Abstract
Cornelia de Lange Syndrome (CdLS) is a rare congenital malformation disorder. About half of the patients with CdLS carry mutations in the NIPBL gene encoding the NIPBL protein, a subunit of the Cohesin loading complex. Recent studies show association of Cohesin with chromatin-remodeling complexes, either by establishing cohesion or by recruiting Cohesin to specific chromosome locations. In yeast two-hybrid assays, we identified an interaction of NIPBL with the histone deacetylases -1 and -3. These interactions were confirmed in mammalian cells by coimmunoprecipitation and a critical region for interaction was defined to a stretch of 163 amino acids of a highly conserved region of NIPBL, which is mutated in patients with CdLS. Utilizing reporter gene assays, we could show that NIPBL fused to the GAL4-DNA-binding domain (GAL4-DBD) represses promoter activity via the recruitment of histone deacetylases. Interestingly, this effect is dramatically reduced by both NIPBL missense mutations identified in CdLS and by chemical inhibition of the histone deacetylases. Our data are the first to indicate a molecular and functional connection of NIPBL with chromatin-remodeling processes via the direct interaction with histone deacetylases.
INTRODUCTION
Following replication, sister chromatids are attached to each other prior to segregation in mitosis and meiosis. This association, called sister chromatid cohesion (SCC), depends on Cohesin, a highly conserved complex of proteins. This complex consists of at least four subunits, the structural maintenance of chromosomes (SMC) proteins SMC1 and SMC3, a member of the kleisin protein family RAD21 and SCC3 (1). Cohesins are thought to form a ring structure that surrounds the replicated sister chromatids, with SMC1 and SMC3 creating a V-shaped heterodimer, bridged by RAD21 (2,3). SCC3 binds to this complex via a C-terminal domain of RAD21. The association of Cohesin complexes with chromatin appears to be facilitated by two additional proteins, SCC2 and SCC4.
Recently, a growing number of Cohesins and its regulatory proteins have been associated with human developmental disorders. Mutations in ESCO2, which encodes a factor essential for the establishment of SCC, result in Roberts syndrome and SC phocomelia (4). In addition, about 50% of patients with Cornelia de Lange syndrome (CdLS) have mutations in the NIPBL gene, the human homolog of the Drosophila Nipped-B and yeast Scc2 gene (5,6). Recently, the first missense mutations in SMC1A and SMC3 were identified in patients with mild variants of CdLS (7,8).
Although Cohesins were initially identified for their role in SCC, Drosphila Nipped-B was isolated as a protein that facilitates transcriptional regulation by remote enhancer sequences (9). Whether SCC2-like proteins have independent functions in cohesion and transcription, or whether these functions are interconnected remains unclear.
Furthermore, increasing evidence indicates that SCC correlates with chromatin remodeling. The Cohesin's SMC1, SMC3 and Scc1/RAD21 have been found to copurify with a chromatin-remodeling complex containing the ATPase SNF2h, presumably due to a direct interaction between Cohesins and chromatin-remodeling complexes (10). Additional links are suggested by mutations of the chromatin-remodeling factors RSC and INO80 that result in defects in SCC (11–13).
To understand the molecular mechanisms underlying CdLS, we used a yeast two-hybrid screen to identify binding proteins of NIPBL. We identified a specific interaction of NIPBL with the histone deacetylases-1 and -3 (HDAC1 and HDAC3) that was verified by coimmunoprecipitation in eukaryotic cells. Luciferase reporter gene assays and chromatin-immunoprecipitation analyses support the model that NIPBL may initiate chromatin-remodeling processes through the recruitment of these HDACs. Furthermore, we could show that missense mutations identified in patients with CdLS influence the interaction of NIPBL with histone deacetylases and result in a decreased activity of this functional interaction.
MATERIAL AND METHODS
Yeast two-hybrid assay
Two overlapping fragments (fragment N, aa 1117–1899 and fragment C, aa 1838–2597) representing the highly conserved region of NIPBL were used as yeast two-hybrid baits. These fragments were PCR-amplified (primers available upon request) from a marathon-ready human fetal brain cDNA library (Clontech-Takara, Saint-Germain-en-Laye, France). Fragments were inserted into the Matchmaker GAL4 Two-Hybrid Sytem 3 (Clontech-Takara) pGBKT7 plasmid. Yeast cells were transformed according to the Matchmaker 3 manual and bait expression was confirmed by western blotting using an anti-GAL4-DBD antibody (Santa Cruz, CA, USA). Human chondrocyte and human ovary cDNA prey libraries were screened according to the manufacturer's protocol (Matchmaker 3, Clontech-Takara). Plasmids from clones obtained under stringent selection were isolated and sequenced. Identity was determined using BlastN and BlastP homology searches. All constructs used were verified by sequencing (PE Applied Biosystems, Darmstadt, Germany).
