Abstract
The Epstein–Barr virus (EBV)-encoded latent membrane protein-1 (LMP1), a functional homologue of the tumor necrosis factor receptor family, substantially contributes to EBV's oncogenic potential by activating nuclear factor-κB (NF-κB). miR-155 is an oncogenic miRNA critical for B-cell maturation and immunoglobulin production in response to antigen. We report that miR-155 expression is much higher in EBV-immortalized B cells than in EBV-negative B cells. LMP1, but not LMP2, up-regulated the expression of miR-155, when transfected in EBV-negative B cells. We analyzed two putative NF-κB binding sites in the miR-155 promoter; both sites recruited NF-κB complex, in nuclear extract from EBV-immortalized cells. The exogenous expression of LMP1, in EBV-negative background, is temporally correlated to induction of p65 with binding on both NF-κB sites and with miR-155 overexpression. The induction of p65 binding together with increased RNA polymerase II binding, confirms that LMP1-mediated activation of miR-155 occurs transcriptionally. In reporter assays, miR-155 promoter lacking NF-κB binding sites was no longer activated by LMP1 expression and an intact AP1 site is needed to attain maximum activation. Finally, we demonstrate that LMP1-mediated activation of miR-155 in an EBV-negative background correlates with reduction of protein PU.1, which is a possible miR target.
INTRODUCTION
Epstein–Barr virus (EBV), which infects over 90% of the adults, appears to have evolved to exploit the normal biology of B-cell development in order to persist as a life-long asymptomatic infection. However, EBV can contribute to oncogenesis. Indeed, it is frequently found in Burkitt's lymphoma, Hodgkin's lymphoma, nasopharyngeal carcinoma and lymphoproliferative diseases in immune-suppressed individuals (1,2). EBV-related oncogenesis is principally associated with latency during which only a limited subset of the full repertoire of viral genes are transcribed. In addition to protein-coding genes, the non-coding RNAs EBER1, EBER2 and a set of EBV-encoded microRNAs (miRNAs) are expressed in all three forms of latency (2). Of the genes expressed during viral latency in EBV-associated diseases, LMP1 is the one most implicated in tumor formation (3,4). LMP1 is invariably expressed in Burkitt's lymphoma, it is required for EBV-mediated transformation of lymphocytes in vitro and it transforms rodent fibroblasts. Moreover, transgenic mice expressing LMP1 under the control of an immunoglobulin promoter develop B lymphocyte tumors more frequently than wild-type mice (3).
LMP1 is a six-transmembrane constitutively active signaling molecule that functionally mimics members of the cellular tumor necrosis factor receptor (TNFR) family. While the transmembrane domains are required for aggregation and constitutive activation of LMP1 (5–8), two cytoplasmic domains, i.e. the C-terminal activator regions 1 and 2 (CTAR-1 and -2), are critical for the transforming properties of LMP1 (9–12). Together, these signaling domains act through cellular TNFR-associated factors (TRAFs) and other cell-signaling molecules to activate, three transcription factors, namely, nuclear factor-κB (NF-κB), activator transcription factor-2 (ATF)-2 and AP-1, via c-Jun N-terminal kinase (JNK) (8–12). By manipulating these cell-signaling pathways, LMP1 affects host cellular processes that regulate cell proliferation, migration and apoptosis, thereby contributing to cellular immortalization and tumorigenesis.
The miR-155 is processed from a primary transcript, known as ‘BIC’ (B-cell integration cluster), whose upstream region was originally found to be a frequent site of integration of the avian leucosis virus in lymphomas (13). Transgenic mouse studies demonstrated that B-cell targeted expression of BIC leads to the development of B-cell malignancies (14). Furthermore, a number of miRNA profiling studies have shown elevated levels of miR-155 in a wide array of cancers including lymphomas (14–19). miR-155 is one of the miRNAs most frequently implicated in cancers. It plays a critical role in lymphocyte activation in vivo (20,21) and is induced by a variety of immune cell stimuli, i.e. toll-like receptor (TLR) ligands, TNF-α, interferon-beta (IFN-β) and antigens [B-cell receptor (BCR) engagement] (16,22). The mechanism by which miR-155 is regulated after TLR and IFN signaling in macrophages is still unknown. Here we show that LMP1 upregulates the expression of miR-155 mainly by activating the NF-κB pathway, which suggests that miR-155 can cooperate in the EBV-mediated transformation of B cells.
MATERIALS AND METHODS
Cell culture
EBV-negative human B cells (DeFew) and EBV-immortalized human B cells MC3 (kindly provided by Prof. Giuseppe Scala, University ‘Magna Grecia’ Catanzaro) Devozione, Cap (kindly provided by Dr Giuseppina Ruggiero, University ‘Federico II’, Naples), were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen) and 2 mM glutamine. Murine embryonic fibroblasts (MEFs) and HEK 293 referred to as LinX, stably transfected with the helper vector, [LinX cell line, Openbiosystem, (http://www.openbiosystems.com/RNAi/LinX/)] were cultured in DMEM medium (GIBCO-Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 2 mM glutamine.
RNA preparation and northern blot
Total RNA was extracted using Trizol reagent (Invitrogen) according to supplier's protocol. Northern blot analysis was carried out on 15 μg of total RNAs. Briefly, all RNA samples were dissolved in loading buffer [0.05% bromophenol blue, 0.05% cyanol, 5% Ficoll (type 400), 80% formamide and 7 M urea], boiled for 5 min at 95°C and loaded onto 15% polyacrylamide gel under denaturizing conditions [15% acrylamide-bis-acrylamide 19/1, 45 mM Tris, 45 mM boric acid, 1 mM EDTA pH 8, 7 M urea, 0.01% N,N,N′,N′-tetramethylethylenediamine (TEMED) and 0.1% ammonium persulfate (APS)]. Samples were resolved by electrophoresis for 90 min at 150 V and transferred onto nylon membranes (Hybond N+, Amersham/GE Healthcare, Little Chalfont, UK) by capillary blot. The nylon membranes were then equilibrated in 1 M NaCl and pre-hybridized in 6× SSC, 5× Denhart's solution (1× Denhart's solution = 0.1% Ficoll, 0.1% polyvinyl pyrrolydone and 0.1% bovine serum albumin), 5 μg/ml of sheared salmon sperm ds-DNA (Ambion, Austin TX, USA) at 42°C for 2 h. After pre-hybridization, 1 × 106 CPM/ml of [γ32P-ATP] radiolabeled oligonucleotide probe was added and the hybridization carried out overnight at 42°C. The membranes were washed twice in 2× SSC, 1% SDS at 42°C for 30 min and exposed either by autoradiography or by phosphorimage screen (Amersham/GE Healthcare). The signals were quantified with image-scanning or by Image-J software analysis.
