Abstract
The elongation phase of transcription by RNA polymerase II (RNAP II) is controlled by a carefully orchestrated series of interactions with both negative and positive factors. However, due to the limitations of current methods and techniques, not much is known about whether and how these proteins physically associate with the engaged polymerases. To gain insight into the detailed mechanisms involved, we established an experimental system for analyzing direct factor interactions to RNAP II elongation complexes on native gels, namely elongation complex electrophoretic mobility shift assay (EC-EMSA). This new assay effectively allowed detection of interactions of TFIIF, TTF2, TFIIS, DSIF and P-TEFb with elongation complexes generated from a natural promoter using an immobilized template. As an application of this assay system, we characterized the association of transcription elongation factor DSIF with RNAP II elongation complexes and discovered that the nascent transcript facilitated recruitment of DSIF. Examples of how the system can be manipulated to address different questions are provided. EC-EMSA should be useful for further investigation of factor interactions with RNAP II elongation complexes.
INTRODUCTION
Transcript elongation by RNAP II is a highly regulated phase of the transcription cycle that is critical not only for generating mature mRNA, but also for regulation of gene expression (1–3). The transcriptional status of the RNAP II elongation complex at any given time is determined by the sequential action of a host of factors that associate with it. In the past two decades, traditional purification techniques have identified numerous proteins that are directly or indirectly involved in controlling elongation by RNAP II. Many of these factors influence elongation either by targeting the movement of the polymerase or by modulating chromatin structure, but many details of the underlying mechanisms are not known. Little is known about how and exactly when the regulatory factors become associated with the elongation complex, whether they interact cooperatively or antagonistically with others, or how the interactions are regulated. Further understanding of factor interactions is hampered by limitations of current methods and techniques. Therefore, novel experimental approaches are needed to analyze factor interactions in a context of functional elongation complexes.
Previously, we showed that elongation complexes could be isolated from a crude extract using immobilized DNA templates (4–9). Recently, the isolation protocol was modified to remove the detergent Sarkosyl and to raise salt in the wash buffers and this modification allowed more efficient function with elongation factors (9). After extensively washing the immobilized templates with 1.6 M KCl, virtually all known factors were stripped from the polymerase and template, and only the highly stable, RNAP II ternary complexes were retained (9). Further analysis of these isolated elongation complexes demonstrated that they were transcriptionally competent and responded properly to purified elongation factors, including DSIF, NELF, P-TEFb and TFIIF (9). Because the factors produced the expected functional consequences, we reasoned that they must be associating with the elongation complexes in an appropriate way. Therefore, meaningful information might be extracted from an analysis of the interactions of the factors with these isolated elongation complexes.
Toward this end, we chose to employ native gel electrophoresis because it is a powerful tool for analyzing protein–nucleic acid interactions. The technique was previously used in the comparison of yeast and human RNAP II elongation complexes formed on promoter containing templates (10) and in a study of the interaction of DSIF with Drosophila RNAP II elongation complexes formed on tailed templates (11). Here, we developed new conditions for analyzing isolated human elongation complexes by native gel electrophoresis. The study builds on the previous functional analysis of isolated elongation complexes (9). The new native gel system efficiently and reproducibly detected isolated elongation complexes and allowed the observation of direct interactions of purified factors with RNAP II elongation complexes.
MATERIALS AND METHODS
Materials
The production of HeLa nuclear extract (8), and purification of human DSIF, TFIIF and P-TEFb (9), TTF2 (12) and TFIIS (13) were previously described.
Preparation of RNAP II elongation complexes on immobilized DNA templates
Isolation of elongation complexes (ECs) was similar to that described earlier (9). Two different immobilized DNA templates were produced by PCR from a plasmid containing the full cytomegalovirus promoter as previously described (7). Both were amplified using a common biotinylated forward primer mapping 838-bp upstream of the transcription start site. Use of different reverse primers resulted in templates that would generate either 183 or 548-nt run-off transcripts. Unless indicated otherwise, the 183-nt run-off template was used. The templates were immobilized on paramagnetic beads (Invitrogen, Carlsbad, CA, USA) (∼0.25 pmol template/reaction) and incubated with HeLa nuclear extract before transcripts were pulse-labeled. Transcription was stopped with 20 mM EDTA and ECs were isolated by washing the beads with a high salt buffer [1.6 M KCl and 20 mM HEPES (pH 7.6)] and eventually resuspended in a low salt buffer [20 mM HEPES (pH 7.6), 60 mM KCl and 200 μg/ml bovine serum albumin]. In some experiments, the isolated ECs were subsequently chased [20 mM HEPES (pH 7.6), 60 mM KCl, 0.5 mM NTPs, 200 μg/ml bovine serum albumin, 3 mM MgCl2, 1 mM DTT and 10 U of RNaseOUT (Invitrogen, Carlsbad, CA, USA)/transcription reaction] and re-isolated by washing with the low salt buffer. When a chase was performed it was for 3 min unless otherwise indicated. In experiments in which ECs were treated with RNase A, 3′-end labeled transcripts were generated. To accomplish this, the same pulse/chase protocol was followed except that 0.5 mM CTP replaced [α-32P]CTP during the pulse. After isolation of the cold ECs, the transcripts were extended for 10 min under the conditions normally used during the pulse [5 μCi of [α-32P]CTP (3000 Ci/mmol), and 0.5 mM ATP, UTP and GTP]. To analyze the length of RNAs, reactions were phenol-extracted and labeled transcripts were analyzed on 9% denaturing gels as described previously (14).
