 0.010 | 0.007 | 0.008 |
| RMSD bond angle (°) | 1.3 | 1.1 | 1.3 |
| Mean B-factor (Å2) protein/ligands | 48 | 37/63 | 32/47 |
Values in parenthesis refer to the highest resolution shell.
aRmerge = ΣiΣh(|Ij,h − <Ih>|)/ΣjΣh(<Ih>), where h are unique reflections indices, Ij,h are intensities of symmetry-related reflections and <Ih> is the mean intensity.
bRfactor.
cRfree are defined by R = Σhkl||Fobs| − |Fcalc||/Σhkl|Fobs|, where h,k,l are the indices of the reflections (used in refinement for Rfactor; 5%, not used in refinement, for Rfree), Fobs and Fcalc are the structure factors, deduced from measured intensities and calculated from the model, respectively.
PDB accession codes are 2jl9, 2jlf, 2jlg.
E491Q-DNA oligonucleotide co-crystals were used for a soaking experiment with GTP and different metal ions, Mg2+ and Mn2+, by the addition of 5 mM MnCl2 to the crystallization drop, followed a few seconds later by the addition of 5 mM MgCl2 and, immediately after, 25 mM GTP (lithium salt).
Data collection and processing
X-ray data from E491Q mutant and the different complexes were collected on station PX 9.6 at the SRS, Daresbury, UK, using an ADSC Q4R CCD detector. Each image covered an oscillation of 1° and exposure times were typically 45–60 s. All data were collected at liquid nitrogen temperatures after briefly washing the crystals with 25% (v/v) glycerol as cryoprotectant. Data were scaled and indexed using HKL-2000 (55).
Crystals of the ϕ6 RdRp E491Q mutant apoprotein diffracted to 3.2 Å resolution and were approximately isomorphous to the wt crystals previously described (6,7,56), belonging to space group P21, with unit cell dimensions a ≈ 106 Å, b ≈ 92 Å, c ≈ 142 Å, β ≈ 102°, whereas the datasets collected for the complexes described (E491Q-RNA-Mn and E491Q-DNA-GTP-Mg2+-Mn2+) were seemingly isomorphous to the P32 SeMet crystal structure determined by Butcher et al. (56), with unit cell dimensions a ≈ b ≈ 109 Å, c ≈ 159 Å. Data processing statistics are given in Table 1.
Structure refinement
The apo structure of the E491Q ϕ6 RdRp mutant was determined by rigid body refinement of the three molecules present in the asymmetric unit with PHENIX (57), using the wt apo structure as a starting model. Visual inspection of the initial model and manual model rebuilding were carried out in COOT (58). Final refinement in PHENIX (57) carried out to a resolution of 3.2 Å, imposing non-crystallographic symmetry restraints as defined for the ϕ6 RdRp-RNA models, resulted in a model with Rfactor = 21.5% and Rfree = 27.2% and good stereochemistry (RMSD bond length = 0.01 Å and RMSD bond angle = 1.3°).
The P32 complex crystals were solved by subjecting the best dataset (E491Q-DNA-GTP-Mg2+-Mn2+) to molecular replacement analysis using the program PHENIX with the wt structure (59,60), revealing three molecules (denoted as I, II and III) in the crystallographic asymmetric unit. Rigid body refinement of this model was then carried out using PHENIX (57). Visual inspection of the electron density maps allowed modelling of a five base DNA oligonucleotide (due to lack of density one nucleotide at the 5′ was not traced) and a GTP molecule bound to two RdRp monomers, and a four base DNA oligonucleotide (two nucleotides at the 5′-end were not modelled) complexed with two GTP molecules and a Mn2+ ion bound to the third monomer in the crystallographic asymmetric unit. Manual model rebuilding followed by refinement in PHENIX (57) to a resolution of 2.8 Å resulted in a final model with Rfactor = 21.6% and Rfree = 29.4% and good stereochemistry (RMSD bond length = 0.008 Å and RMSD bond angle = 1.3°).
The final protein model for the E491Q-DNA-GTP-Mg2+-Mn2+complex was used for rigid body refinement of the other P32 dataset. A similar strategy was then used to trace the different ligands in each complex and final refinements were also carried out using PHENIX (see Table 1 for details for both complexes).
In all models of the complexes, clear differences in the strength and position of the electron density in the DNA- and GTP-binding regions were detectable between the three molecules in the crystallographic asymmetric unit. Therefore, no internal averaging was carried out and the ligands observed in each molecule were modelled and refined separately. Due to the relatively weak electron density levels for the DNA and GTP molecules in molecules I and II, these were assigned an occupancy of 0.5.
Unless otherwise, stated molecular figures were drawn and rendered with Pymol (61).
Structure analysis of viral RdRps
SHP was used to align structures of WNV, FMDV, NV, DV, PV and RV RdRps to ϕ6 RdRp (62). Coot was used to download the molecular models and where possible, electron density maps from the EDS, to check the quality of the fit of the models to the maps (58,63).