Western blot analysis
The cell pellets were resuspended in 1 ml of lysis buffer [10 mM HEPES, pH 7.4, 150 mM KCl, 1 mM EDTA pH 8, 1% Triton X-100, 1 mM DTT, 1 mM orthovadanate, 1 mM NaF and protease inhibitor cocktail (Roche, Basel, Switzerland)] at 4°C for 20 min. Then, 40 μg of proteins, of each sample, were resuspended in Laemmli buffer (23) and resolved by SDS–PAGE (running gel: 0.4 M Tris, pH 8.8, 12% acrylamide/bis-acrylamide 37/1, 0.1% SDS, 0.01% TEMED, 0.1% APS; staking gel: 0.07 M Tris pH 6.8, 5% acrylamide/bis-acrylamide 37/1, 0.1% SDS, 0.01% TEMED, 0.1% APS). The proteins were transferred onto nitrocellulose membranes (Hybond ECL, Amesham/GE Healthcare) by electroblotting. Membranes were blocked in 5% of ‘non-fat milk’ (BioRad, Hercules CA, USA) 0.5% BSA in phosphate buffer solution for 2 h at room temperature and immunoblotted with monoclonal antibody against LMP1 (kindly provided by Dr Dong Yun Lee, McArdle Laboratory for Cancer Research, University of Wisconsin-Madison), mouse polyclonal antibody against γ-tubulin (Sigma Chemical Company, St Louis MO, USA) mouse monoclonal antibody against p65 (SantaCruz, CA, USA), rabbit polyclonal antibody against IκBα (SantaCruz), rabbit polyclonal against H3 (SantaCruz) or rabbit polyclonal antibody against PU.1 (Santa Cruz). Specific secondary antibodies (Sigma Chemical Company), horse-radish peroxidase conjugated, were used for protein detection by enhanced chemiluminescence (ECL, Amesham/GE Healthcare) followed by autoradiography (Hyperfilm ECL, Amesham/GE Healthcare). We used the Image-J software for densitometric analysis.
Cloning and mutagenesis of BIC/miR-155 promoter
The human BIC/miR-155 promoter region extending from 1783 to 1, from 1380 to 1, from 1065 to 1 relative to the start site was isolated from MC3 genomic DNA by PCR using the following primers:
Fw AGCTGAGCTCGAAAGGTCACCCTAGAATTG (−1783),
Fw AGCTGAGCTCGATCTGGCACATGGTAAATG (−1380),
Fw AGCTGAGCTCCAGTCACATGTTGATGAGGC (−1065),
Rev CATGAAGCTTATCCGCTCCCTTCCCGAG (+1).
The isolated fragments were digested with SacI and HindIII and cloned into SacI and HindIII cut pGL3basic (Promega, Madison, WI, USA). The entire promoter region was sequenced and no discrepancies were identified relative to the Genebank genomic sequence (http://www.ncbi.nlm.nih.gov/Genbank/).
Mutagenesis of the pLuc1380 and pLuc1783 reporter plasmid was carried out with the ‘Expand Long Template PCR system’ (Roche) and the following oligonucleotides: 5′-GTA AAT TAA GTA CTA TGC TCG AGC CAG CTC TGA CAT G-3′ and 5′-CAT AGT ACT TAA TTT ACA GAT GGC TCA GGT TGG TTA AG-3′ (NF-kB1), Fw 5′-CAACCTAGAATGAGAAATGCTCGAGTCAGAAAGGCATTGTAGG-3′ and Rev 5′-CATTTCTCATTCTAGGTTGAACTATACCTCCCTTCTCCCAGTG-3′ (NF-κB2), 5′-GGC GCC TGG TCG GTT ATC TCG AGC AAG TGA GTT ATA AAA-3′ and 5′-ATA ACC GAC CAG GCG CCT TTT CTG CAA CCC-3′ (AP-1). In each case, the core transcription factor binding site was replaced by a XhoI restriction site. Mutations were initially screened by digesting with XhoI and then verified by sequence analysis.
Reporter analysis
For each reporter plasmid, 2 × 106 MEFs were distributed in 6 cm dish plates containing 5 ml RPMI1640 (Invitrogen) 10% FBS (Cambrex, East Rutherford, NJ, USA), 2 mM glutamine. FuGene (Roche) of 6 μl were added to 300 μl of DMEM and incubated for 5 min at room temperature. Each reporter vector of 0.5 μg plus 0.2 μg of β-galactosidase vector were then added to the mixture, the tubes were shacked and incubated at room temperature for 15 min. For each transfection, the mixtures were then added to each plate and plates were incubated at 37°C, 5% CO2 for 48 h. Cells were harvested 48 h later and assayed for luciferase activity. Results are presented as average of three independent experiments.
For each reporter plasmid, 2 × 106 DeFew were distributed in 6 cm dish plates containing 5 ml RPMI1640 (Invitrogen) 10% FBS (Cambrex), 4 mM glutamine. FuGene (Roche) of 8 μl were added to 300 μl of DMEM and incubated for 5 min at room temperature. Each reporter vector of 1 μg plus 0.5 μg of β-galactosidase vector were then added to the mixture, the tubes were shacked and incubated at room temperature for 15 min. For each transfection, the mixtures were then added to each plate and plates were incubated at 37°C, 5% CO2. After 6 h each sample was split in two, and one of them was treated with 10 ng/ml of phorbol myristoylated acetate (PMA). Cells were harvested 48 h later and assayed for luciferase activity. Results are presented as average of three independent experiments.
Chromatin immunoprecipitation assay
MC3 or DeFew cells (5 × 106 cells) were exposed to 1% formaldehyde for 10 min at 37°C to obtain protein–DNA cross-linking. The nuclear fraction was sonicated to yield chromatin fragments of 200–1000 bp; 5% of the total volume was removed from each sample and used as the input fraction. Chromatin was pre-cleared by pre-incubation with a DNA salmon sperm/protein A-agarose 50% slurry (Upstate, Temecula, CA, USA) for 1 h at 4°C. The agarose was centrifuged, and the pre-cleared chromatin supernatant was incubated with the antibodies (3 μg) against RNA pol II (mAb anti RNA polymerase II, Active Motif, Rixensart, Belgium), p50 or p65 (pAb anti p50 or pAb anti p65, kindly provided by Dr N. Rice, Frederick Cancer Research and Development Center, Frederick, MD, USA), against histone 3 (pAb anti H3, SantaCruz), overnight at 4°C. The protein–DNA-antibody complexes were collected by the addition of the salmon sperm DNA–protein A-agarose (2 h at 4°C) and washed, and protein–DNA cross-linking was reversed (4 h at 65°C). DNA was purified by phenol/chloroform extraction and ethanol precipitation and aliquots (25%) of the purified materials underwent PCR (5 min at 94°C, 1 min at 94°C, 1 min at 52°C, 1 min at 72°C for 40 cycles, 5 min at 72°C).