β-Galactosidase assay
Liquid β-galactosidase assays were performed as per the manufacturer (Matchmaker 3, Clontech). Briefly, NIPBL-subfragments 4–8 were generated from fragment C and inserted into the pGBKT7 plasmid. Full-length HDAC1, HDAC3 and HDAC6 were amplified from human fetal brain cDNA (HDAC1) or full-length ORFs containing plasmids (InvivoGen, San Diego, USA), restriction sites added by PCR and inserted into the pGADT7 plasmid (Clontech-Takara). Yeast cells (AH109) were cotransformed with the NIPBL-fragments 4–8 and all HDAC containing plasmids, respectively. Proper expression was verified by western blotting using specific anti-GAL4-DBD and anti-GAL4-AD antibodies (Santa Cruz). The overnight cultures were measured at 600 nm, cells were harvested and resuspended in buffer Z (60 mM, Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0). An aliquot (V) was taken and o-nitrophenyl-β-d-galactopyranoside was added. After a defined incubation period (t) reaction was stopped by the addition of 1 M Na2CO3. The solutions were cleared of insoluble material by centrifugation and the OD was measured at 420 nm. The β-galactosidase activity (U) was calculated by the following equation: U = 1000 × OD400/(t × V × OD600). All β-galactosidase activities (U) listed represent the result of at least six independent yeast transformants.
Immunoprecipitations
A volume containing 1 mg of total protein extracts from HeLa cells was dissolved in 1 ml of incubation buffer (20 mM HEPES pH 7.9, 75 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 0.1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4) and Proteinase Inhibitor Cocktail (Roche). The solutions were precleared with 30 µl of preequilibrated Protein G PLUS/Protein A-Agar Suspension (Calbiochem, Darmstadt, Germany) for 60 min. The supernatant was then incubated with 10 µl of anti-delangin (anti-NIPBL) antibody (Absea, Beijing, China) for 100 min and 60 µl of preequilibrated A/G-suspension slurry for 12–16 h. The loaded suspensions were precipitated, washed three to five times with incubation buffer and resuspended in SDS-gel loading buffer [62 mM Tris pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol, 0.005% (w/v) bromophenol blue]. All incubations were carried out at 4°C with constant motion using an end-over-end rotor. Precipitates were analyzed by SDS–PAGE and western blotting using the anti-delangin (Absea), anti-HDAC1 (Santa Cruz) and anti-HDAC3 antibodies (Santa Cruz).
Reporter gene assays
NIPBL fragment 4 was inserted into a pcDNA-GAL4-DBD expression plasmid to obtain the GAL4-NIPBL fusion construct. Full-length HDAC1 was amplified and inserted into pcDNA3.1. HDAC3 and HDAC6 expression plasmids were obtained from InvivoGen. The GAL4-TATA-Luc and the GAL4-tk-Luc reporters have been described (14,15).
Transient transfection assays of COS-7 and CHO cells were performed in 96 half-area well plates (Corning, Munich, Germany) with Lipofectamine2000 reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instruction. The phRG-TK Renilla luciferase expression vector (Promega, Mannheim, Germany) was used as a transfection control. Activity of Firefly and Renilla luciferase was measured after 48 h incubation with the Dual Luciferase Reporter Assay System (Promega) in a GeniosPro Luminometer (Tecan, Crailsheim, Germany). All measurements were performed in triplicates. Relative luciferase activity was determined as rate of the average firefly:renilla luciferase activity. To inhibit histone deacetylases, cells were treated with 100 nM Trichostatin A (Merck) and 5 µM Sodium butyrate (Sigma, Taufkirchen Germany), respectively for 24 h prior to measurement. DMSO was used as negative control for Trichostatin A treated cells. All assays had a minimum of six replicates.
Chromatin immunoprecipitation assays
As adapted from Wei et al. (18), COS7 cells were transfected with GAL4-tk-reporter and either the NIPBL-fragment4 GAL4-DBD or the empty GAL4-DBD expression plasmids as described. Chromatin immunoprecipitation (ChIP) assays were performed according to the manufacturer's recommendation (Upstate Biotechnology, Lake Placid, NY, USA). Proteins were cross-linked to DNA by adding formaldehyde to a 1% final concentration. Chromatin was incubated with anti-acetylated histone 3, anti-HDAC1 or anti-HDAC3 antibodies (Upstate Biotechnology; Millipore, Schwalbach, Germany; Santa Cruz) overnight at 4°C. Precipitated immunecomplexes were treated with proteinase K, and DNA was purified by phenol/chloroform extraction for PCR detection with primers flanking the tk promoter region of the GAL4-tk-luc reporter (5′-agcgtcttgtcattggcg-3′ and 5′-ttaagcgggtcgctgcag-3′). Amplified fragments (102 bp) were analyzed on a 1% agarose gel. An SV40 promoter fragment was amplified as an internal control (130 bp; 5′-ttagtcagcaaccaggtg-3′ and 5′- gttaggggcgggactatg-3′).