RESULTS
A structurally equivalent site is found in other viral RdRps
The structures of a subset of other viral RdRps deposited in the PDB database were objectively compared to ϕ6 RdRp, by automated structural superposition (Table 2). In the majority of cases the selected structures represented the RdRps in a resting apo state, however in two cases the structures represented the polymerases undergoing elongation (3BSO and 1N35). As expected the invariant catalytic aspartates are structurally conserved across these polymerases; more surprising however was the conservation of the structural equivalent of the high affinity manganese-binding site in ϕ6 RdRp. In the majority of structures a magnesium ion is reported to be present, however in the case of PV and WNV RdRps a Ca2+ ion has been modelled, whilst in RV a Mn2+ ion has been built. In all cases these ions are within 3 Å of the high affinity manganese site in ϕ6 RdRp, after automated superposition based on only the protein structure. Based on the available electron density maps the published interpretation of ions looks plausible, except in the two cases where a calcium ion has been modelled (2HCN and 1RDR1). The coordination of these calciums does not agree with the well established pentagonal bipyramid coordination geometry of calcium (64), and it is possible that in these cases a well-ordered magnesium ion (picked up within the cell or during purification and crystallization) is present (see Supplementary Data, Figure 1).
Table 2.
Comparision of the equivalent cations bound in viral RNA polymerases to the structural manganese in ϕ6 RdRP
| Virus | PDB ID | RMSD distance (Å)a | Percentage of ϕ6b | Published ion | Resolution (Å) | Distance from ϕ6 Mn2+c | Comments |
|---|---|---|---|---|---|---|---|
| WNV | 2hcn | 2.7 | 40 | Ca2+ | 2.4 | 1.6 | From map and coordination geometry it is unlikely to be Ca2+ |
| WNV | 2hfz | 2.9 | 50 | Mg2+ | 2.8 | 1.3 | Map and coordination geometry consistent with Mg2+ |
| FMDV | 1wne | 2.8 | 51 | Mg2+ | 3.0 | 1.6 | Structure factors not available |
| RHDV | 1khv | 2.8 | 52 | (H2O) | 2.5 | – | Lu3+ ion 6.0 Å from Mn2+ in ϕ6. Evidence for Mg2+ in place of H2O at Mn2+ site |
| RHDV | 1khw | 2.7 | 52 | – | 2.7 | – | Poor density in region of active site makes interpretation difficult |
| NV | 1sh3 | 2.8 | 52 | Mg2+ | 3.0 | 0.9 | Map and coordination geometry consistent with Mg2+ |
| NV | 3bso | 2.8 | 52 | (H2O) | 1.7 | 1.4 | Water molecule present in equivalent position in the elongation complex |
| DV | 2j7u | 2.8 | 50 | Mg2+ | 1.9 | 1.8 | Map and coordination geometry consistent with Mg2+ |
| PV | 1rdr | 2.9 | 33 | Ca2+ | 2.4 | 2.1 | From map and coordination geometry it is unlikely to be Ca2+ |
| RV | 1mwh | 2.9 | 42 | Mn2+ | 2.5 | 1.0 | Cap bound structure with Mn2+ion bound in equivalent site |
| RV | 1n35 | 2.8 | 42 | – | 2.5 | – | No ion present in equivalent position |
aRMSD of structurally matched Cα atom positions.
bPercentage of ϕ6 RdRp structurally equivalenced.
cDistance of the metal ion binding site from the Mn2+-binding site of ϕ6 RdRp. Based on superposition of Cα atoms using SHP (62) (see Materials and methods section).
Figure 1.
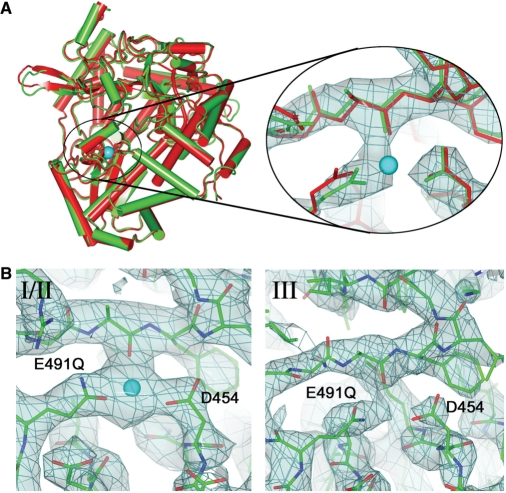
Mn2+ ion binding to ϕ6 wt and E491Q polymerases. (A) Cartoon representation of the overall superposition of the apo E491Q mutant model (green) with the wt model (red). The Mn2+ ion bound to the wt is shown as a cyan sphere. The zoom view shows the manganese-binding site for the same molecule, with the electron density shown in light blue. The wt enzyme is shown in red showing the difference in the side chain position for E491Q. The manganese ion from the wt model is again shown in cyan. (B) The manganese-binding site in the E491Q-RNA-Mn2+ complex, showing electron density for molecule I in the left panel (molecule II is essentially indistinguishable) and for molecule III. The Mn2+ ion, present only in molecules I and II is shown in cyan.
Mutation at the Mn2+-binding site decreases the affinity for structural manganese
A ϕ6 RdRp mutant (E491Q) was constructed, in which one of the three amino acids coordinating the strongly bound manganese ion (7), E491, was substituted with glutamine. Of the three Mn2+ coordinating amino acids (6) E491 was chosen as mutations in D454 (one of the conserved catalytic aspartates) would most likely result in an inactive enzyme whereas mutation in A495 would probably have no effect (interaction coordinated by carbonyl oxygen). The E491Q and wt enzymes were expressed and purified according to published protocols (9) and subjected to biochemical and structural analyses. To illuminate the role of manganese and the effects of the mutation at its binding site, structures of the E491Q mutant alone and in complex with oligonucleotides and/or GTP and/or divalent cations were determined (Table 1).