Preparation of nuclear extracts and electrophoresis mobility shift assay
MC3 cells (5- to 6 × 106 cells) were washed with cold phosphate buffered saline and harvested. The cell pellet was resuspended in extraction buffer containing 10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.5 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin, passed through a needle, kept on ice for 45 min, and centrifuged (15 min at 14 000 r.p.m. at 4°C). The nuclear pellet was then resuspended in high salt extraction buffer containing 10 mM HEPES, pH 7.9, 0.4 mM NaCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 10 µg/ml aprotinin, 10 µg/ml leupeptin and incubated for 45 min at 4°C. The nuclear extract supernatant was obtained by centrifugation (30 min at 14 000 r.p.m. at 4°C), protein concentration was determined and 5 µg aliquots were stored at −80°C until used. Double-stranded synthetic oligonucleotides were radiolabeled using [γ32P]ATP (3000 Ci mmol–1; Amersham Biosciences) and T4 polynucleotide kinase (Fermentas, Burlington, ON, Canada). The binding reaction was carried out for 20 min at room temperature with 5 µg of nuclear proteins in 10% glycerol, 60 mM KC1, 1 mM EDTA, 1 mM dithiothreitol and 0.250 µg/µl poly[dI–dC] containing 50 000 cpm of radiolabeled probe and a 80-fold molar excess of unlabeled competitor oligonucleotide when indicated. For supershift experiments, 4 µg of specific anti-p65 or anti-p50 were added to the binding reaction and incubated for 30 min, after which we added the radiolabeled probe. DNA–protein complexes were separated by 5% non-denaturing polyacrylamide gel and revealed by either autoradiography or phosphorimage screen (Amersham Biosciences).
Transient transfection
DeFew cells (3 × 106 cells) were centrifuged, washed in PBS and resuspended in 300 μl of electroporation medium (RPMI 1640 medium, 20% FBS and 2 mM Glutamine). Cells were mixed with 20 μg of plasmid expressing LMP1 (pcDNA3-LMP1), or LMP2 (pcDNA3-LMP2) or both and subsequently transferred in electroporation cuvette. Cells were electroporated twice at 220 V, 960 μFa and incubated 5 min on ice. Cells were plated in 10 ml of RPMI 1640 medium, 10% FBS and 2 mM glutamine for 2 days. After 2 days, they were harvested and assayed for miR-155 and LMP1 expression.
Viral generation, infection and time course
The retroviral vector LMP1pBABE-puro was kindly provided by Prof. Eiji Hara (University of Tokushima, Japan). LinX cell line, stably transfected with the helper vector, were grown in DMEM medium 10% FBS, 2 mM glutamine and 100 μg/ml hygromicin. Transfections with LMP1pBABEpuro or pBABEpuro (empty vector) were as follows: 6 cm plate of semi-confluent (about 1 × 106 cells) cells were transfected and 7.5 μg of DNA vector incubated in 20 μl of FuGene and incubated in 4 ml of medium without antibiotic for 24 h. After incubation, the medium was harvested, replaced with 4 ml of fresh medium and the LinX cells incubated for 24 h for a second round of viral generation.
DeFew cells (4 × 106 cells) were centrifuged, washed in PBS and resuspended in 4 ml of virus containing medium plus 4 μg/ml of Polybrene (Sigma). The infection was performed by spinoculation, namely by centrifugation at 3500 r.p.m. for 90 min at 20°C. After centrifugation, the cells were resuspended in 10 ml of RPMI and incubated at 37°C, 5% CO2. Twenty-four hours later, the cells underwent a second round of infection. After 48 h, cells were treated with 1 μg/ml of puromicine to start selection of positive clones.
For the time course, DeFew cells (4 × 106 cells) were centrifuged, washed in PBS and resuspended in 12 ml of virus containing medium plus 12 μg/ml of Polybrene (Sigma). The infection was performed by spinoculation, namely by centrifugation at 3500 r.p.m. for 90 min at 20°C. After centrifugation, the cells were resuspended in 10 ml of RPMI and incubated at 37°C, 5% CO2. Twenty-four hours later, 1 μg/ml of puromocine was added to cells to start selection. Cells were harvested at 6, 24, 48 and 72 h after infection and assayed for miR-155 and LMP1 expression.
Transient transfection of MC3
MC3 cells (3 × 106 cells) were centrifuged, washed in PBS and resuspended in 300 μl of electroporation medium (RPMI 1640 medium, 20% FBS and 2 mM glutamine). Cells were mixed in a tube with 20 μg of pRc/CMV-HA-IκBα-S32/36A (kindly provided by Prof. Ileana Quinto, University Magna Grecia, Catanzaro) and subsequently put in electroporation cuvette. Cells were electroporated twice at 220 V, 960 μFa and incubated 5 min on ice. Cells were plated in 10 ml of RPMI 1640 medium, 10% FBS and 4 mM glutamine for 2 days. After 2 days, they were harvested and assayed for miR-155, p65 and IκBα expression.
RESULTS
The expression of mir-155 is regulated by LMP1
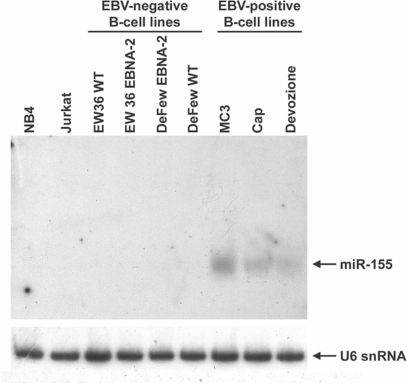
We measured miR-155 expression in three EBV-immortalized cell lines (MC3, Devozione and Cap) and in two EBV-negative B-cell lines (EW36 and DeFew), a T-cell lymphoma cell line (Jurkat) and NB4 from acute promyelocytic leukemia. As shown in Figure 1, miR-155 was expressed at relevant levels only in the EBV-positive cells according to Mrazek et al. (24). In the EBV-negative B cells, miR-155 expression was not affected by stable transfection of the EBV nuclear factor 2 (EBNA2) (Figure 1), a viral transacting factor that activates the expression of such cellular and viral promoters as the HIV-1 long terminal repeat (25) and the interleukin-6 promoter (26).
Figure 1.
miR-155 is upregulated in EBV-immortalized cell lines. Northern blot analysis on total RNAs obtained from 9 cell lines. The blot was analyzed for the expression of mature miR-155 and normalized by the ubiquitous small nuclear RNA U6. Note that basal levels of miR-155 expression in DeFew cells are not visible in this blot due to the brief exposure.