RESULTS
Identification of HDAC1 and HDAC3 as NIPBL binding proteins
The Cohesin complex consists of multiple protein subunits. In order to identify direct binding proteins of NIPBL, we used the GAL4-Matchmaker III yeast two-hybrid system (Clontech). In silico analyzes of the NIPBL protein predict multiple HEAT-repeats, a glutamine-rich region in the N-terminal part and a bipartite nuclear localization signal (Figure 1A) (5,6,16). Furthermore, at least two different isoforms of NIPBL exist in humans; a small isoform [2697 amino acids (aa) and a calculated molecular weight (mw) of 304 kDa], and a large isoform (2804 aa and mw 316 kDa). Because of the large protein size, we were unable to express full-length NIPBL in yeast cells and created two overlapping fragments (fragment N and fragment C). The latter includes the C-terminal half of NIPBL, which is highly conserved during evolution (Figure 1A and B). Both fragments were used to screen human chondrocyte and ovary libraries. Using fragment C as bait, we isolated more than 250 clones encoding 56 putative NIPBL-binding proteins, which are currently under investigation. Five of these clones encoded three overlapping fragments of the histone deacetylases 1 (HDAC1). Three clones coded for the entire open reading frame of the histone deacetylases 3 (HDAC3), two of which are independent and only differ in the length of the 3′ untranslated region (UTR) (Table 1).
Figure 1.
Predicted structure of the entire NIPBL protein and all fragments used. (A) The two described isoforms (A and B) of NIPBL differ in the very C-terminal part of the protein only. The domains predicted by in silico analyses are indicated, including the five HEAT repeats (H1–H5), the Glutamine (Gln)-rich region (418–462), the bipartite nuclear localization signal [NLS; 1108–1124 and the HP1-interacting motif (22)]. The two missense mutations R1856G and R1895T are indicated by asterisks. (B) Fragments ‘N’ and ‘C’ were fused to the GAL4-DNA-binding domain and used as baits in yeast two-hybrid assays. Fragments 4–8 represent overlapping sub-fragments of fragment C and were used in β-galactosidase assays to narrow down the smallest region necessary for the interaction of NIPBL with HDAC1 and HDAC3. The expression of all constructs was verified by the use of an anti-GAL4-DBD antibody. (C) Fragment 4 (columns 1–3) as well as fragment 5 (columns 4–6) interact with HDAC1 (columns 1 and 4) and HDAC3 (columns 2 and 5), whereas no interactions with fragments 6–8 were detectable (data not shown). As additional control, interaction of HDAC6 with any of the fragments used could be excluded (columns 3 and 6). (D) The two human missense mutations R1856G and R1895T were generated by site-directed mutagenesis and the binding capacities to HDAC1 and HDAC3 were analyzed and compared to the wild-type protein, respectively. While both amino acid substitutions do not alter the binding capacities of NIPBL to HDAC1 (compare columns 1 with 2 and 3), the interaction with HDAC3 resulted in a reduction of β-galactosidase activity to 50% for both mutations compared to the wild-type construct (columns 4–6). Equal expressions of the HDAC1- and HDAC3-GAL4-AD fusion constructs were verified by western blotting.
Table 1.
Clones encoding HDAC1/HDAC3 identified by yeast two-hybrid
| Clone | Protein | Insert (position bp) | cDNA library |
|---|---|---|---|
| 1 | HDAC 1 | 236 –3′UTR + 26 | Chondrocyte |
| 2 | HDAC 1 | 236 − 3′UTR + 26 | Chondrocyte |
| 3 | HDAC 1 | 3 – 1417 | Chondrocyte |
| 4 | HDAC 1 | 53 – 1440 | Ovary |
| 5 | HDAC 1 | 53 – 1440 | Ovary |
| 6 | HDAC 3 | −4 – 3′UTR + 13 | Chondrocyte |
| 7 | HDAC 3 | −4 – 3′UTR + 13 | Chondrocyte |
| 8 | HDAC 3 | 1 – 3′UTR + 56 | Chondrocyte |
The clones encoding HDAC1/HDAC3 are numbered 1–8. The encoding regions of the isolated plasmids are listed by base pair position in the ORF of HDAC1/HDAC3. The minus (−) defines the position in the 5′UTR, 3′UTR positions are defined with plus (+).
To confirm our yeast two-hybrid results and to narrow down the interaction region within NIPBL, we performed liquid β-galactosidase assays using fragment C and five additional constructs (fragments 4–8) encoding overlapping parts of fragment C (Figure 1B). As prey constructs, we used the full-length open reading frames (ORFs) of HDAC1 and HDAC3, each fused to the GAL4-activating domain (GAL4-AD). As shown in Figure 1C, fragments 4 (encoding aa 1838–2000) and 5 (aa 1838–2380) interact with HDAC1 as well as HDAC3, whereas no interactions with fragments 6–8 (6: aa 2000–2380; 7: 2000–2598; 8: aa 2200–2597) were detectable. Thus, the HDAC-interacting region of the NIPBL protein could be narrowed down to 163 aa (1838–2000). This domain includes the highly conserved HEAT repeats H2 and H3. In addition, we analyzed the interaction of NIPBL with HDAC6, which was not identified in our yeast two-hybrid screens. HDAC6 did not interact with the NIPBL-fragments.