Analysis of elongation complexes by native gel electrophoresis
ECs were liberated from the paramagnetic beads by restriction enzyme digestion before analysis on native gels. ECs were digested for 15 min at 37°C with 10 U of the indicated restriction enzyme in the presence of 20 mM HEPES (pH 7.6), 60 mM KCl, 200 μg/ml bovine serum albumin, 8 mM MgCl2, 1 mM DTT and 10 U of RNaseOUT/transcription reaction. In most experiments, SacI which cuts −17 bp upstream of the start point of transcription was utilized. Where indicated BamHI and NdeI, which cut at −798 and −352, respectively were used. Usually, the ECs were digested in bulk with a total volume equal to the number of transcription reactions times 6 μl and then the beads were removed by magnetic concentration. Binding reactions (18 μl) contained an aliquot of the released ECs (6 μl) and the indicated added proteins and maintained the conditions used above in the digestion. After 10 min at room temperature, the samples were supplemented with glycerol to 10% and directly loaded onto a 4% acrylamide (1:50 bisacrylamide) gel cast and run in 0.5× Tris/glycine [12.5 mM Tris (pH 8.3) and 96 mM glycine]. The gels were run at 6 W for 2.5 h at 4°C, before being dried and subjected to autoradiography.
Digestion of ECs with RNase or DNase
Isolated ECs were treated with the indicated amounts of RNase-free DNase I (NEB) for 10 min or 100 ng/ul of RNase A (Fermentas, Glen Burnie, MD, USA) or 20 min at 37°C under the same conditions for the restriction enzyme digestion except that for RNase A treatment RNaseOUT was omitted. After DNase I treatment, the supernatant was analyzed directly, but after RNase A treatment, the ECs were washed with 1.6 M KCl and 20 mM HEPES (pH 7.6) to remove RNase A and eventually released from the beads by restriction digestion before being analyzed.
Transcript cleavage by TFIIS
ECs after a 5-min chase were either mock treated or incubated with 100 ng of purified TFIIS for indicated times in a 12 μl mixture containing 20 mM HEPES (pH 7.6), 60 mM KCl, 200 μg/ml bovine serum albumin, 8 mM MgCl2, 1 mM DTT and 10 U of RNaseOUT. The cleavage reactions were terminated by adding EDTA to 20 mM. To remove TFIIS after the treatment, the ECs were washed and resuspended with the low salt buffer.
Phosphorylation of RNAP II-CTD or DSIF by P-TEFb
In most experiments, transcription was initiated in the presence of 1 μM flavopiridol, which inhibits phosphorylation of the CTD by P-TEFb but not TFIIH. In some experiments, CTD phosphorylation was completely blocked by the addition of DRB to 1.5 mM during initiation (15). Phosphorylation of the RNAP II-CTD in the isolated ECs by P-TEFb was carried out as described earlier (9). In some cases, indicated in the text (Figure 7C), the treated ECs were then washed with 1.6 M KCl and 20 mM HEPES (pH 7.6) to remove P-TEFb. Phosphorylation of DSIF was carried out by an incubation of indicated amount of P-TEFb with DSIF for 10 min at room temperature under the same conditions for the restriction enzyme digestion described above. After the incubation, the activity of P-TEFb was inhibited by adding flavopiridol to 1 µM.
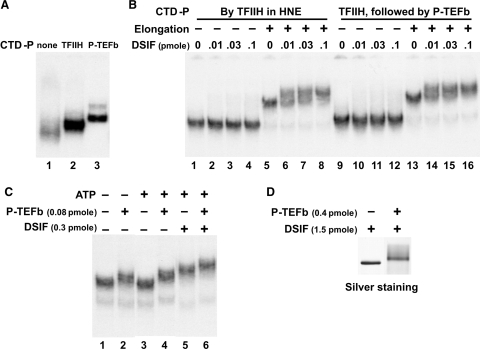
Figure 7.
Impact of phosphorylation of the CTD and DSIF on the DSIF•EC interaction. (A) Effects of CTD phosphorylation on the mobility of ECs. ECs were generated under conditions that resulted in the CTD being unphosphorylated or phosphorylated by TFIIH or TFIIH and P-TEFb as indicated in the text and Materials and methods section. Then the three different ECs were analyzed on a native gel. (B) Effects of CTD phosphorylation on the DSIF•EC interaction. ECs with the CTD being phosphorylated by TFIIH or TFIIH and P-TEFb were generated under the same conditions as in (A). After subsequent elongation (3 min) or not, the ECs were released with SacI (−17) and analyzed by EC-EMSA for their ability to interact with indicated amounts of DSIF. (C) Effects of phosphorylation of both the CTD and DSIF on the DSIF•EC interaction. ECs containing long transcripts (elongated for 3 min) were generated and incubated with the indicated factors and 0.5 mM ATP for 10 min followed by analysis on a native gel. (D) Phosphorylation of DSIF by P-TEFb. The same amounts of DSIF and P-TEFb were incubated with ATP under the same conditions as in (B) and 5× of the reactions was analyzed by SDS–PAGE followed by silver staining.
RESULTS
Establishment and validation of EC-EMSA
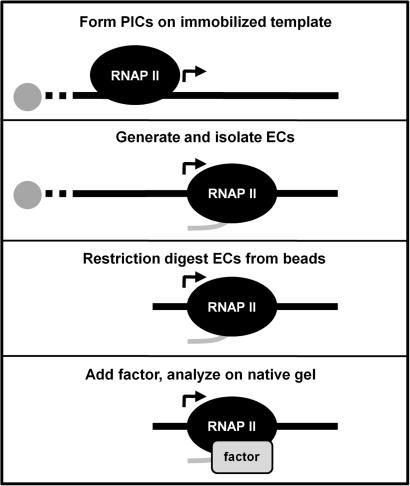
To directly analyze factor associations with RNAP II elongation complexes (ECs), an elongation complex electrophoretic mobility shift assay (EC-EMSA) was developed. The core of this technique is to use labeled ECs to probe interactions with factor(s) of interest. An outline of the method is shown in Figure 1. Transcription preinitiation complexes (PICs) are formed on the CMV promoter encoded in an immobilized DNA template in the presence of HeLa nuclear extract. Upon the addition of nucleotides including limiting [α-32P]CTP, RNAP II initiates and generates labeled, nascent transcripts that are about 20 nt in length within 30 s. The early elongation complexes are halted upon the addition of EDTA and are then extensively washed with a buffer containing 1.6 M salt, which removes all proteins except engaged RNAP II. The isolated ECs are then released from beads through restriction enzyme digestion, incubated with purified transcription factor(s) and analyzed on a low-percentage native gel. The capability of the factor(s) of interest to interact with ECs is evaluated by monitoring the mobility shift of ECs. As estimated in our early studies (9), the amount of labeled ECs used in each binding reaction is about 0.01 pmole.