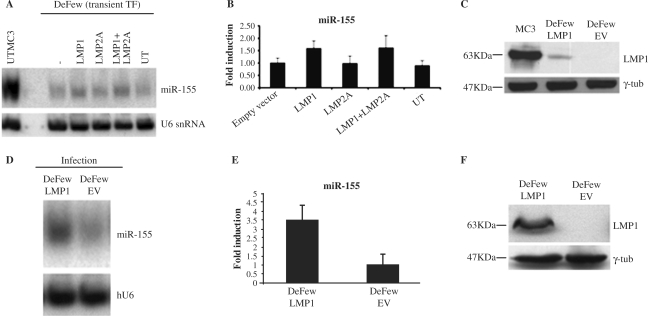
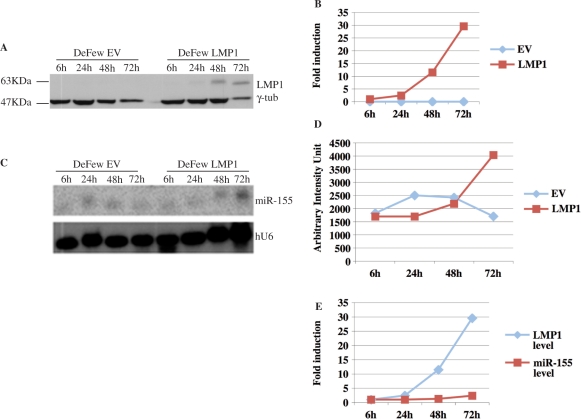
To evaluate whether LMP1 induces miR activation, we transiently transfected a vector expressing LMP1, LMP2 or both in DeFew cells (EVB-negative B cells). There was a 1.6-fold induction of miR-155 expression in LMP1-transfected cells, no induction in LMP2-transfected cells and a 1.7-fold induction in cells transfected with LMP1 and LMP2 (Figure 2A and B). The significance of this finding is underestimated because only a few cells were transfected by LMP1 expression vector. Indeed, LMP1 was much lower in transiently transfected DeFew cells than in MC3 cells (Figure 2C). To overcome this problem, we infected the DeFew cells with a retroviral vector expressing LMP1 (pBABEpuroLMP1). As shown in Figure 2D, we obtained a higher expression of LMP1, at 72 h post-infection, resulting in about 3.5-fold induction of miR-155 expression compared to DeFew cells infected with the empty virus (Figure 2E and F).
Figure 2.
LMP1 enhances the expression of miR-155 in an EBV-negative B-cell line. (A) A representative northern blot analysis on total RNAs obtained from DeFew cells transfected (TF) as indicated or untransfected (UT). The blot was analyzed for the expression of mature miR-155 and normalized by the ubiquitous small nuclear RNA (U6). (B) Graphic representation of the northern blot results as average of three independent experiments. (C) Western blot analysis of LMP1 expression in the transfected cells compared to MC3 cell immortalized by EBV. Input proteins were equalized by detecting the endogeneous γ-tubulin. (D) Northern blot analysis of total RNAs obtained from DeFew cells at 72 h post-infection with retroviral vector expressing LMP1 or empty vector. The blot was analyzed for the expression of mature miR-155 and normalized by U6. (E) Graphic representation of the northern blot results as average of three independent experiments. (F) Western blot analysis of the expression of LMP1 upon infection with the retroviral vector. Input proteins were equalized by detecting the endogeneous γ-tubulin.
miR-155 expression can be driven through activation of two NF-κB elements in the BIC/miR-155 promoter
The foregoing results suggested that the LMP1-mediated induction of miR-155 occurred via a transcriptional mechanism. A recent study showed that the miR-155 promoter contains an AP-1 active site, located at 40 nt upstream from the TATA box and a NF-κB site located at 1150 nt upstream from the transcription starting site (27). The AP-1 active site plays a key role in activation of miR-155 expression induced by BCR (27). To identify potential cis-elements active on miR-155, we isolated a promoter region of ∼1.9 kb of the BIC gene, which encodes miR-155 (28), from MC3 cells and cloned it into a luciferase reporter plasmid. We looked for cis-element homologies using TESS (http://www.cbil.upenn.edu/cgi-bin/tess) or TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html), and identified some potential transcription factor binding sites, among which, an additional NF-κB site located at 1697 nt upstream from the transcription starting site (Supplementary Material Figure 1). Because NF-κB signaling plays a central role in B-cell physiology, we focused on the two NF-κB cis-elements.
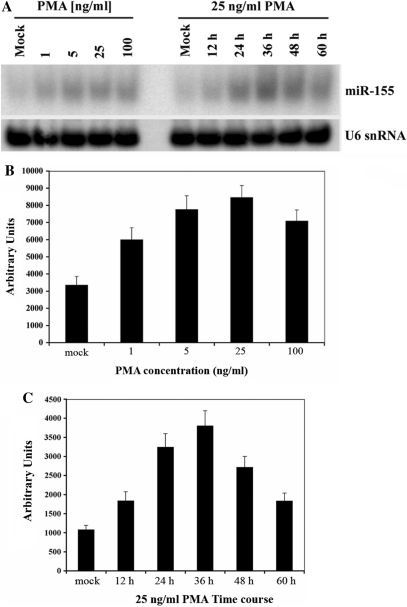
We next examined by northern blot the induction of miR-155 expression in DeFew cells treated with PMA, which induces NF-κB by activating protein kinase C (PKC) and related phosphorylation of inhibitor-κBα (IκBα), thereby leading to its degradation (29). PMA (25 ng/ml) increased miR-155 expression by about 3-fold (Figure 3A and B); miR-155 expression peaked 36 h after treatment and decreased thereafter (Figure 3A and C). The latter finding is in accordance with the pattern of PKC-activated NF-κB expression (29).
Figure 3.
PMA induces miR-155 expression in a dose- and time-dependent manner. (A) Left: northern blot analysis of RNAs from DeFew cells treated with different concentrations of PMA; Right: northern blot analysis of RNAs from DeFew cells at different times upon PMA treatment. The blots were analyzed for the expression of mature miR-155 and normalized by U6. (B and C) Graphic representation of the average of three independent northern blot experiments.