Missense mutations affect the interaction of NIPBL with histone deacetylases
In separate studies, we identified a novel de novo missense NIPBL mutation (R1895T) predicted to result in an amino acid exchange within the critical region of NIPBL interaction with HDAC1 and HDAC3. Another missense mutation (c.5566A>G; p.R1856G) within this region was recently described by Selicorni et al. (17). Both patients show classical CdLS facial features and growth but no limb anomalies. We used site-directed mutagenesis to create NIPBL-expression plasmids including the amino acid exchanges R1895T and R1856G, respectively. To test whether these mutations have any influence on the binding capacity of NIPBL to HDAC1 and HDAC3, we performed β-galactosidase assays. Equal expression levels of the NIPBL and HDAC fusion proteins were monitored by western blotting (Figure 1D). While these mutations do not significantly alter the binding of NIPBL to HDAC1, a 2-fold decrease in β-galactosidase activity suggests a reduction in NIPBL interaction with HDAC3 (Figure 1D).
NIPBL forms stable complexes with endogenous HDAC1 and HDAC3 in mammalian cells
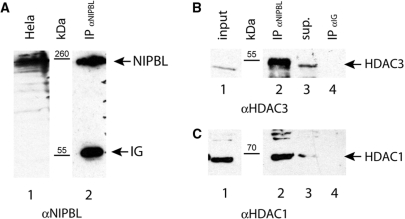
Since the above assays were performed in transformed yeast cells, we analyzed whether endogenous NIPBL protein interacts with HDAC-1 and -3 in mammalian cell lines. Because HeLa cells express detectable amounts of NIPBL (18), we used a monoclonal anti-NIPBL antibody to a C-terminal motif of NIPBL (18) for immunoprecipitation. Western blotting of the anti-NIPBL precipitates with anti-HDAC1 and anti-HDAC3-specific antibodies confirms interaction of endogenous NIPBL and HDAC-1 and -3 (Figure 2). No HDAC1- or HDAC3-specific signal was detected in control samples using A/G sepharose loaded with anti-Ig antibodies for precipitation.
Figure 2.
NIPBL coimmunoprecipitates with HDAC1 and HDAC3 in HeLa cells. (A) Expression of NIPBL was detected with an anti-NIPBL antibody in extracts of HeLa cells (lane 1). The anti-NIPBL antibody was used for immunoprecipitation of NIPBL from HeLa extracts and specific precipitation of NIPBL was monitored (lane 2). (B) HDAC3 was shown with an anti-HDAC3 antibody in HeLa cell extracts (lane 1). The anti-NIPBL antibody and a control anti-IG antibody were used for immunoprecipitation (IP), respectively. HDAC3 was found to be coprecipitated with NIPBL (lane 2), whereas no HDAC3-specific signal could be detected in the control IP (lane 4). Noncoprecipitated HDAC3 was visible in the supernatant (lane 3). (C) Proper expression of HDAC1 was monitored in HeLa cell extracts (lane 1). HDAC1 could only be identified in the anti-NIPBL precipitates (lane 2), while no HDAC1-specific signal was detectable in the IPs using the anti-IG antibody (lane 4). Nonprecipitated HDAC1 could be found in the supernatants of the IPs (lane 3).
NIPBL-mediated recruitment of histone deacetylases modifies promoter activity
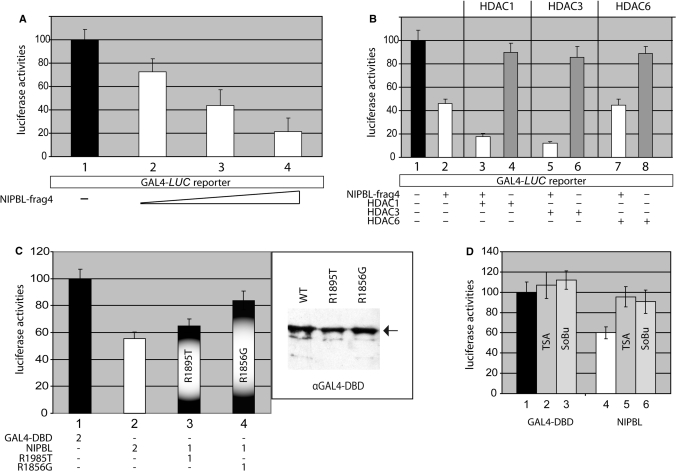
To further investigate a functional association of NIPBL with histone deacetylases, we utilized luciferase reporter gene assays. To accomplish this, the minimal HDAC-interacting region of NIPBL, fragment 4, was fused to the GAL4-DBD containing plasmid to obtain a GAL4-DBD-NIPBL fusion construct. This fusion protein is able to bind to the promoter regions of reporter plasmids containing GAL4-binding sites 5′ to the luciferase cDNA under the control of the TATA box (TATA-Luc) or the tk promoter, respectively. Baseline luciferase activities of mammalian COS7 and CHO cell lines transfected with TATA-Luc or the tk promoter plasmids were arbitrarily set as 100% (Figure 3A, column 1). Notably, cotransfection with NIPBL results in a concentration-dependent decrease of reporter activity (Figure 3A, columns 2–4). To elucidate whether this NIPBL-mediated trans-repressional activity may be due to the recruitment of endogenous histone deacetylases, we cotransfected HDAC1 and HDAC3 expression plasmids. Both HDACs-1 and -3 result in additional decrease of the promoter activity, whereas cotransfection with HDAC6 shows no functional effect (Figure 3B). In control samples, overexpression of each histone deacetylase (HDAC-1, -3 or -6) in the absence of the GAL4-DBD-NIPBL construct has no significant influence on reporter activities (Figure 3B, columns 4, 6 and 8).