Figure 1.
EC-EMSA. The diagram indicates the important steps of the elongation complex EMSA. PICs are formed on an immobilized template by incubation with HeLa nuclear extract. ECs are generated by initiation in the presence of NTPs including 32P-labeled CTP and the ECs are isolated by washing with 1.6 M salt. The ECs are removed from the paramagnetic beads by digestion with a restriction enzyme. These complexes are incubated with factor(s) of interest (factor) and analyzed on a native gel.
To validate this system, RNAP II ECs before and after addition of transcription factors known to interact with elongating RNAP II were analyzed on a native gel. A strong radioactive signal with a distinct migration was detected with intact ECs, but not with the RNA extracted from these ECs (Figure 2A, lanes 1 and 2), suggesting that the signal visualized represents the ECs. Earlier results from functional transcription assays indicated that the initiation and elongation factor TFIIF could stably associate with isolated ECs (9). Inclusion of TFIIF in the binding reaction resulted in a complete shift of the ECs to higher position on the native gel (Figure 2A, lane 3). TTF2, an ATP-dependent termination factor, was also tested for its interaction with ECs. In the absence of ATP, TTF2 caused a portion of the signal from ECs to have a lower mobility, indicating that only a portion of ECs were bound with TTF2 at the concentration of the factor used (Figure 2A, lane 4). Addition of ATP in the binding reaction with TTF2 resulted in the disruption of the elongation complexes (Figure 2A, lane 5). The fact that the band attributed to ECs was sensitive to the action of TTF2 further confirmed that it represented elongation complexes. Taken together, these results indicate that the EC-EMSA system developed here can be used to analyze factor binding to ECs.
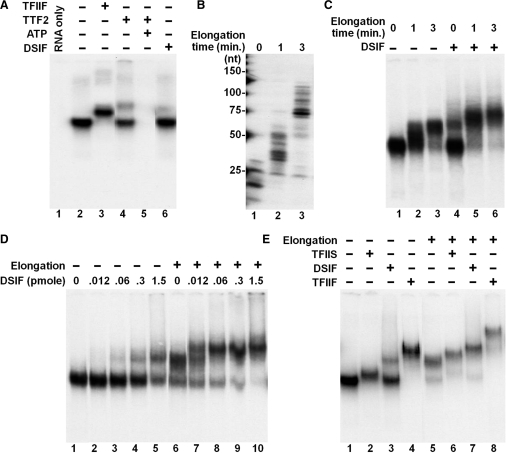
Figure 2.
Detection of factor interactions with isolated elongation complexes using EC-EMSA. (A) Initial validation of EC-EMSA. Isolated ECs were incubated with no factors or with 0.4 pmol of TFIIF, 0.25 pmol of TTF2 (± 0.5 mM of ATP) or 0.3 pmol of DSIF before analysis by native gel electrophoresis. As a negative control, transcripts associated with isolated ECs were extracted and analyzed (RNA only). (B and C) Interactions between DSIF with ECs containing various lengths of RNAs. The transcripts in isolated ECs were further extended for indicated times. The transcripts in the resultant ECs were extracted and analyzed on a denaturing RNA gel (B) or the ECs were incubated with or without 0.3 pmol of DSIF and then subjected to analysis on a native gel (C). (D) Dose response of DSIF. Isolated ECs were left untreated or further elongated for 3 min and then incubated with indicated amounts of DSIF before analysis on a native gel. (E) Interactions of TFIIS, DSIF and TFIIF to isolated elongation complexes containing short or long transcripts. Isolated ECs were left untreated or further elongated for 3 min before being released from the beads by SacI (−17), and then incubated with no factors or with 0.4 pmol of TFIIS, 0.3 pmol of DSIF or 0.4 pmol of TFIIF. The samples were analyzed on a native gel.
Characterization of the detailed binding mechanisms for DSIF to ECs using EC-EMSA
DSIF preferentially binds to ECs with long transcripts
Human elongation factor DSIF is composed of two subunits that are homologues of yeast proteins Spt5 and Spt4 (16). It was reported that the large subunit, hSpt5, contains an RNAP II-interacting region (17,18). Because DSIF negatively controls RNAP II transcription in the early stage of elongation (7), a mobility shift of isolated ECs is expected to be observed when the ECs are incubated with DSIF. However, very little shift was detected when isolated ECs were incubated with an excess amount of DSIF (0.3 pmol) (Figure 2A, lane 6). Given that the isolated ECs only contained very short nascent transcripts (∼20 nt) and all the endogenous RNAP II-binding factors were removed during high salt isolation, we reasoned that an efficient DSIF•EC interaction may occur upon further elongation or may require other RNAP II-interacting factor(s).