E491Q mutant complexes crystallized in space group P32, with three molecules (denoted as I, II and III) in the crystallographic asymmetric unit. Surprisingly the crystals have identical space group and similar cell dimensions to the SeMet crystals previously used to solve the wt apoprotein structure (6). However, the wt crystals only possess two molecules in the asymmetric unit (Table 1), whose position and orientation correspond to two of the three molecules in the mutant complex crystals. Close examination of the crystallographic contacts of the wt SeMet P32 crystal form reveal a loose packing of the RdRp molecules with a large gap that, in the case of the mutant enzyme, can accommodate a third molecule, with only small rearrangements at the protein surface of the first two molecules. Nevertheless, there were structural differences between the three molecules in the E491Q crystals, most strikingly at the template, substrate and manganese-binding sites. For the complexes described here, the third molecule (denoted III) exhibits more flexibility, whilst I and II are stabilized by more rigid crystallographic contacts and exhibit similar features to each other. Furthermore, electron density for Q491 was very weak in molecule III, indicating a high degree of flexibility of the side chain, which is presumably linked to the 1.5 Å displacement of loop 66–77 in the thumb domain away from loop 489–493, leading to a relaxation of the thumb and finger domains.
Overall, the E491Q apoenzyme has no significant differences from the wt (rms deviation in Cα positions of 0.4 Å; Figure 1A). However, in contrast to the wt, where bound manganese was seen at high occupancy even when purified in the presence of 0.1–1 mM EDTA and crystallized in the absence of Mn2+ (data not shown), no Mn2+ was detected in the E491Q crystal. As a consequence, Q491 assumes a different conformation, to establish direct interactions with A495 and D454 (these are mediated by the Mn2+ ion in the wt protein). Strikingly, however, when crystals were grown in the presence of 2 mM MnCl2 and an RNA oligonucleotide (complex denoted E491Q-RNA-Mn; see Table 1), a Mn2+ ion was bound in molecules I and II, but not III, indicating that the binding of Mn2+ by the E491Q mutant enzyme is probably dependent on stabilization of the protein by crystal contacts (Figure 1B). Mn2+ was absent in all the three E491Q molecules when crystals were soaked in 25 mM GTP, 5 mM Mg2+ and 5 mM Mn2+ (denoted E491Q-DNA-GTP-Mg2+-Mn2+; Table 1), suggesting that the high concentration of GTP had effectively scavenged the ion.
Manganese binding affects the thermal stability of ϕ6 RdRp
Thermal denaturation assays were used to probe the stability of the wt and E491Q RdRps under different divalent cations conditions. The effect of the E491 to Q mutation on the temperature-dependent unfolding was reflected in these thermofluor measurements (65). The melting curves obtained for E491Q and wt polymerases (Figure 2) revealed that the mutant has a significantly higher melting temperature (50°C) than the Mn2+ bound wt enzyme (46.5°C), suggesting that the structural Mn2+ in the wt apoenzyme reduces its structural stability. In line with this, the stability of the wt enzyme was enhanced in the presence of 5 mM EDTA (Figure 2) which strips off the bound Mn2+ (Salgado, P.S., Stuart, D.I. and Grimes, J.M., unpublished data). Likewise, E491Q polymerase was somewhat destabilised in the presence of excess of MnCl2 (Figure 2).
Figure 2.
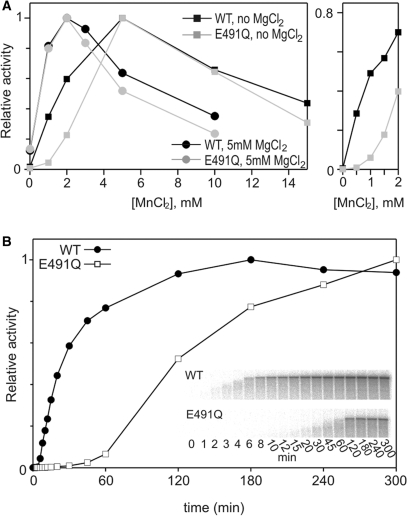
Thermal stability of ϕ6 wt and E491Q RdRps. Thermal melting curves for wt and E491Q polymerases are shown, along with calculated melting temperature (Tm). Wild-type or E491Q were mixed with SYPRO Orange (Molecular Probes, Invitrogen) in starting buffer (50 mM Tris–HCl, 50 mM NaCl) alone and with the addition of either 5 mM EDTA, 10 mM MnCl2 or both 5 mM EDTA and 10 mM MnCl2. The Tm was calculated from the maximum value of the negative first derivative of fluorescence intensity versus temperature; this is approximately the midpoint of the unfolding transition (68). Any shift in Tm under different conditions to that of the starting buffer (i.e. in the presence of an additive), indicates a change in protein stability. An increase in Tm indicates a stabilization of the protein by an increase in structural order and a reduction in conformational flexibility, while a decrease in Tm indicates a destabilization (65).
The mutation at the manganese-binding site affects multiple steps in RNA polymerization
In order to define the optimal conditions for RNA synthesis for the E491Q enzyme, this activity was measured for both mutated and wt protein in varying concentrations of MnCl2, both in the presence and absence of MgCl2 (Figure 3A). When 5 mM MgCl2 was present, the optimal MnCl2 concentration was 2 mM (Figure 3A), whereas in the absence of magnesium, the optimum was 5 mM (Figure 3A) for both enzymes. The specific activity of E491Q was about 75% of the wt in the optimal MnCl2 concentration both in the presence and absence of MgCl2 (data not shown). Higher concentrations of Mn2+ were inhibitory, as has been previously observed for the HCV enzyme as well as for ϕ6-related bacteriophage RdRps (11,12,17,18). Interestingly, there was a marked difference in the stimulatory effect of manganese between the wt and mutant polymerases in the absence of MgCl2 (Figure 3A; right panel), which was not detected in the presence of MgCl2 (resembling conditions within the cell). A basal activity in the presence of MgCl2 only (Figure 3A, first data point, circles) was approximately 1/8 of the optimal activity for both enzymes.