The NF-κB elements in the BIC/miR-155 promoter bind NF-κB factors in vitro and in vivo in MC3 cells
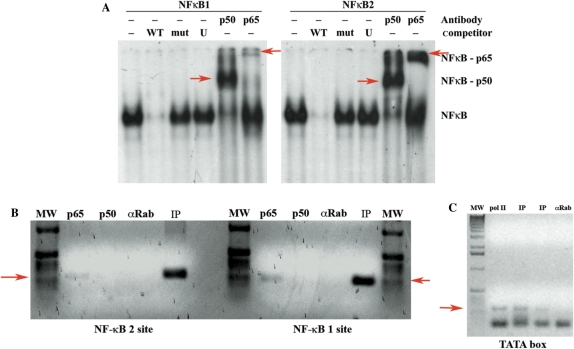
To determine the activity of the two NF-κB elements identified in the miR-155 promoter, we performed EMSA using nuclear extracts from MC3 cells. Briefly, the nuclear extracts were incubated with dsDNA oligonucleotides identical to the sequence of the NF-κB site 1 (−1150) or 2 (−1697). Both sites underwent an electrophoretic mobility shift, which disappeared when we used a 80-fold molar cold competitor, but not when we used the same amount of a mutant competitor that is not bound by NF-κB complexes (Figure 4A). There was a supershift when the nuclear proteins were pre-incubated with antibodies against p65/RelA or p50, two members of the NF-κB family (Figure 4A). Because LMP1 could activate the p52/p65 NF-κB complex (non-canonical pathway) (30,31), we repeated the experiment using an anti p52 antibody, and found no supershift under these conditions (data not shown). In line with the hypothesis that the canonical NF-κB pathway is involved in the LMP1-mediated activation of miR-155, we found that miR-155 expression was inhibited in MC3 cell expressing an exogenous, undegradable form of IκBα (32), which is an inhibitor of the canonical NF-κB pathway (Supplementary Figure 2).
Figure 4.
The two putative NF-κB binding sites are bound by NF-κB complexes in vitro and in vivo in MC3 cells. (A) Electrophoretic mobility shift assay using nuclear extracts from MC3 cells (EBV-positive cells) on the two NF-κB binding sites. Left panel: NF-κB site 1 (−1150). Right panel: NF-κB site 2 (−1697). WT = 80-fold molar cold wild-type competitor; mut = 80-fold molar cold mutant competitor; U = 80-fold molar cold unspecific competitor; p50 = supershift with anti NF-κB p50 antibody; p65 = supershift with anti NF-κB p65 antibody. Supershift bands are indicated with red arrows. (B) ChIP assay of chromatin extracted from MC3 cells using polyclonal antibodies directed against the NF-κB p65 or NF-κB p50. After immunoprecipitation, the DNA samples were purified and subjected to PCR with specific primers amplifying the NF-κB site 1 region (right panel) or NF-κB site 2 region (left panel). MW = molecular weight marker; p65 = pAB anti NF-κB p65; p50 = pAb anti NF-κB p50; αRab = unspecific polyclonal antibody anti rabbit immunoglobulin; IP = input. The red arrows indicate the specific amplified band. (C) ChIP assay of chromatin extracted from MC3 cells using monoclonal antibody directed against the RNA pol II. After immunoprecipitation, the DNA samples were purified and subjected to PCR with specific primers amplifying the TATA box region. MW = molecular weight marker; pol II = mAB anti RNA pol II; IP = input. The two lanes represent two different amounts of input sample used for the PCR; αRab = unspecific polyclonal antibody anti rabbit immunoglobulin. The red arrow indicates the specific amplified band.
To determine whether the NF-κB p50/p65 complex is recruited in vivo on the BIC/miR-155 promoter, we examined the two NF-κB sites by chromatin immunoprecipitation assay (ChIP). We show that p65 but not p50 is engaged on the two NF-κB sites (Figure 4B). However, we cannot rule out that p50 contributes to the active complex because it could simply be covered by DNA and p65, and thus inaccessible to the antibody. RNA pol II drives miR-155 transcription (33). Therefore, we checked, in the same experiment if the BIC promoter was also engaged by the RNA pol II. As shown in Figure 4C we found the RNA polymerase on the TATA box of BIC promoter together with p65 (Figure 4C).
LMP1 requires intact NF-κB cis-elements to enhance miR-155 expression
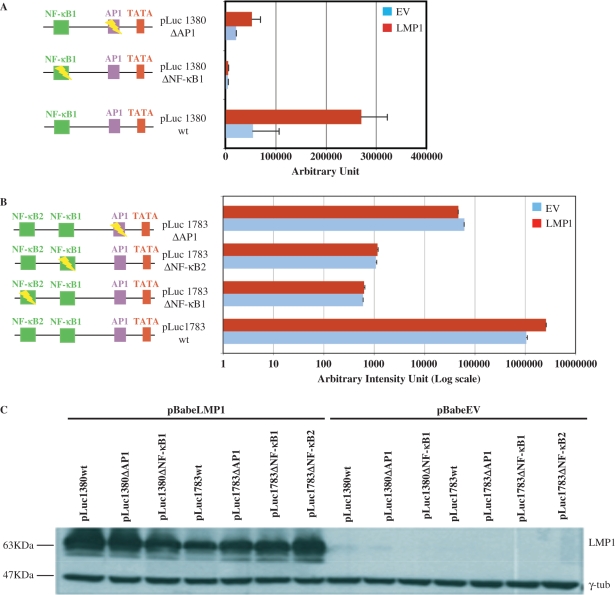
The foregoing experiments provide compelling evidence that LMP1 drives miR-155 upregulation and that the miR promoter responds to NF-κB transacting factors. However, LMP1 can activate two different pathways: the NF-κB pathway and the Jun/Fos (AP-1) pathway (6,8–10) and the miR-155 promoter has a cis-element responsive to AP-1 40 nt upstream from the start site (27). Using truncated forms of BIC promoter cloned upstream the luciferase gene reporter, we observed that only the first 1380 nt drive the maximum activation of the promoter by PMA (Figure 5) or LMP1 (Figure 6). This could indicate that the NF-κB site 2 is irrelevant in the LMP1-mediated activation of miR-155. To verify the effectiveness of the two NF-κB cis-elements in LMP1-enhanced miR-155 promoter, we used both the promoters cloned upstream luciferase gene reporter [pLuc1783 wild-type (wt) and pLuc1380 wt]. Subsequently, we mutated NF-κB site 1 (pLuc1783ΔNF-κB1 and pLuc1380ΔNF-κB), the AP-1 site (pLuc1783ΔAP-1 and pLuc1380ΔAP-1) and the NF-κB site 2 (pLuc1783ΔNF-κB2) using site-directed mutagenesis. All vectors were used for transfection and reporter assay in EBV-negative MEF. As shown in Figure 6A, pLuc1380 wt was upregulated by about 5-fold when cotransfected with a vector expressing LMP1. In the presence of LMP1, pLuc1380ΔNF-κB expression was about 80-fold less, and the expression of pLuc1380ΔAP-1 about 6-fold less than pLuc1380 wt expression (Figure 6A). These data would suggest that NF-κB site 1 plays a pivotal role in LMP1 activation and that substantial cooperation by the AP-1 site is required to attain maximum activation. Consistent with these results, NF-κB seems to be central also for basal levels of miR-155 expression. Indeed, without LMP1, pLuc1380ΔNF-κB expression was about 17-fold lower than pLuc1380 wt expression, whereas pLuc1380ΔAP1 was downregulated by about 3-fold (Figure 6A). When the reporter assays were repeated using the full-length vector pLuc1783 wt, induction was reduced (about 3-fold), in the presence of LMP1 compared to pLuc1380 wt (compare Figure 6A and B), which is in line with the results reported in Figure 5. Surprisingly, all mutated forms of this vector (pLuc1783ΔNF-κB2, pLuc1783ΔNF-κB1 and pLuc1783ΔAP-1) were no longer activated by LMP1 expression (Figure 6B). The graph in Figure 6B is presented in logarithmic scale in order to show that the mutations in both NF-κB sites dramatically reduced the basal level of miR-155 promoter-driven luciferase expression, whereas the AP-1 mutation reduced basal levels to a much lesser extent. The latter observation is consistent with our data obtained with the truncated promoter. The expression of LMP1 was checked in each sample (Figure 6C) for a further normalization of the luciferase assays.