Figure 3.
NIPBL-mediated recruitment of HDACs induces transcriptional repression. (A) The HDAC-interacting region of NIPBL, fragment 4, was fused to GAL4-DBD to obtain a GAL4-DBD-NIPBL-frag4 construct (NIPBL-frag4). Cells carrying a luciferase reporter with GAL4-consensus binding sequences 5′ to a LUC reporter gene were transfected with increasing amounts of NIPBL-frag4. The luciferase activity of cells transfected with an empty GAL4-DBD plasmid was set as 100% (column 1). NIPBL-frag4 reduces the reporter activity in a concentration-dependent manner (compare columns 1 with 2–4). (B) Cotransfections with HDAC1- as well as HDAC3-encoding plasmids enhance the transcriptional repression mediated by NIPBL-frag4 (columns 3 and 5), while transfection with HDAC6 does not alter the activity (column 7). As control, the effects of the individual HDAC on the reporter (HDAC1, 3, 6 and columns 4, 6, 8) were monitored in the absence of NIPBL. (C) Whereas missense mutation R1895T only slightly decreases the NIPBL-mediated activity (column 3), mutation R1856G severely reduces the activity (column 4). Equal expression of wild-type and mutant constructs was monitored by the use of an anti-GAL4-DBD antibody in western blots. (D) The effect of TSA and sodium-butyrate (SoBu) on the trans-repressive activity of the NIPBL-frag4 construct was assessed in COS7 cells. Columns 1–3 show the activities of cells tansfected with the empty GAL4-DBD plasmid. Here, TSA and SoBu only very slightly increase the activities. Columns 4–6 represent the results of cells transfected with the NIPBL construct. While the transfection with NIPBL-frag4 decreases reporter gene activity down to 50% in nontreated cells (column 4), treatments with TSA and SoBu, respectively, almost completely abolish the trans-repressional effect of NIPBL (columns 5 and 6). Experiments were done at least in triplicates and the standard deviations are indicated by error bars.
To analyze whether the missense mutations R1895T and R1856G within the HDAC-interacting region of NIPBL have an effect on NIPBL-mediated repression, we generated the constructs GAL4-DBD-R1895T and GAL4-DBD-R1856G. These constructs were used to transfect different cell lines carrying the TATA-Luc or tK promoter containing reporter plasmid. While R1895G only slightly decreases NIPBL-mediated repression (55.4–64.9%), R1856T has a more dramatic effect (55.5–83.8%; Figure 3C). Equal expression of NIPBL constructs was verified by western blotting of extracts with an anti-GAL4-DBD antibody (Figure 3C).
The activity of NIPBL is sensitive to chemical inhibition of histone deacetylases
The direct binding of NIPBL with HDAC1 and HDAC3 as shown in our coimmunoprecipitations and the transcriptional repression demonstrated in the luciferase reporter assays, strongly suggest a functional correlation of NIPBL and histone deacetylation mechanisms. To further characterize whether the NIPBL-mediated repression was dependent upon histone deacetylase activity, we utilized sodium butyrate (SoBu) and trichostatin A (TSA), chemical inhibitors of histone deacetylases. Both inhibitors have been shown to result in hyperacetylation of histones H3 and H4, and result in enhanced promoter activities (19). In their presence, the activity of the GAL4-DBD-NIPBL fusion construct was nearly abolished (Figure 3D), providing further support between the function of NIPBL and histone deacetylation. Notably, TSA- or SoBu-treatment in the presence of the empty GAL4-DBD construct, demonstrate only a minimal increase in reporter gene activity, suggesting the effect is directly on the transcriptional repression by NIPBL, rather than on global up-regulation of transcription.