To further characterize the association of DSIF with ECs, the effect of further elongation was examined. The transcripts in isolated ECs were extended upon the addition of NTPs for 1 or 3 min. The lengths of the transcripts were determined by analysis of extracted RNA on a denaturing gel (Figure 2B). As previously determined, without the effect of any elongation factors, the intrinsic elongation rate of the polymerases in our system is about 20–25 nt/min (9). After 1 or 3 min of elongation, the average length of the transcripts was increased from 20 to 35 nt and 75 nt, respectively (Figure 2B). Simultaneously, the electrophoretic mobility of these ECs in the absence or presence of DSIF was analyzed on a native gel (Figure 2C). Interestingly, in the absence of factor supplementation, the mobility of ECs was gradually reduced as the transcripts became longer (Figure 2C, lanes 1–3). The reason for this mobility change will be addressed below. More importantly, upon incubation with a constant amount of DSIF, the fraction of the ECs shifted was significantly increased as the nascent transcripts became longer (Figure 2C). This indicated that DSIF interacted more efficiently with the ECs containing long transcripts (35 and 75 nt) than with the ECs containing short transcripts (20 nt). To further confirm this result, DSIF was titrated into binding reactions containing ECs with short or long transcripts (0 or 3 min chase). As seen in Figure 2D, again, 0.3 pmol of DSIF exhibited a weak association with the isolated ECs with short RNAs. Even when 1.5 pmol of DSIF was used (in a great excess to the estimated amount of ECs) only about a half of the ECs shifted (Figure 2D, compare lanes 5 and 1). Similar to that shown in Figure 2C, the additional 3 min extension of the transcripts changed the mobility of ECs on the native gel and produced a lower mobility signal (the upper band) (Figure 2D, lane 6). The lower band in lane 6 had the same mobility as that shown in lane 1 and likely represented a fraction of the ECs that failed to extend transcripts. Usually, about 10% of the halted polymerases do not restart transcription, both in our system (9) and in others (19) (Figure 2B). As shown in lanes 6–10, the mobility of the two populations of ECs was affected differently upon the incubation with increasing concentrations of DSIF. When as little as 0.012 pmol of DSIF was used, about half of the elongation complexes containing long transcripts were shifted (Figure 2D, compare lanes 6 and 7). A total of 0.06 pmol of DSIF was enough to cause a complete shift of the upper band (Figure 2D, lane 8). In contrast, the mobility shift of the lower band was not complete even when 1.5 pmol of DSIF was used. These data demonstrate that a further extension of the transcripts in the isolated early elongation complexes dramatically increases the binding affinity of DSIF. For comparison, we also tested two other RNAP II-interacting factors, TFIIS and TFIIF. Both factors bound efficiently with the ECs and the extent for their interactions was not impacted by the change of the length of the transcripts (Figure 2E). We conclude that DSIF preferentially associates with ECs that have nascent transcripts that are 35 nt or longer.
Nascent transcripts facilitate the efficient association of DSIF with ECs
The finding that extension of the nascent transcripts caused a change in the electrophoretic mobility of ECs (Figure 2C, lanes 1–3) that correlated with enhanced binding of DSIF suggested that there might be a conformational change in the ECs that was recognized by DSIF. Such a conformational change was suggested by earlier work demonstrating an alteration in the properties of elongation complexes after the synthesis of 50 nt of RNA (19). Additionally, it was not obvious how the lowering of the mobility of ECs could be explained by the addition of a relatively small mass of negatively charged RNA. Extra negative charge should have increased the mobility of the ECs. Therefore, we performed several experiments to address if there was a conformational change as the transcripts were extended and, if so, if it was responsible for the preferential association of DSIF with ECs containing long transcripts.
The position of the polymerase on the template affects the mobility of ECs
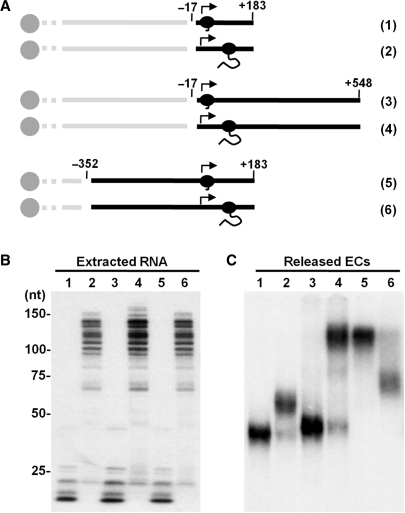
To determine if the location of RNAP II on the template had an effect on the mobility of ECs, ECs with different lengths of template and with the polymerase at different relative positions were analyzed. Three sets of ECs were generated by using different downstream primers (generating 3′-ends at +183 or +548) or by using different restriction enzymes to liberate the ECs (at positions −17 or −352) (Figure 3A). For each set, ECs were either not chased and contained nascent transcripts <25 nt or were chased for 5 min and contained transcripts 100–150 nt in length (Figure 3B). For the ECs with the least DNA, extension of the nascent transcripts caused a shift to lower mobility as was seen earlier (Figure 3C, lanes 1 and 2). Addition of 365 bp of downstream DNA had almost no effect on the mobility of ECs with short transcripts (Figure 3C, lane 3), but when the transcripts were extended a dramatic reduction of the mobility of ECs was observed (Figure 3C, lane 4). Extension of the DNA to −352 upstream of the transcription start site had an equally dramatic lowering of the mobility (Figure 3C, compare lanes 1 and 5). In this case, extension of the transcript actually increased the mobility of the ECs (Figure 3C, lane 6). Analysis of these results suggests that the relative position of the polymerase on the DNA template had a great impact on the electrophoretic mobility of elongation complexes. In each pair, the mobility of the ECs increased if the polymerases moved toward the end of the DNA and decreased if they moved toward the middle of the DNA. The ECs with the lowest mobility had long DNA ‘arms’ both upstream and downstream and the ECs with highest mobility had at least one short DNA arm. A similar phenomenon has been observed for the mobility of protein•DNA complexes on native gels when the binding site for the protein is located near the end or in the middle of the DNA (20). Therefore, this result suggested that the mobility shifts seen when transcripts were elongated was very likely due to the position of the polymerase on the DNA.
Figure 3.