Figure 5.
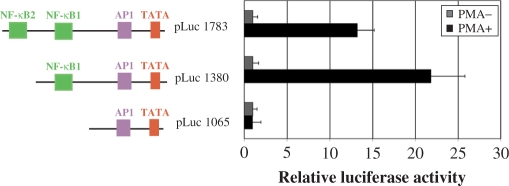
Truncated form of the BIC promoter (pLuc1380) is the most inducible by PMA. Luciferase assay performed in De Few cells using the full-length miR-155 promoter and two truncated forms. The transfected cells were treated or mock treated with 25 ng/ml of PMA. In presence of PMA the truncated form of the BIC promoter (pLuc1380) reaches the maximum of luciferase activity. The graphic representation of the luciferase assay is the average of three independent experiments.
Figure 6.
NF-κB cis-elements are essential for basal and LMP1-mediated activation of miR-155 pomoter. (A and B) The indicated miR-155 promoter luciferase vectors were co-transfected with pBABEpuroLMP1 or empty vector and incubated for 48 h. Luciferase expression was normalized by β-gal/μg of total protein and represented as average of three independent experiments. (C) Western blot analysis of LMP1 expression in the co-transfected MEF cells, normalized by γtubulin.
These data indicate that both NF-κB cis-elements play a central role in LMP1-mediated activation of the promoter and may suggest the possible presence of a negative regulatory region between the two cis-elements.
The binding of p65/RelA to both NF-κB cis-elements of miR-155 promoter is temporally correlated to LMP1 expression
To determine whether the exogenous expression of LMP1, in DeFew cells, is correlated temporally with the upregulation of the endogenous miR-155, we performed a time-course experiment in DeFew cells infected with pBABEpuroLMP1 or with the empty vector as control. Each time point was split into three for western blot, northern blot and ChIP analyses. LMP1 expression was observed 24 h post-infection and peaked at 72 h (Figure 7A and B), whereas the upregulation of miR-155 occurred at 48 h post-infection and increased at 72 h (Figure 7C and D). These results indicate that the upregulation of miR-155 is temporally dependent on the expression of LMP1 (Figure 7E).
Figure 7.
Time-course of LMP1-mediated activation of miR-155. (A) Western blot analysis of protein extracts obtained at indicated times from DeFew cells infected with pBABEpuroLMP1 or empty vector. The blot was analyzed for the expression of LMP1 and normalized by γ-tub. (B) Graphic representation of the western blot results. (C) Northern blot analysis of RNAs obtained at indicated times from DeFew cells infected with retroviral vector expressing LMP1 or empty vector. The blot was analyzed for the expression of miR-155 and normalized by U6. (D) Graphic representation of the northern blot results. (E) Graphic representation of the correlation of miR-155 and LMP1 expression at indicated times.
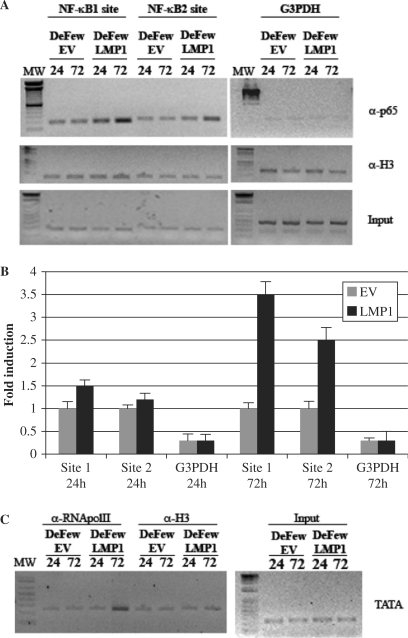
To further clarify the function of the two NF-κB cis-elements in the endogenous miR-155 promoter, we performed ChIP analysis followed by semi-quantitative PCR. There was constitutive binding of p65/RelA on both NF-κB sites (Figure 8A, left panel and Figure 8B). This binding did not increase in the cells infected with the empty vector (Figure 8A, left panel). In contrast, in the cells expressing LMP1, the binding of p65/RelA on NF-κB 1 site increased 1.5-fold at 24 h and 3.5-fold at 72 h (Figure 8A, left panel and Figure 8B), whereas the binding of p65/RelA on the NF-κB 2 site increased 1.3-fold at 24 h and 2.5-fold at 72 h (Figure 8A, left panel and Figure 8B). At the same time points, the binding of RNA pol II on the TATA box of BIC promoter increased in the LMP1-infected cells but not in the control cells (Figure 8C left panel). G3PDH was used as negative control and the assay was normalized against histone H3 (Figure 8A and C, right panel). Taken together, these data and the results reported in Figure 7 strongly suggest that LMP1 activates p65 binding on miR-155 promoter and this lead to upregulation of miR expression.
Figure 8.
Semi-quantitative ChIP on the BIC promoter at indicated times in DeFew cells after retroviral infection with a vector expressing LMP1 or empty vector. ChIP assay of chromatin extracted at the indicated times from DeFew cells infected with pBABEpuroLMP1 or empty vector. (A) left panel: After immunoprecipitation with the indicated antibodies, the DNA samples were purified and subjected to PCR with specific primers amplifying the NF-κB site 1 region or NF-κB site 2 region. MW = molecular weight marker; p65 = pAB anti NF-κB p65; ti NF; H3 = pAb anti Histone H3. Right panel: After immunoprecipitation, the DNA samples were purified and subjected to PCR with specific primers amplifying the G3PDH promoter region. (B) Graphic of the results showed in panel (A; C) After immunoprecipitation with mAb directed against the RNA pol II, the DNA samples were purified and subjected to PCR with specific primers amplifying the TATA box region. MW = molecular weight marker; pol II = mAB anti RNA pol II; H3 = pAb anti Histone H3.