NIPBL-mediated recruitment of endogenous HDACs induces histone deacetylation
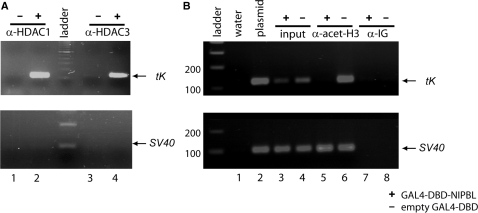
To verify that the promoter activity of the GAL4-DBD-NIPBL fusion is a direct consequence of NIPBL-mediated recruitment of endogenous HDAC1 and HDAC3, ChIP assays were conducted. COS7 cells were cotransfected with tK reporter and the GAL4-DBD-NIPBL construct. Following formaldehyde cross-linking, anti-HDAC1 and anti-HDAC3 antibodies were used for immunoprecipitation. Purified DNA fragments were amplified by PCR and analyzed in electrophoresis (Figure 4A). A tK promoter-specific fragment of 130 bp was identified in both anti-HDAC1 and anti-HDAC3 precipitates, indicating an interaction of both endogenous histone deacetylases with the tK promoter (Figure 4A, upper panel, lanes 2 and 4). In contrast, no signal was detectable in precipitates of cells transfected with an empty GAL4-DBD plasmid (Figure 4A, upper panel, lanes 1 and 3). Furthermore, HDAC1 and HDAC3 do not interact with the SV40 promoter, which regulates the expression of the GAL4-DBD-NIPBL construct (Figure 4, lower panel). These data clearly support a role for NIPBL-mediated recruitment of endogenous histone deacetylases.
Figure 4.
Chromatin-immunoprecipitation (ChIP) assays. (A) The NIPBL-mediated recruitment of HDAC1 (lane 2) and HDAC3 (lane 4) was shown by specific PCR analyses of the purified anti-HDAC1- and anti-HDAC3-precipitates, whereas no signal was detectable in the precipitates of cells transfected with the empty GAL4-DBD plasmid (lanes 1 and 3). As control, the association of HDAC1 and HDAC3 with the SV40 promoter was excluded (lower panel). (B) ChIP assays were used to demonstrate histone deacetylation. The histone deacetylation occurs as the result of the GAL4-DBD-NIPBL (+) expression (lane 5), but not if the GAL4-DBD is expressed alone (−) (lane 6). The upper panel of the figure shows a GAL4-tK promoter specific fragment of ∼130 bp. In the bottom part, a 120 bp PCR product indicating acetylation of the SV40 promoter is shown. Water (lane 1), plasmid DNA (lane 2) as well as input DNA before immunoprecipitation (lanes 3 and 4) were used as controls. As additional control a nonspecific antibody was used for precipitation (α-IG; lanes 7 and 8).
To further analyze if these interactions have an effect on chromatin acetylation, we used an assay (20), where chromatin is precipitated with an antibody that specifically recognizes acetylated lysines-9 and -14 of histone H3. The precipitated DNA fragments were PCR-amplified and analyzed by agarose gel electrophoresis. An expected 130 bp tK promoter-specific fragment can be amplified from control plasmid DNA (Figure 4B, lane 2) and from chromatin prior to precipitations (Figure 4B, lanes 3 and 4). Interestingly, no PCR product is seen for the precipitates of cells transfected with the GAL4-DBD-NIPBL construct, indicating a specific deacetylation of the tK promoter (lane 5). However, the tk promoter PCR product was detectable in controls using the empty GAL4-DBD plasmid (Figure 4B, lane 6). The acetylation status of the SV40 promoter that regulates the expression of the NIPBL construct was monitored in parallel (lower panel of Figure 4B). As expected, since no GAL4-binding sites are present in this vector, H3 acetylation is present, regardless of the presence or absence of the GAL4-DBD-NIPBL construct (Figure 4B, lanes 5 and 6).
DISCUSSION
NIPBL is a member of the highly conserved Scc2-protein family. These proteins, required for the loading of Cohesin onto chromatin, have been implicated in double-strand DNA repair and in the regulation of gene expression. Although both isoforms described for the human NIPBL are fairly large, only a few functional domains have been predicted by in silico analyses. These include a putative bipartite nuclear localization signal, a glutamine-rich region, an HP1-interacting motif and several HEAT repeats, which have been implicated in protein–protein interactions (21). These HEAT repeats are located within the C-terminal part of NIPBL, spanning about amino acids 1300–2500, which is highly conserved in different species and expected to be functionally significant. Moreover, a majority of CdLS-associated missense mutations map to this domain.
In a yeast two-hybrid assay to identfiy NIPBL-binding proteins, we isolated the histone deacetylases, HDAC1 and HDAC3, important cofactors that regulate chromatin structure by deacetylating histone proteins. Through the use of truncated NIPBL constructs, we refined the critical region of NIPBL interaction with HDAC1 and HDAC3 to a stretch of 163 aa (1838–2000), which contains two of the five conserved HEAT repeats. As the use of partial fragments of NIPBL could result in altered protein binding, we analyzed the interaction of full-length NIPBL with HDAC1 and HDAC3. In immunoprecipitation assays, we were able to verify the interaction of the endogenous NIPBL with both HDAC1 and HDAC3 in mammalian cells.