Figure 3.
Optimal divalent cation conditions and polymerization rates for ϕ6 wt and E491Q RdRps. (A) Manganese dependence of the wt and E491Q polymerases in replication reactions with 5 mM MgCl2 and without magnesium. Reaction mixtures containing increasing concentrations of MnCl2, 50 μg/ml sΔ+13 ssRNA template and 270 nM polymerase were incubated at 30°C for 2 h. The reactions were analysed by standard gel electrophoresis and quantified with a phosphoimager (Fuji BAS1500). (A) The graph is normalized setting the highest observed value within each plot to 1. (Right Panel) A phosphorimager analysis of the reactions with 0–2 mM MnCl2 and without MgCl2. The graph is normalised setting the value obtained with 5 mM MnCl2 to 1 within each plot. (B) Time-dependent accumulation of wt- and E491Q-directed replication products. Standard reaction mixtures containing 2 mM MnCl2, 5 mM MgCl2, 270 nM polymerase and 110 μg/ml l+ ssRNA template were incubated at 30°C and 10 μl aliquots, sampled at the time points indicated, were analysed on a standard agarose gel. Inserts show the original gels used for the phosphoimager quantification.
More detailed biochemical characterization of the E491Q polymerase under optimal Mn2+ concentration (in the presence of Mg2+) revealed that the overall rate of polymerization was compromised due to the mutation (Figure 3B). Polymerization reactions were carried out using a 6374-nt long ϕ6 l+ RNA template and aliquots sampled at different time points. The accumulation of full-length L dsRNA over time was plotted for both wt and E491Q polymerases (Figure 3B). Full-length dsRNA accumulated in ∼120 min for wt whereas no plateau was reached for E491Q during the 300 min reaction time. The average polymerization rate was estimated to be 27 bp/s for the wt and 2.5 bp/s for the E491Q enzymes (calculations based on the size of the L segment, see Materials and Methods section for details). The difference in the elongation rates applied also when reactions were carried out in varying MnCl2 concentrations (Supplementary Data, Figure 2). This reduction could reflect difficulties in the template or NTP binding, assembly of an initiation complex or a general slow down of the catalysis during RNA chain elongation. The different components of the polymerization reaction are addressed separately below.
Template RNA binding
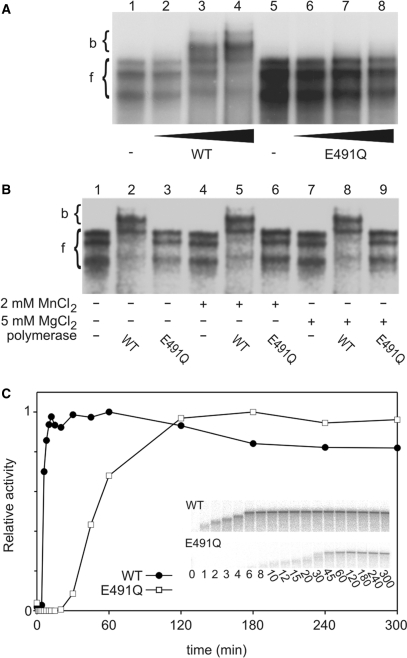
To test whether the reduction in the overall polymerization rate relates to a change in the nucleic acid affinity of E491Q, ssRNA binding was assessed by a gel retardation assay (see Materials and Methods section). The highly structured 5′-terminal region of the ϕ6 genome segment s+ (66) was used for the analysis. This RNA may adopt different folds, which is reflected as mobility differences in non-denaturing polyacrylamide gel (Figure 4A, lanes 1 and 5). Whilst slowly migrating wt complexes were detected in a non-denaturing polyacrylamide gel in the presence of ssRNA template and increasing amount of polymerase (Figure 4A, lanes 1–4), no complexes were detected for E491Q polymerase (Figure 4A, lanes 5–8), suggesting a reduction in template binding. This was not due to different template preferences since E491Q showed no significant difference in specificity for the template 3′-terminal nucleotide compared to wt in replication reactions (data not shown). From the gel depicted in Figure 4A it was possible to deduce an average dissociation constant for the RNA·wt complex, which was in the range of 10−6 M.
Figure 4.
The mutation at the manganese-binding site affects multiple steps in RNA polymerization. (A and B) ssRNA binding to wt and E491Q polymerases. wt and E491Q polymerases were incubated on ice for 10 min with spac ssRNA template and the reaction mixtures were analysed in gel retardation assay. Positions of free (f) and polymerase-bound (b) ssRNA are marked on the left. (A) Template RNA binding in the absence of divalent cations. Lanes: free spac ssRNA (1); wt protein added to 0.02, 0.1 and 0.2 mg/ml (lanes 2–4); free spac ssRNA (5); E491Q added to 0.02, 0.1 and 0.2 mg/ml (lanes 6–8). (B) Effect of MnCl2 and MgCl2 on ssRNA binding. The polymerase–ssRNA mixtures were supplemented with 2 mM MnCl2 or 5 mM MgCl2 as indicated below the gel. (C) Time-dependent accumulation of replication elongation products. In heparin trap assay preincubation mixtures, containing 110 μg/ml l+ ssRNA template, 1 mM ATP and GTP (initiation of 10 nt), 2 mM MnCl2, 5 mM MgCl2 and wt or E491Q polymerase, were incubated for 20 min before the addition of heparin. After heparin incubation [α-32P]UTP and the missing NTPs were added and 10 μl aliquots, sampled at the time points indicated, were analysed on a standard agarose gel. Inserts show the original gels used for the phosphoimager quantification.