Effect of position of RNAP II on DNA template on electrophoretic mobility. (A) Diagram of templates and released ECs. Six different ECs (black) were generated and released by restriction enzyme digestion at the position indicated (relative to the transcription start site) as described in the text and in Materials and methods section. (B and C) Analysis of ECs. Transcripts associated with the indicated ECs were extracted and analyzed on a denaturing gel (B) and ECs were analyzed on a native gel (C).
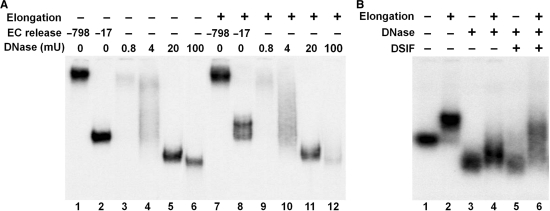
To eliminate the effect of the DNA arms on the electrophoretic mobility of the ECs, DNase I digestion of ECs was performed. Such ECs with limited DNA content should still run into a native gel because the surface charge of RNAP II is highly negative (21). ECs were first generated on the +183 template. When ECs were released from the beads by restriction digestion at a site near the attachment point of the template with the beads (−798), very low mobility ECs were generated compared to ECs generated by digestion at −17 (Figure 4A, compare lane 1 to 2). When the immobilized complexes were treated with increasing amounts of DNase I, the first ECs liberated had the mobility similar to that of the ECs with long DNA arms (Figure 4A, compare lane 3 to 1). As the amount of DNase I was increased, more complexes were released and the mobility increased dramatically. At the highest extent of digestion, the ECs were beginning to be disrupted as evidenced by a decrease in the EC signal seen on the gel (Figure 4A, lane 6). An identical set of reactions was performed on complexes containing transcripts that were elongated for 3 min. As was seen above, a significant fraction of these ECs had a lower mobility after restriction digestion at −17 (Figure 4A, compare lane 8 to 2). As DNase I was added to the ECs with long RNAs, a very similar pattern of ECs was detected (Figure 4A, compare lanes 9–12 to 3–6). The RNase free-DNase I used in this experiment did not act on RNA (data not shown). Importantly, at the high concentrations of DNase I, the mobility of complexes with short transcripts was identical to that of the complexes with long transcripts (Figure 4A, compare lanes 5 and 6 to lanes 11 and 12). This further supports the idea that the aberrant mobility of complexes with long transcripts was due an effect of the position of the ECs within the template. The ability of ECs to bind DSIF was then tested after digestion with DNase I. DSIF did not bind well with the DNase I-treated ECs that had short transcripts (Figure 4B, compare lane 5 to 3), but did bind to DNase I-treated ECs containing long transcripts (Figure 4B, compare lane 6 to 4). These experiments strongly suggest that the differential association of DSIF to ECs observed is due to differences in the length of the nascent RNA. However, a conformational change due to extension of the nascent transcripts is still a possible explanation.
Figure 4.
Interaction of DSIF with DNase I-treated ECs. (A) DNase I treatment of isolated ECs. ECs with short or long transcripts (3 min of elongation) were released from the beads either by BamHI (−798), SacI (−17) or the indicated amounts of DNase I and then analyzed on a native gel. (B) EC-EMSA analysis DSIF interaction. ECs with short or long transcripts were digested with DNase I (20 mU per reaction), incubated with or without 0.3 pmol of DSIF and analyzed on a native gel. As a control, ECs were released by SacI in lanes 1 and 2.
Treatment of ECs with TFIIS does not affect binding of DSIF
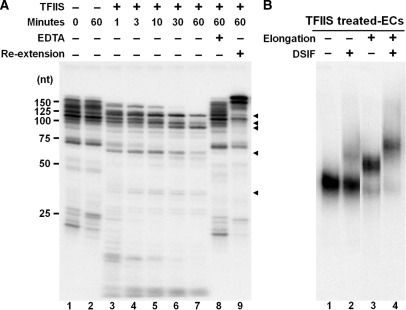
Another possible explanation for differential DSIF binding to ECs is that the elongation complexes are in different transcriptional states before and after transcript extension, and somehow DSIF differentiates between the two states. ECs with short transcripts are generated by limiting CTP during a short pulse and, therefore, polymerases are stalled at sites that would normally support rapid elongation. ECs with long transcripts are generated by transcript extension in the presence of high concentrations of all four NTPs and because of this all polymerases are stopped at naturally occurring pause sites. Because it is possible that DSIF recognizes this paused state found predominately in the ECs with long transcripts, treatment of these complexes with TFIIS was carried out to convert them back into the elongation competent state. A time course of TFIIS mediated transcript cleavage was performed on ECs and the expected pattern of shortened transcripts (22) was observed (Figure 5A). At these ‘reverse pause sites’ further backtracking of polymerases is disfavored and TFIIS-mediated cleavage of the RNAs is inhibited (Figure 5A, lanes 3–7). Transcript cleavage required TFIIS and Mg2+, and after extensive shortening, addition of NTPs allowed efficient re-extension up to the run-off site at +183 (Figure 5A, lane 9). ECs with short or long transcripts were treated with TFIIS for 10 min and then re-isolated to remove TFIIS. Without further elongation following TFIIS treatment, polymerases would be in the elongation state and would remain that way after removal of TFIIS. For these polymerases to re-enter backtracked mode, they would have to further extend their transcripts to return to a site at which backtracking could occur more readily. When complexes with shortened transcripts were analyzed by EC-EMSA, DSIF was found to bind well only to complexes with long transcripts even though these ECs were not in the paused state (Figure 5B). This experiment strongly suggests that differential DSIF binding is not due to potential differences in the elongation competence of the ECs.
Figure 5.