To assess relevance of the critical region for the interaction of NIPBL with HDAC1 and HDAC3 to CdLS, we tested two newly identified NIPBL missense mutations. One of these we recently identified as a de novo mutation (R1895T) and the other was recently described by Selicorni and colleagues (R1856G). Both patients have classical CdLS phenotype without limb abnormalities (17). Each mutation results in decreased binding of NIPBL to HDAC3, whereas the interaction with HDAC1 seems to be unaffected. Both mutations are separated by 39 amino acids and are localized within (R1856G) or adjacent (R1895T) to HEAT-repeat 2.
In luciferase reporter gene assays we show that this region of NIPBL when fused to the GAL4-DBD modifies reporter gene expression. Using ChIP assays we show that this NIPBL-mediated activity is due to direct interaction with endogenous histone deacetylases. Furthermore, cotransfection of HDAC1 and HDAC3 result in a further decrease of promoter activity, whereas HDAC6, which does not directly associates with the NIPBL fragment, does not modify the reporter gene expression significantly. Interestingly, the two missense mutations analyzed have similar effects on HDAC1 and HDAC3 binding affinities, but exert different functional activities. Mutation R1895T only slightly alters this function, whereas mutant R1856G significantly decreases the activity mediated by NIPBL. Lastly, chemical inhibition of histone deacetylation by treatment of the cell cultures with TSA or SoBu almost completely abolishes the NIPBL-mediated activity. These data are the first to clearly point to a direct functional connection of the mammalian SCC2-like protein NIPBL with the recruitment of histone deacetylating enzymes.
Histone acetylation, chromatin remodeling and Cohesin
Although the interplay of chromatin structure and histone modification with Cohesin has been frequently described, the precise function is poorly understood. It has been hypothesized that chromatin structure may play an important role in determining whether, and where, Cohesin binds to chromosomes in eukaryotic cells (22).
It is known that histone modifications affect the recruitment of Cohesin to particular chromosomal loci (23). In our ChIP analyses, we could show that aa 1838–2000 of NIPBL are able to initiate the deacetylation of lysine 9 of histone 3 (H3K9), which would allow its methylation. The heterochromatin binding protein 1 (HP1), which was also described to bind NIPBL (24), recognizes methylated lysine 9 on histone H3 (H3K9) and deletion of Swi6, the yeast ortholog of HP1, causes a loss of Cohesin binding (25). This is consistent with earlier observations that the interaction between the Scc3 Cohesin subunit with Swi6 is needed to recruit Cohesin to centromeric regions in yeast (23,26). On the other hand, it was shown very recently, that the Suv39h-HP1 pathway is not essential for the enrichment of Cohesin at centromeres in mammals (27). These observations clearly indicate differences in the functional connection of HP1/Swi6 and Cohesins in yeast and mammalian cells.
In addition to the above literature, it has been shown that diminishing HDACs by TSA-treatment or mutation promotes abnormal sister chromatid separation. Sister chromatids do not separate when cells enter mitosis with hyperacetylated histones. Thus, the presence of acetylated histones in mitosis induces both aberrant chromosome numbers (i.e. aneuploidy) and defects in chromosome structure (28). Recently, Kimata and colleagues (29) described TSA-hypersensitivity of a mutant Mis4, a Cohesin loading adherin protein. TSA-treatment of mis4 mutant cells results in decreasing Cohesin-binding to chromatin in the chromosome arm regions. Furthermore, the correlation of various histone modifications and Cohesin is the subject of a continuously growing number of publications. In human cells, a chromatin-remodeling complex containing the ATPase SNF2h was found to copurify with RAD21 and SMC proteins, presumably due to a direct interaction between Cohesin and this complex (10). Mutations in the yeast chromatin-remodeling complex RSC cause modest defects in sister-chromatid cohesion (11,12). Furthermore, deletions of Rsc2, encoding a further alternative variant of the RSC complex, significantly reduces SCC by reducing the amount of chromosome-associated Cohesins (30).