Since E491Q had a mutated manganese-binding site, possible effects of divalent cations on the template binding were studied in more detail. As apparent from the autoradiogram, binding of the wt protein clearly shifted the RNA template regardless of the divalent cation conditions (no divalent cations, 2 mM MnCl2 or 5 mM MgCl2; Figure 4B, compare lanes 2, 5 and 8 to 1, 4 and 7) whereas E491Q protein showed no retardation of the RNA template in any of the conditions employed (Figure 4B, lanes 3, 6 and 9). Thus addition of manganese could not reverse the effect of the E491 to Q mutation on template binding. Overall these results indicate that the strong affinity binding site for Mn2+ is required for stable RNA binding.
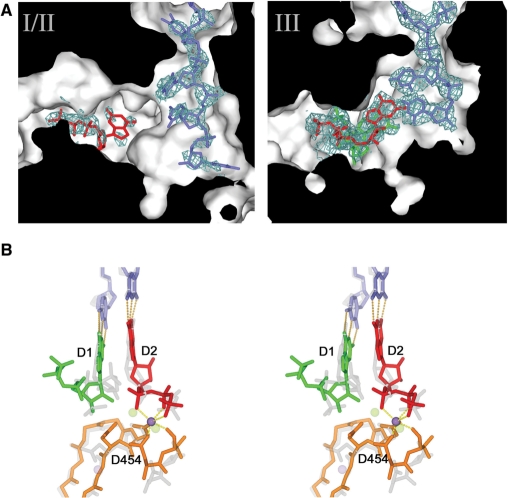
The reduced affinity for RNA was also reflected in the electron density maps of the co-crystals of E491Q with RNA (denoted E491Q-RNA-Mn2+; Table 1). Weak electron density for RNA was visible in the template tunnel in all three molecules. The density was weak, particularly deeper into the template tunnel, indicating that the template may not completely penetrate the tunnel. Whilst in molecule III the electron density was detectable further into the tunnel (data not shown), even here the template did not seem to reach the template specificity pocket [S pocket (6)], assuming an intermediate conformation, similar to that observed previously for a short RNA oligonucleotide (7). In the E491Q-DNA-GTP-Mg2+-Mn2+ crystals, the DNA oligonucleotide was better ordered (Figure 5A), reflecting differences in the binding of DNA and RNA to ϕ6 RdRp.
Figure 5.
NTP and template binding to E491Q polymerase. (A) Cut through of molecules I and III in the E491Q-DNA-GTP-Mg2+-Mn2+ complex to show the substrate and template tunnel, revealing weak density for DNA and GTP in molecules I/II and strong density for an inhibited complex of the DNA oligo (in blue) and 2 GTPs (D1 in green and D2 in red) in molecule III. (B) Stereo view of the inhibited complex compared to the initiation complex described in reference 14. The DNA oligonucleotide and the two GTPs of the inhibited complex are coloured as molecule III (right hand panel), and the manganese ion is coloured in purple. The protein components are in orange. The corresponding components of the active initiation complex (14) are drawn in semi-transparent grey, and the structural manganese ion and the catalytic magnesiums are drawn in semi-transparent purple and green, respectively. The two GTPs are labelled D1 and D2.
NTP binding
Molecules I and II of the E491Q-DNA-GTP-Mg2+-Mn2+ crystals (Table 1) revealed a single GTP molecule in the substrate channel (Figure 5A, left panel). The phosphate backbone of this GTP interacts with the key ‘interrogating’ residues (R225, R268 and R270) at the I site, whilst the base stacks with Y630 from the initiation platform loop. The GTP protrudes further into the substrate tunnel than previously reported for the NTP-bound state (6), providing a snapshot of a further step in the formation of the initiation complex.
Formation of an initiation complex
Whilst molecules I and II in the E491Q-DNA-GTP-Mg2+-Mn2+ crystals (Table 1) have a GTP molecule bound in the substrate tunnel, molecule III has formed an inactive initiation complex, where the mutation at the manganese coordination site causes a number of structural changes. As with the wt initiation competent stage (6), the template DNA established Watson–Crick base-pair interactions with two incoming GTP molecules (Figure 5). However, in the absence of the manganese ion, the C-terminal domain is displaced away from the body of the protein. This displacement of the C-terminal domain moves Y630, the key residue that primes initiation by packing against the NTP complementary to the 3′-terminal nucleotide (T1) of the template, D1 (6), over 4 Å out of the active site. In addition, the side chain of the D454 reorients, establishing hydrogen bonds with the sugar of D1 and the phosphate backbone of the second NTP, D2. As a result, the phosphate backbone of D1 is positioned some 6 Å away from the phosphate backbone of D2. In this conformation, catalysis cannot occur; a situation similar to that observed in the calcium-inhibited complex previously described (7). In addition, unlike the wt initiation complex, where two magnesium ions are found in the active site (6), only one divalent cation (which we believe to be manganese due to the presence of a peak in an anomalous difference Fourier; f″ ≈ 1 eV at 0.87 Å), is found coordinated to the phosphate backbones.