TFIIS-mediated transcript cleavage does not affect the association of DSIF with ECs. (A) Cleavage of the transcripts associated with ECs by TFIIS. Transcripts in isolated ECs were elongated for 5 min and then treated with 100 ng of TFIIS for indicated times. Incubation without TFIIS (lane 2) or with TFIIS and 20 mM EDTA (lane 8) served as negative controls. In lane 9, the ECs that had been treated with TFIIS for 60 min were washed with a low salt buffer to remove TFIIS, followed by a 3 min ‘re-extention’. The transcripts were extracted from each sample and analyzed on a denaturing gel. Arrowheads indicate ‘reverse pause sites’ resulting from extensive TFIIS cleavage. (B) Interaction of DSIF with TFIIS-treated ECs. ECs containing short or long transcripts were treated with 100 ng of TFIIS for 10 min followed by a low salt wash. The ECs were released with SacI and analyzed by EC-EMSA. A total of 0.3 pmol of DSIF was used where indicated.
A long nascent transcript facilitates DSIF binding
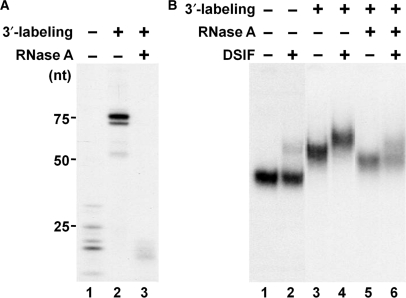
After ruling out the above possibilities, it seemed more likely that the extended nascent transcripts might be directly responsible for enhancement of the association of DSIF with ECs. To test this idea, three types of ECs were generated. The first contained the standard 5′-end labeled short transcripts (Figure 6A, lane 1). The second contained 3′-end labeled transcripts that were predominately 75 nt in length (Figure 6A, lane 2) and RNase A treatment of these ECs generated the third type that contained transcripts <25 nt in length (Figure 6A, lane 3). The 3′-end labeling was necessary to avoid removal of the radioactive signal during RNase A treatment. EC-EMSA was then used to examine binding of DSIF to all three types of complexes after release from the beads by restriction digestion at −17. Again, DSIF did not interact well with ECs containing 5′-end labeled short transcripts (Figure 6B, lanes 1 and 2). Also, DSIF strongly interacted with ECs containing long transcripts (Figure 6B, lanes 3 and 4). Importantly, after RNase A digestion, that removed all accessible RNA, the complexes lost their ability to interact well with DSIF (Figure 6B, lanes 5 and 6). Similar results were obtained on the three types of ECs after DNase I treatment (data not shown). We conclude that the RNase-sensitive portion of nascent transcripts is required for efficient association of DSIF with ECs. Supporting this result we also found that DSIF bound directly to RNA using standard RNA EMSA (data not shown).
Figure 6.
Removal of accessible RNA weakens the association of DSIF and ECs. (A) RNase A treatment of ECs. ECs with short 5′-end labeled transcripts were generated as usual (lane 1). ECs with long 3′-end labeled transcripts were generated as described in Materials and methods section. ECs were treated with 1 μg of RNase A per reaction where indicated and were then released from beads by SacI digestion. Transcripts were then extracted and analyzed on a denaturing gel. (B) The effect of RNase A treatment on DSIF•EC interaction. ECs obtained from the same experiment shown in (A) were incubated without or with 0.3 pmol of DSIF and analyzed on a native gel.
Phosphorylation of the CTD or DSIF by P-TEFb does not affect the DSIF•EC interaction
The carboxy-terminal domain (CTD) of the largest subunit of RNAP II is regulated by sequential phosphorylation/dephosphorylation events during transcription. This is essential for properly integrating transcription and mRNA processing by modulating the recruitment of factors to the CTD (23). Previously, the Handa group performed co-IP experiments using HeLa nuclear extract and showed that DSIF co-precipitated better with hypophosphorylated RNAP II than with hyperphosphorylated RNAP II, and suggested that phosphorylation of the CTD might lead to the dissociation of DSIF from the elongation complex (24). More recently, the Gilmour group showed that the CTD had no effect on the interaction of DSIF with ECs generated with purified Drosophila RNAP II (11).
To help resolve the potentially conflicting results, we examined the effect of CTD-phosphorylation on DSIF binding. ECs were generated that contained no phosphorylation, phosphorylation acquired during initiation (TFIIH) or phosphorylation during initiation followed by further phosphorylation with recombinant P-TEFb. To obtain ECs with unphosphorylated RNAP II, initiation was carried out in the presence of 1.5 mM of DRB, which inhibits the kinase activity of TFIIH and P-TEFb (15). Normal initiation conditions allowed TFIIH phosphorylation and in the presence of flavopiridol inhibited phosphorylation by P-TEFb (25). Hyperphosphorylation of the CTD was accomplished by incubating the isolated normal ECs with P-TEFb and ATP. Then P-TEFb was removed by a high salt wash and the hyperphosphorylated ECs were analyzed by EC-EMSA. Blocking phosphorylation of the CTD during initiation led to production of ECs with slightly higher mobility than that normally seen with ECs phosphorylated by TFIIH (Figure 7A, compare lanes 1 and 2). Phosphorylation of the ECs by both TFIIH and P-TEFb further lowered the mobility (Figure 7A, compare lanes 2 and 3). All these mobility shifts seen upon phosphorylation suggest that the CTD is indeed phosphorylated as expected. Because addition of more negative charge should have caused an increase in mobility, these results suggest that phosphorylation of the CTD may lead to a more extended conformation of the CTD that reduces the mobility of the ECs. Next, the association of DSIF to the ECs with their CTD being phosphorylated by either TFIIH only or by TFIIH plus P-TEFb was analyzed using EC-EMSA. DSIF did not associate strongly with ECs containing short transcripts regardless of the amount of phosphorylation (Figure 7B, lanes 1–4 and 9–12). The reduction in mobility upon transcript extension was detected for both types of ECs (Figure 7B, lanes 5 and 13), and DSIF bound to both ECs equally well regardless of the phosphorylation status of the CTD (Figure 7B, lanes 5–8 and 13–16). We conclude that the phosphorylation status of the CTD of RNAP II has no direct effect on DSIF binding.