The protein levels of transcription factor PU.1 correlates with LMP1-induced miR-155 expression
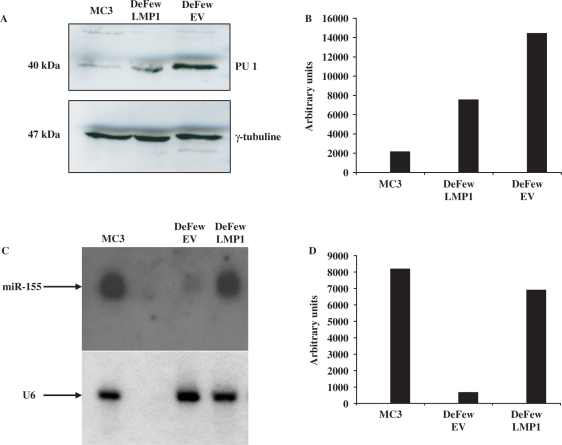
It was recently reported that miR-155 targets PU.1 mRNA thereby affecting its translation (35). Hence, we checked whether the levels of protein PU.1 could be lowered by LMP1 activation of miR-155. To this aim, we infected DeFew cells with the retroviral vector expressing LMP1 or with the empty vector as control. The results show a 2-fold decrease of PU.1 protein levels in the DeFew cells infected with LMP1-expressing vector versus empty vector (Figure 9A and B). In the same experiment, we ran a northern blot analysis to evaluate miR-155 expression. As shown in Figure 9C and D, miR-155 expression in LMP1-expressing DeFew cells was 10-fold higher than in the empty vector. Interestingly, PU-1 levels were very low in EBV-immortalized MC3 cells overexpressing miR-155
Figure 9.
PU.1 protein levels correlate with LMP1-mediated activation of miR-155 expression (A) western blot analysis of PU.1 expression in EBV-immortalized B cells (MC3) EBV-negative B cells (DeFew) infected with pBABEpuroLMP1 (DeFew LMP1) or pBABEpuro, and selected with 1 mg/ml of puromycin for 7 days (DeFew Empty Vector); (B) Graph of densitometric analysis of A normalized by γ-tubuline; (C) northern blot analysis of mir-155 expression in EBV-immortalized B cells (MC3) EBV-negative B cells (DeFew) infected with pBABEpuroLMP1 (DeFew LMP1) or pBABEpuro (DeFew Empty Vector) and selected with 1 mg/ml of puromycin for 7 days; (D) Graph of densitometric analysis of C normalized by U6.
DISCUSSION
microRNAs, like transcription factors, regulate the expression of several genes post-transcriptionally, and are thus important players in such relevant cellular processes as cell growth, differentiation, cell signaling and apoptosis [see (11) for a review]. miRNAs themselves are subjected to fine expression regulation, their transcription being driven by RNA pol II responsive promoters (33). It is generally recognized that miRNAs play a central role in the molecular etiology and maintenance of cancer.
In this study, we describe the activation of miR-155, which is a potential oncomiR (14), by LMP1, a trans-membrane EBV protein considered to be the major oncoprotein of EBV. LMP1 is expressed during viral latency and it activates the signaling pathways NF-κB and AP-1 (12). Using miRNA array, it has been reported that LMP1-induced the expression of numerous miRNAs, such as miR-146, whose expression is upregulated by the LMP1-induced NF-κB pathway (36,37). Here we show that EBV-infected cells constitutively express high levels of miR-155, and that the exogenous expression of LMP1 in EBV-negative B cells enhances miR-155 expression in a time- and dose-dependent manner (Figure 7). Yin et al. (27) reported that the promoter of the BIC gene encoding miR-155 contains a conserved AP-1 element 40 bases upstream from the transcription start site that is essential for the induction of miR-155 mediated by activation of the BCR (27). They also identified a putative NF-κB site at −1150 nt on the BIC promoter whose mutation did not affect miRNA enhancement by activation of the BCR (27). We found an additional NF-κB putative binding site at –1697 nt on the BIC promoter. We show that the endogenous promoter is responsive to PMA (Figure 3), a well recognized activator of the canonical inhibitor kinase kinase-dependent (IKK) (29) NF-κB pathway. Moreover, both putative NF-κB sites were able to bind in vitro the NF-κB proteins p50 and p65 in nuclear extract from MC3 cells (Figure 4A).
Because LMP1 induces both the canonical IKK-dependent (30) and non-canonical NF-κB-induced kinase-dependent (NIK) (31) NF-κB pathways, we tested whether p52 (an NF-κB protein activated by a non-canonical pathway) was part of the binding complex, but we did not detect it. Accordingly, inhibition of the canonical NF-κB pathway by exogenous expression of IκBα led to reduced expression of miR-155 in MC3 (Supplementary Figure 2). This could indicate that the non-canonical pathway is activated only at the beginning of EBV infection when the virus needs to maximize this signaling pathway.
Furthermore, both the NF-κB sites bound to p65 in vivo as did the RNA pol II on the TATA box of the BIC promoter, in MC3, which indicates ongoing miR-155 transcription (Figure 4B and C) suggesting that the two NF-κB sites are active in an EBV-positive background. However, PMA- and LMP1-driven induction was higher with a truncated form of the BIC promoter lacking the distal site (NF-κB site 2) than with the full-length promoter (Figures 5 and 6). To shield light on the effectiveness of the two NF-κB cis-elements, we performed reporter assays using the full length (pLuc1783) or a truncated form (pLuc1380) of miR-155 promoters either wild-type or mutated in the NF-κB site 1, NF-κB site 2 or AP-1. Our results indicate that both NF-κB sites are pivotal for basal and LMP1-induced expression of miR-155 (Figure 6). Conversely, mutation of the AP-1 site in the pLuc1380 reduced the constitutive expression of the promoter, which in fact was still inducible by LMP1 albeit to a lesser extent (Figure 6). However, mutation of the AP-1 site in the pLuc 1783 abolished the LMP1-induced expression. Yin et al. [unpublished data in (34)] suggested that miR-155 promoter contains one or more CpG islands. The presence of putative CpG as well as cis-elements bound by negative regulatory protein(s) or promoter secondary structures could explain why the truncated form of the promoter, lacking this putative NRE, is inducible at higher extent by both PMA and LMP1.