RNA chain elongation
Heparin is a competitive inhibitor of RNA synthesis, preventing initiation but not affecting elongation (47,67) and thus allows analysis of elongation separately. With limiting nucleotides, wt or E491Q enzymes were allowed to extend 10 nt from the 3′-end of the l+ template before addition of heparin to inhibit the formation of new initiation complexes in the pre-incubation mixtures. Subsequently, missing NTPs and [α-32P] labelled UTP were supplied to allow elongation to proceed and aliquots sampled at different time points. The accumulation of full-length dsRNA over time was plotted for both wt and E491Q polymerases (Figure 4C). The E491Q mutant required approximately seven times as long for the complete synthesis of the double-stranded product as the wt enzyme, indicting reduction in the rate of nucleotide incorporation. The elongation rate was estimated to be 31 bp/s for the wt and 4.3 bp/s for the E491Q enzymes (with ϕ6 l+ segment ssRNA template, calculations based on the size of the L segment, see Materials and methods section for details). Despite the decreased elongation rate of the E491Q enzyme, there was no decrease in processivity (Figures 3B and 4E, insets); shorter, apparently incomplete, dsRNA products synthesized by E491Q polymerase were detected until ∼45 min, when full-length dsRNA products appeared (Figure 4E, inset).
DISCUSSION
The detailed biochemical and structural studies of the E491Q mutant form of ϕ6 RdRp reported here reveals that the bound structural manganese ion plays a critical role in the catalytic properties of the enzyme, affecting both initiation and elongation, explaining its stimulatory effect on catalysis. In addition, these structural studies of the mutant have, due to the altered properties of the molecule, allowed the capture of an additional point on the priming pathway as well as an inhibited initiation complex. This binding site is conserved across the vast majority of viral RNA polymerase structures deposited in the PDB databank (Table 2), and reflects the fundamental importance of this divalent cation-binding site to the polymerase activity. In the case of FMDV, NV and DV, a magnesium ion has been seen bound at this conserved site. In the case of WNV and PV polymerases the ion bound has been interpreted as a calcium ion. From the electron density maps calculated from the observed data (63) and the known coordination geometry of calcium, we suggest the ions may be magnesium (the octahedral coordination geometry and electron density levels support this) sequestered from the environment (see Supplementary Data, Figure 1). This would be consistent with the observed mechanism of inhibition of RdRp by calcium, which does not involve binding at this site (7). Indeed we observed that for ϕ6 RdRp, manganese ions sequestered from the environment can occupy this site and that magnesium can replace the bound manganese (Salgado, P.S., Stuart, D.I. and Grimes, J.M., unpublished data).
Key to the effect on initiation and elongation is the structural destabilization on manganese binding, as seen in the relative increase in melting temperature of the E491Q mutant compared to wt enzyme in the thermal shift assays. This increased melting temperature of the mutant can be driven back down to close that of the wt enzyme, by the addition of a great excess of manganese to the buffer. Confirmation that the mutation of E491 to Q reduces the binding affinity of ϕ6 RdRp for this structural manganese comes from the different states of the molecule observed when manganese is present in the soaking buffer. The crystal contacts for molecules I and II partly stabilize a manganese bound form, whilst molecule III is manganese free. This increase in the structural order or rigidity of the mutant on loss of manganese has a direct impact on template binding and initiation, as well as reducing, by close to an order of magnitude, the rate of elongation of the purified enzyme.
Thus it appears that the RdRp requires a degree of structural fluidity for efficient initiation as well as the transition from initiation to elongation, and that the binding of manganese is sufficient to generate this destabilizing effect. The diminished manganese affinity of the E491Q mutant means that only at higher concentrations does it play this destabilizing role, at which point it is approaching inhibitory levels (presumably the inhibition arises from manganese substituting magnesium in the catalytic site).
The mutation also has an effect on the positioning of the C-terminal domain, when an initiation complex forms. This domain contributes the platform essential for priming the initiation reaction relating the effect of the manganese ion to initiation. However this ion also locks the catalytic D454 away from the incoming GTPs (D1 and D2), facilitating the correct positioning of their phosphate backbones (Figure 5), so that in the presence of the catalytic magnesium ions (as observed in the wt initiation complex) the correct geometry is established for productive catalysis to occur. In contrast, in the absence of the manganese ion the side chain of D454 flips up towards the phosphate backbones of D1 and D2, and disrupts their positioning.
Our crystallographic data reveal that the binding of manganese ion plays a key role in establishing the correct geometry for catalysis, essential to both initiation and elongation. We have shown biophysically and biochemically that the binding of manganese is sufficient to destabilize the structure of ϕ6 RdRP, leading to structural fluidity, which enables efficient RNA binding, initiation and elongation. From our analyses of viral polymerases it is clear that this manganese-binding site is conserved as a divalent cation-binding site across a large number of viral RNA polymerases, suggesting that these functions may be broadly conserved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research grants (1213992 and 1213467 to D.H.B.); Finnish Centre of Excellence Program (2006-2011); Human Frontier Science Program (RGP0320/2001-M); SPINE2-COMPLEXES project LSHG-CT-2006-031220; Helsinki Graduate School in Biotechnology and Molecular Biology (to M.R.L.K.); Kuopio Naturalists’ Society (to M.M.P.); the Royal Society (to J.M.G.); the Medical Research Council, UK (to D.I.S.). Funding for open access charge: Medical Research Council, UK and the Academy of Finland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to S. Vehma, J. Turunen and R. Tarkiainen for technical assistance. M. Lehti constructed plasmid pNL18 and Dr E. Makeyev plasmid pEM33. pLM plasmids were a gift from Dr L. Mindich.