Because both DSIF and the CTD are phosphorylated by P-TEFb during the transition into productive elongation (9,26), the interaction of DSIF with ECs was examined after phosphorylation of the CTD and DSIF by P-TEFb. For this experiment, the ECs were chased to produce long RNAs, but as shown before a small fraction of the complexes (the light lower band) did not exhibit the reduction in mobility caused by RNA extension (Figure 7C, lane 1). Addition of P-TEFb caused a further reduction in mobility indicating that P-TEFb interacts with the ECs (Figure 7C, lane 2). Phosphorylation of the ECs alone caused the expected, slight shift in ECs with short and long transcripts (Figure 7C, compare lanes 4 to 2). The mobilities seen suggest that P-TEFb maintains its interaction with ECs containing long RNAs, but this interpretation is made more difficult because of the simultaneous shift caused by CTD phosphorylation. DSIF lowered the mobility of ECs containing long transcripts even after phosphorylation of the CTD and DSIF by P-TEFb (Figure 7C, lanes 4–6). Under the conditions used DSIF was fully phosphorylated as detected by a shift in migration when analyzed by SDS–PAGE with silver staining (Figure 7D). We conclude that DSIF remains associated with ECs containing long transcripts even when the RNAP II-CTD and DSIF are phosphorylated by P-TEFb.
DISCUSSION
We developed a method that utilizes native gel electrophoresis to analyze factor interactions with RNAP II elongation complexes generated by initiation from a physiological promoter. The new system detected interactions of DSIF, TFIIF, TTF2, TFIIS and P-TEFb with isolated ECs.
Because it was possible to manipulate the length of the nascent RNA, it was discovered that of all the factors examined only DSIF showed preference for the ECs with long transcripts. In addition, the ECs could be treated with DNase or RNase while maintaining their integrity. DNase treatment was useful in explaining mobility shifts of ECs upon RNA extension and RNase treatment was used to further document the facilitation of DSIF binding by long nascent RNA. Finally, we were able to investigate the influence of phosphorylation of the CTD and DSIF by P-TEFb on the interaction between DSIF and ECs. Together with the recently described defined transcription system (9) it is now possible to correlate binding with functional consequences of the factors.
A variety of systems have been developed to analyze RNAP II ECs and factor interactions and each has advantages and disadvantages. Reines used a monoclonal antibody that recognized RNA to pull down RNAP II ECs and showed that these complexes did not contain one of the transcription initiation factors (27). While all polymerases isolated this way were in active ECs, the requirement for an antibody pull down, a defined set of factors and long nascent transcripts made this system less useful than the one described here. The advent of templates immobilized to agarose beads (28,29) and eventually to paramagnetic beads (6) enabled the development of new approaches to study RNAP II ECs. Immobilized templates have been used to examine factor interactions by western blot detection of proteins surviving washing of the ECs. Using such a system DSIF was shown to interact with ECs containing 22-nt long nascent transcripts (30). In that study shortening of the transcripts to 15 nt had no effect on the association of DSIF (31). Unfortunately, due to the nature of the assay it was not possible to know what fraction of ECs were associated with factors and this complicates the interpretation of the results. Analysis of RNAP II transcription complexes using native gels has proven useful for examination of the stepwise formation of human initiation complexes and elongation complexes (32,33) and for comparison of yeast and human elongation complexes (10). Native gel analysis was also performed to examine the association of DSIF with Drosophila ECs. In that study ECs were generated with purified RNAP II on a soluble tailed template (11).
The system described here has the advantage that the elongation complex was formed on a natural promoter under standard initiation conditions and because the templates are initially immobilized, the ECs can be easily isolated. The high salt washes of ECs in our system lead to an efficient removal factors bound specifically to ECs or nonspecifically to DNA, which allows stronger interpretations experiments in which factors are added back to isolated ECs. In addition, the position and phosphorylation status of the polymerase, and RNA content of the ECs could be easily manipulated using our system. As demonstrated, whether the mobility shift of ECs upon factor incubation is due to the direct association of factor with RNAP II, DNA template or nascent RNA can be elucidated by carrying out DNase and/or RNase treatment on the isolated ECs before factor addition. The impact of RNAP II-CTD on factor binding can also be directly tested by modifying the CTD prior to factor addition or by removal of the CTD through chymotrypsin treatment (34,35). The conditions of generating transcription competent ECs by TFIIS treatment were also worked out and this would be useful for identification of factors that can differentiate the ECs with different transcription status. This system could also be useful for find out particular region(s) in a factor of interest that is involved in the association to ECs. We have been using this technique to compare the wild type factors and their mutants with specific regions deleted to narrow down the EC-binding domains. Even though the resolution of ECs from ECs with one or two factors bound was high, our system is limited when the number of factors is increased or when modifications of the polymerase or factors are involved. For instance, results from EC-EMSA using chromatinized templates are likely to be difficult to evaluate. Our previous studies have shown that DSIF and NELF work together and that TFIIF and NELF compete for positive and negative effects in the presence of DSIF (7,9). Unfortunately, attempts to study NELF using EC-EMSA were thwarted due to NELF's propensity to interact with elongation complexes nonspecifically. Although there are currently some limitations in the use of EC-EMSA when looking at many factors at once, the methods can provide very useful information about individual factors and the effects of nascent RNA or phosphorylation of the CTD on the binding of those factors. In addition, EC-EMSA should be very useful when examining the effect of specific mutations or modifications on the binding of individual factors. For example, it should be possible to discover mutations in DSIF that reduce the enhancement of binding by nascent RNA.