Subsequently, we performed time-course experiments in EBV-negative cells infected with an exogenous LMP1. Our data show that the expression of LMP1 is temporally correlated with the activation of p65 binding on both NF-κB sites (Figure 8), recruitment of RNA pol II and miR-155 induction (Figures 7 and 8). All data reported give a clear indication that LMP1 enhances miR-155 expression, mainly, via activation of the canonical NF-κB pathway.
Our results, together with the previous data reported by Yin et al. (27), illustrate the exquisite regulation of this miR by various stimuli, and highlight its importance in B-cell physiology.
PU.1 is a downstream effector of miR-155 thanks to the presence of a miR-155 complementary seed sequence in the 3′ UTR of PU.1 mRNA (35). In the latter report, B cells lacking miR-155 generated reduced extrafollicular and germinal center responses and did not produce high-affinity IgG1 antibodies. PU.1 is highly expressed in miR-155-deficient B cells and its overexpression in wild-type B cells results in reduced numbers of IgG1-switched cells, which indicates that miR-155 plays a key role in antigen-driven B-cell maturation. PU.1 is a master gene of the Ets transcriptional factor family that promotes cell growth, differentiation and apoptosis thereby playing an important role in hematopoiesis [see (38) for a review]. It is both an oncogene (38) and a tumor suppressor gene that promotes apoptosis. Here we show that LMP1-mediated activation of miR-155 is correlated with diminished levels of the PU.1 protein. Many mRNAs were predicted to be direct targets of miR-155. A recent work reported that miR-155 plays an important role in EBV-regulated gene expression of the infected cells (34). It is thus possible that the phenotypic alterations and the oncogenic potentialities observed in miR-155-overexpressing mice (14) are the result of deregulation of other targets in addition to PU.1. In silico analysis indicated that miR-155 may target SOCS1 (39) and WEE1 (40) (G.G. and M.M., unpublished data), which would better account for its oncogenic role. Probably, a miRNA can have two or more targets or even different targets depending on the cellular context. Studies to elucidate the molecular targets of miR-155 after LMP1 induction, and to determine their reciprocal role during EBV infection and EBV-mediated transformation are underway in our laboratory.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Education and Research (MIUR) PRIN 05; Funding for open access charge: Department of Biochemistry and Medical Biotechnologies, University of Naples Federico II, Naples, Italy.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Ileana Quinto and Prof. Giuseppe Scala for supplying cells, antibodies and other materials and for all the critical suggestions, and Mrs Jean Gilder for the help in editing.
REFERENCES
- 1.Kieff ED, Rickinson AB. Epstein-Barr virus and its replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virol. 5th. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2603–2654. [Google Scholar]
- 2.Young LS, Murray PG. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 3.Kulwichit W, Edwards RH, Davenport EM, Baskar JF, Godfrey V, Raab-Traub N. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl Acad. Sci. USA. 1998;95:11963–11968. doi: 10.1073/pnas.95.20.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liebowitz D, Wang D, Kieff E. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J. Virol. 1986;58:233–237. doi: 10.1128/jvi.58.1.233-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatzivassiliou E, Miller WE, Raab-Traub N, Kieff E, Mosialos GA. Fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-kappa B, and stress-activated protein kinase. J. Immunol. 1998;160:1116–1121. [PubMed] [Google Scholar]
- 7.Izumi KM, Kaye KM, Kieff ED. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl Acad. Sci. USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumi KM, Kieff ED. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl Acad. Sci. USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaye KM, Izumi KM, Mosialos G, Kieff E. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J. Virol. 1995;69:675–683. doi: 10.1128/jvi.69.2.675-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eliopoulos AG, Blake SM, Floettmann JE, Rowe M, Young LS. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 1999;73:1023–1035. doi: 10.1128/jvi.73.2.1023-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliopoulos AG, Young LS. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 12.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 13.Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas, evidence for stage-specific events. Mol. Cell Biol. 1989;9:2657–2664. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 16.den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Gene Chromosome Canc. 2003;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam W, Hughes SH, Hayward WS, Besmer P. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J. Virol. 2002;76:4275–4286. doi: 10.1128/JVI.76.9.4275-4286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Mrázek J, Kreutmayer SB, Grässer FA, Polacek N, Hüttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scala G, Quinto I, Ruocco MR, Mallardo M, Ambrosino C, Squitieri B, Tassone P, Venuta S. Epstein-Barr virus nuclear antigen 2 transactivates the long terminal repeat of human immuno-deficiency virus type 1. J. Virol. 1993;67:2853–2861. doi: 10.1128/jvi.67.5.2853-2861.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scala G, Quinto I, Ruocco MR, Arcucci A, Mallardo M, Caretto P, Forni G, Venuta S. Expression of an exogenous interleukin 6 gene in human Epstein Barr virus B cells confers growth advantage and in vivo tumorigenicity. J. Exp. Med. 1990;172:61–68. doi: 10.1084/jem.172.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell Receptor Activation Induces BIC/miR-155 Expression through a Conserved AP-1 Element. J. Biol. Chem. 2008;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 29.Kang CD, Han CS, Kim KW, Do IR, Kim CM, Kim SH, Lee EY, Chung BS. Activation of NF-kappaB mediates the PMA-induced differentiation of K562 cells. Cancer Lett. 1998;132:99–106. doi: 10.1016/s0304-3835(98)00165-7. [DOI] [PubMed] [Google Scholar]
- 30.Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc. Natl Acad. Sci. USA. 2004;101:141–6. doi: 10.1073/pnas.2237183100. . Epub 2003 Dec 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eliopoulos AG, Caamano JH, Flavell J, Reynolds GM, Murray PG, Poyet JL, Young LS. Epstein-Barr virus-encoded latent infection membrane protein 1 regulates the processing of p100 NF-kappaB2 to p52 via an IKKgamma/NEMO-independent signalling pathway. Oncogene. 2003;22:7557–7569. doi: 10.1038/sj.onc.1207120. [DOI] [PubMed] [Google Scholar]
- 32.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo. J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 2008;82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motsch N, Pfuhl T, Mrazek J, Barth S, Grässer FA. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 38.Oikawa T, Yamada T, Kihara-Negishi F, Yamamoto H, Kondoh N, Hitomi Y, Hashimoto Y. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 1999;6:599–608. doi: 10.1038/sj.cdd.4400534. [DOI] [PubMed] [Google Scholar]
- 39.Wormald S, Hilton DJ. The negative regulatory roles of suppressor of cytokine signaling proteins in myeloid signaling pathways. Curr. Opin Hematol. 2007;1:9–15. doi: 10.1097/00062752-200701000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Kellogg DR. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J. Cell Sci. 2003;116(Pt 24):4883–4890. doi: 10.1242/jcs.00908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.