REFERENCES
- 1.Lai MM. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 2.Agol VI, Paul AV, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 3.Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 4.Lawton JA, Estes MK, Prasad BV. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 2000;55:185–229. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 6.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 7.Salgado PS, Makeyev EV, Butcher SJ, Bamford DH, Stuart DI, Grimes JM. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure. 2004;12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Makeyev EV, Grimes JM. RNA-dependent RNA polymerases of dsRNA bacteriophages. Virus Res. 2004;101:45–55. doi: 10.1016/j.virusres.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Makeyev EV, Bamford DH. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage ϕ6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurila MR, Salgado PS, Stuart DI, Grimes JM, Bamford DH. Back-priming mode of ϕ6 RNA-dependent RNA polymerase. J. Gen. Virol. 2005;86:521–526. doi: 10.1099/vir.0.80492-0. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Makeyev EV, Bamford DH. Comparison of polymerase subunits from double-stranded RNA bacteriophages. J. Virol. 2001;75:11088–11095. doi: 10.1128/JVI.75.22.11088-11095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Makeyev EV, Butcher SJ, Gaidelyte A, Bamford DH. Two distinct mechanisms ensure transcriptional polarity in double-stranded RNA bacteriophages. J. Virol. 2003;77:1195–1203. doi: 10.1128/JVI.77.2.1195-1203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranjith-Kumar CT, Gutshall L, Kim MJ, Sarisky RT, Kao CC. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 2002b;76:12526–12536. doi: 10.1128/JVI.76.24.12526-12536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi GH, Zhang CY, Cao S, Wu HX, Wang Y. De novo RNA synthesis by a recombinant classical swine fever virus RNA-dependent RNA polymerase. Eur. J. Biochem. 2003;270:4952–4961. doi: 10.1046/j.1432-1033.2003.03897.x. [DOI] [PubMed] [Google Scholar]
- 15.Ranjith-Kumar CT, Santos JL, Gutshall LL, Johnston VK, Lin-Goerke J, Kim MJ, Porter DJ, Maley D, Greenwood C, Earnshaw DL, et al. Enzymatic activities of the GB virus-B RNA-dependent RNA polymerase. Virology. 2003;312:270–280. doi: 10.1016/s0042-6822(03)00247-2. [DOI] [PubMed] [Google Scholar]
- 16.Alaoui-Lsmaili MH, Hamel M, L'Heureux L, Nicolas O, Bilimoria D, Labonte P, Mounir S, Rando RF. The hepatitis C virus NS5B RNA-dependent RNA polymerase activity and susceptibility to inhibitors is modulated by metal cations. J. Hum. Virol. 2000;3:306–316. [PubMed] [Google Scholar]
- 17.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari E, Wright-Minogue J, Fang JW, Baroudy BM, Lau JY, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijk AA, Frilander M, Bamford DH. Differentiation between minus- and plus-strand synthesis: polymerase activity of dsRNA bacteriophage ϕ6 in an in vitro packaging and replication system. Virology. 1995;211:320–323. doi: 10.1006/viro.1995.1409. [DOI] [PubMed] [Google Scholar]
- 21.Ojala PM, Bamford DH. In vitro transcription of the double-stranded RNA bacteriophage ϕ6 is influenced by purine NTPs and calcium. Virology. 1995;207:400–408. doi: 10.1006/viro.1995.1099. [DOI] [PubMed] [Google Scholar]
- 22.Poranen MM, Paatero AO, Tuma R, Bamford DH. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell. 2001;7:845–854. doi: 10.1016/s1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- 23.Poranen MM, Tuma R. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 2004;101:93–100. doi: 10.1016/j.virusres.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Van Dijk AA, Huismans H. The in vitro activation and further characterization of the bluetongue virus-associated transcriptase. Virology. 1980;104:347–356. doi: 10.1016/0042-6822(80)90339-6. [DOI] [PubMed] [Google Scholar]
- 25.Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, et al. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 2007;282:10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 26.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 27.Ng KK, Pendas-Franco N, Rojo J, Boga JA, Machin A, Alonso JM, Parra F. Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 2004;279:16638–16645. doi: 10.1074/jbc.M400584200. [DOI] [PubMed] [Google Scholar]
- 28.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85Å resolution. J. Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 30.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage–structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 31.Ranjith-Kumar CT, Kim YC, Gutshall L, Silverman C, Khandekar S, Sarisky RT, Kao CC. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 2002;76:12513–12525. doi: 10.1128/JVI.76.24.12513-12525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shim JH, Larson G, Wu JZ, Hong Z. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 2002;76:7030–7039. doi: 10.1128/JVI.76.14.7030-7039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laurila MR, Makeyev EV, Bamford DH. Bacteriophage ϕ6 RNA-dependent RNA polymerase: Molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 2002;277:17117–17124. doi: 10.1074/jbc.M111220200. [DOI] [PubMed] [Google Scholar]