Much of our effort here was in characterizing the interaction of DSIF with the elongation complex because this important factor plays a major role in the regulation of RNAP II elongation. We found: (i) DSIF by itself does not bind well to the ECs containing transcripts shorter than 25-nt, while its binding affinity to ECs is dramatically increased when the nascent transcripts are extended to 35-nt or longer, (ii) the phosphorylation status of the CTD or DSIF does not affect the interaction of DSIF with ECs. The differential binding of DSIF to ECs containing long versus short transcripts was not seen earlier in an assay that used western blotting to analyze DSIF transfer from a nuclear extract to immobilized elongation complexes (30). In that study DSIF associated with ECs containing RNA that was as short as 15 nt. Perhaps other factors in the nuclear extract facilitated DSIF binding or the binding seen was not stoichiometric with the ECs. Using our system a strong positive effect of long nascent transcripts on the direct, stoichiometric association of DSIF was detected. In an earlier study, DSIF binding to Drosophila RNAP II was observed by mobility shift of RNAP II elongation complexes formed on a tailed template, and in that study the nascent RNA was about 110-nt long (11). Early evidence suggested that the CTD phosphorylation might negatively influence DSIF interaction with RNAP II (24). However, DSIF was shown to interact with Drosophila RNAP II regardless of the presence of the CTD (11). The finding that DSIF interaction with ECs is not influenced by P-TEFb-mediated phosphorylation of the CTD, supports other studies in which DSIF was found by ChIP to map throughout the body of transcribed genes (36–38).
FUNDING
National Institutes of Health (GM35500 to D.H.P.). Funding for open access charge: NIH (GM35500).
Conflict of interest statement. None declared.
REFERENCES
- 1.Marshall RM, Grana X. Mechanisms controlling CDK9 activity. Front Biosci. 2006;11:2598–2613. doi: 10.2741/1994. [DOI] [PubMed] [Google Scholar]
- 2.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 4.Peng J, Liu M, Marion J, Zhu Y, Price DH. RNA polymerase II elongation control. Cold Spring Harbor Symp. Quant. Biol. 1998;63:365–370. doi: 10.1101/sqb.1998.63.365. [DOI] [PubMed] [Google Scholar]
- 5.Xie Z, Price DH. Purification of an RNA polymerase II transcript release factor from Drosophila. J. Biol. Chem. 1996;271:11043–11046. doi: 10.1074/jbc.271.19.11043. [DOI] [PubMed] [Google Scholar]
- 6.Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 8.Adamson TE, Shore SM, Price DH. Analysis of RNA polymerase II elongation in vitro. Methods Enzymol. 2003;371:264–275. doi: 10.1016/S0076-6879(03)71019-2. [DOI] [PubMed] [Google Scholar]
- 9.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 10.Pardee TS, Ghazy MA, Ponticelli AS. Yeast and human RNA polymerase II elongation complexes: evidence for functional differences and postinitiation recruitment of factors. Eukaryot. Cell. 2003;2:318–327. doi: 10.1128/EC.2.2.318-327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Wu CH, Gilmour DS. Analysis of polymerase II elongation complexes by native gel electrophoresis. Evidence for a novel carboxyl-terminal domain-mediated termination mechanism. J. Biol. Chem. 2004;279:23223–23228. doi: 10.1074/jbc.M402956200. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Liu M, Spencer CA, Price DH. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol. Cell. 2004;14:375–385. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- 13.Palangat M, Renner DB, Price DH, Landick R. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl Acad. Sci. USAmerica. 2005;102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price DH, Sluder AE, Greenleaf AL. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J. Biol. Chem. 1987;262:3244–3255. [PubMed] [Google Scholar]
- 15.Moteki S, Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 16.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 19.Ujvari A, Pal M, Luse DS. RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 2002;277:32527–32537. doi: 10.1074/jbc.M201145200. [DOI] [PubMed] [Google Scholar]
- 20.Wu HM, Crothers DM. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 21.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 22.Guo H, Price DH. Mechanism of DmS-II-mediated pause suppression by Drosophila RNA polymerase II. J. Biol. Chem. 1993;268:18762–18770. [PubMed] [Google Scholar]
- 23.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 24.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 26.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 27.Reines D. RNA polymerase II elongation complex. Elongation complexes purified using an anti-RNA antibody do not contain initiation factor alpha. J. Biol. Chem. 1991;266:10510–10517. [PMC free article] [PubMed] [Google Scholar]
- 28.Arias JA, Peterson SR, Dynan WS. Promoter-dependent phosphorylation of RNA polymerase II by a template-bound kinase. Association with transcriptional initiation. J. Biol. Chem. 1991;266:8055–8061. [PubMed] [Google Scholar]
- 29.Arias JA, Dynan WS. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J. Biol. Chem. 1989;264:3223–3229. [PubMed] [Google Scholar]
- 30.Ping YH, Rana TM. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 2001;276:12951–12958. doi: 10.1074/jbc.M006130200. [DOI] [PubMed] [Google Scholar]
- 31.Ping YH, Chu CY, Cao H, Jacque JM, Stevenson M, Rana TM. Modulating HIV-1 replication by RNA interference directed against human transcription elongation factor SPT5. Retrovirology. 2004;1:46. doi: 10.1186/1742-4690-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores O, Lu H, Killeen M, Greenblatt J, Burton ZF, Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc. Natl Acad. Sci. USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 34.Adamson TE, Shutt DC, Price DH. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 2005;280:32262–32271. doi: 10.1074/jbc.M505532200. [DOI] [PubMed] [Google Scholar]
- 35.Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc. Natl Acad. Sci. USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 38.Fujita T, Ryser S, Tortola S, Piuz I, Schlegel W. Gene-specific recruitment of positive and negative elongation factors during stimulated transcription of the MKP-1 gene in neuroendocrine cells. Nucleic Acids Res. 2007;35:1007–1017. doi: 10.1093/nar/gkl1138. [DOI] [PMC free article] [PubMed] [Google Scholar]