- 34.Palmenberg A, Kaesberg P. Synthesis of complementary strands of heterologous RNAs with Qβ replicase. Proc. Natl Acad. Sci. USA. 1974;71:1371–1375. doi: 10.1073/pnas.71.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabor S, Richardson CC. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl Acad. Sci. USA. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van de Sande JH, Loewen PC, Khorana HG. Studies on polynucleotides. CXVIII. A further study of ribonucleotide incorporation into deoxyribonucleic acid chains by deoxyribonucleic acid polymerase I of Escherichia coli. J. Biol. Chem. 1972;247:6140–6148. [PubMed] [Google Scholar]
- 37.Wang TS, Korn D. Specificity of the catalytic interaction of human DNA polymerase beta with nucleic acid substrates. Biochemistry. 1982;21:1597–1608. doi: 10.1021/bi00536a021. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Beaudry A, McSwiggen J, Sousa R. Determinants of ribose specificity in RNA polymerization: effects of Mn2+ and deoxynucleoside monophosphate incorporation into transcripts. Biochemistry. 1997;36:13718–13728. doi: 10.1021/bi971609o. [DOI] [PubMed] [Google Scholar]
- 39.Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 40.Gottlieb P, Strassman J, Mindich L. Protein P4 of the bacteriophage ϕ6 procapsid has a nucleoside triphosphate-binding site with associated nucleoside triphosphate phosphohydrolase activity. J. Virol. 1992;66:6220–6222. doi: 10.1128/jvi.66.10.6220-6222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage ϕ6. Virology. 1994;202:258–263. doi: 10.1006/viro.1994.1341. [DOI] [PubMed] [Google Scholar]
- 42.Poranen MM, Koivunen MRL, Bamford BH. Non-templated terminal nucleotidyltransferase activity of dsRNA bacteriophage ϕ6 RNA-dependent RNA polymerase. J. Virol. 2008;82:9254–9264. doi: 10.1128/JVI.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frilander MJ, Turunen JJ. RNA ligation by T4 DNA ligase. Weinheim, Germany: Wiley-VCH; 2004. [Google Scholar]
- 44.Frilander M, Bamford DH. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage ϕ6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- 45.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 46.Pagratis N, Revel HR. Detection of bacteriophage ϕ6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology. 1990;177:273–280. doi: 10.1016/0042-6822(90)90480-f. [DOI] [PubMed] [Google Scholar]
- 47.Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 48.Konarska MM, Sharp PA. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989;57:423–431. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- 49.Walter TS, Diprose J, Brown J, Pickford M, Owens RJ, Stuart DI, Harlos K. A procedure for setting up high-throughput nanolitre crystallization experiments. I. Protocol design and validation. J. Appl. Cryst. 2003;36:308–314. [Google Scholar]
- 50.Brown J, Walter TS, Carter L, Abrescia NGA, Aricescu AR, Batuwangala TD, Bird LE, Brown N, Chamberlain PP, Davis SJ, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. II. Crystallization results. J. Appl. Cryst. 2003;36:315–318. [Google Scholar]
- 51.Walter TS, Diprose JM, Mayo CJ, Siebold C, Pickford MG, Carter L, Sutton GC, Berrow NS, Brown J, Berry IM, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Cryst. 2005;D61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGraw T, Mindich L, Frangione B. Nucleotide sequence of the small double-stranded RNA segment of bacteriophage ϕ6: novel mechanism of natural translational control. J. Virol. 1986;58:142–151. doi: 10.1128/jvi.58.1.142-151.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gottlieb P, Metzger S, Romantschuk M, Carton J, Strassman J, Bamford DH, Kalkkinen N, Mindich L. Nucleotide sequence of the middle dsRNA segment of bacteriophage ϕ6: placement of the genes of membrane-associated proteins. Virology. 1988;163:183–190. doi: 10.1016/0042-6822(88)90245-0. [DOI] [PubMed] [Google Scholar]
- 54.Frilander M, Poranen M, Bamford DH. The large genome segment of dsRNA bacteriophage ϕ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Macromol. Cryst. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Butcher SJ, Makeyev EV, Grimes JM, Stuart DI, Bamford DH. Crystallization and preliminary X-ray crystallographic studies on the bacteriophage ϕ6 RNA-dependent RNA polymerase. Acta Cryst. 2000;D56:1473–1475. doi: 10.1107/s0907444900010702. [DOI] [PubMed] [Google Scholar]
- 57.Adams PD, Grosse-Kunstleve RW, Hung L.-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Cryst. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 60.Collaborative Computational Project,N. Acta Cryst. 1994;D50:760–763. [Google Scholar]
- 61.DeLano WL. 2004. DeLano Scientific LLC, San Carlos. [Google Scholar]
- 62.Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 63.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala Electron-Density Server. Acta Cryst. 2004;D60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 64.Harding MM. Geometry of metal-ligand interactions in proteins. Acta Cryst. 2001;D57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 65.Geerlof A, Brown J, Coutard B, Egloff MP, Enguita FJ, Fogg MJ, Gilbert RJ, Groves MR, Haouz A, Nettleship JE, et al. The impact of protein characterization in structural proteomics. Acta Cryst. 2006;D62:1125–1136. doi: 10.1107/S0907444906030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pirttimaa MJ, Bamford DH. RNA secondary structures of the bacteriophage ϕ6 packaging regions. RNA. 2000;6:880–889. doi: 10.1017/s1355838200992598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dynan WS, Burgess RR. In vitro transcription by wheat germ ribonucleic acid polymerase II: effects of heparin and role of template integrity. Biochemistry. 1979;18:4581–4588. doi: 10.1021/bi00588a019. [DOI] [PubMed] [Google Scholar]
- 68.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol. Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.