The function of NIPBL in regulating gene expression
Although Scc2-like proteins are necessary to recruit the Cohesin complex to chromosomes, patients with CdLS usually do not show significant cohesion defects (31,32), suggesting that the diverse developmental deficits are caused by gene-expression changes similar to those in Drosophila, where heterozygous Nipped-B mutations also do not result in severe cohesion defects. In yeast, Cohesin binds almost exclusively between genes and most binding sites are between convergent transcription units. Coupled with the finding that Scc2 does not colocalize with Cohesin, this led to the idea that Cohesin loads onto chromosomes at Scc2-binding sites and is subsequently pushed to the end of the genes by RNA polymerase (33,34). This contrasts sharply from the data obtained in Drosophila. In Drosophila, Nipped-B was discovered in a genetic screen for factors that facilitate long-range transcriptional activation of the cut and Ultrabithorax (Ubx) homeobox genes by enhancers positioned some 80 and 50 kb away from the promoters (9,35). It has been shown previously that Nipped-B and Cohesin bind to the same sites throughout the entire nonrepetitive Drosophila genome. Here, Nipped-B seems to preferentially bind to transcribed regions and overlap with RNA polymerase II. According to these results, high transcriptional activity does not seem to prevent Cohesin binding (36). The colocalization of Nipped-B with Cohesin supports the idea that it dynamically regulates binding of Cohesin and suggests additional mechanisms by which Cohesin might affect transcription (36). Although we have identified NIPBL-mediated recruitment of histone deacetylases to chromatin, which results in local alterations of the chromatin structure, the precise function of this interaction is still unknown. The numerous genomic loci where Nipped-B has been identified suggest a more general role in transcriptional regulation. Whether NIPBL directly regulates specific promoter activities or may participate in creating a platform for the recruitment of specific transcription (co-) factors needs to be analyzed in future experiments.
In this report, we show for the first time a direct interaction and functional consequence of histone deacetylating enzymes with the Cohesin-loading protein NIPBL. Although the precise function of this interplay is still unknown, our data presented here give new insight into the molecular pathology of CdLS and further contribute to the hypothesis that it is likely not the result of defects in cohesion; but rather, it is likely due to changes on chromatin structure and effects on gene expression.
FUNDING
Medical Faculty of Lübeck (E15-2008).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are thankful to Matthew Deardorff (Division of Genetics, Department of Pediatrics, the University of Pennsylavnia, USA) and Stefan Weger (Institut für Infektionsmedizin, Charité Campus Benjamin Franklin in Berlin, Germany), for the critical and fruitful discussion of the article.
REFERENCES
- 1.Uhlmann F. The mechanism of sister chromatid cohesion. Exp. Cell. Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Gruber S, Haering CH, Nasmyth K. Chromosomal Cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 3.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J. Structure and stability of Cohesin's Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 4.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 5.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 7.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 8.Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, et al. Mutations in Cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads Cohesin onto human chromosomes. Nature. 2002;418:994–948. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 11.Baetz KK, Krogan NJ, Emili A, Greenblatt J, Hieter P. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodelling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 2004;24:1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Hsu JM, Laurent BC. The RSC nucleosome-remodeling complex is required for Cohesin's association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 13.Ogiwara H, Enomoto T, Seki M. The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle. 2007;6:1090–1095. doi: 10.4161/cc.6.9.4130. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser FJ, Lüdecke HJ, Weger S. SUMOylation modulates transcriptional repression by TRPS1. Biol. Chem. 2007;388:381–390. doi: 10.1515/BC.2007.051. [DOI] [PubMed] [Google Scholar]
- 15.Böhm J, Kaiser FJ, Borozdin W, Depping R, Kohlhase J. Synergistic cooperation of Sall4 and cyclin D1 in transcriptional repression. Biochem. Biophys. Res. Commun. 2007;356:773–779. doi: 10.1016/j.bbrc.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 16.Strachan T. Cornelia de Lange syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr. Opin. Genet. Dev. 2005;15:258–264. doi: 10.1016/j.gde.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, Bentivegna A, Masciadri M, Domi A, Divizia MT, et al. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin. Genet. 2007;72:98–108. doi: 10.1111/j.1399-0004.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- 18.Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han S, Lu J, Zhang Y, Cheng C, Li L, Han L, Huang B. HDAC inhibitors TSA and sodium butyrate enhanced the human IL-5 expression by altering histone acetylation status at its promoter region. Immunol. Lett. 2007;108:143–150. doi: 10.1016/j.imlet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Wei LN, Hu X, Chandra D, Seto E, Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 2000;275:40782–40787. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 21.Neuwald AF, Hirano T. HEAT repeats associated with condensins, Cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riedel CG, Gregan J, Gruber S, Nasmyth K. Is chromatin remodeling required to build sister-chromatid cohesion? Trends Biochem. Sci. 2004;29:389–392. doi: 10.1016/j.tibs.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of Cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell. Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 24.Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher F.J., III The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 26.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 27.Koch B, Kueng S, Ruckenbauer C, Wendt KS, Peters JM. The Suv39h-HP1 histone methylation pathway is dispensable for enrichment and protection of Cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- 28.Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell. 2003;14:3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimata Y, Matsuyama A, Nagao K, Furuya K, Obuse C, Yoshida M, Yanagida M. Diminishing HDACs by drugs or mutations promotes normal or abnormal sister chromatid separation by affecting APC/C and adherin. J. Cell Sci. 2008;121:1107–1118. doi: 10.1242/jcs.024224. [DOI] [PubMed] [Google Scholar]
- 30.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 31.Kaur M, DeScipio C, McCallum J, Yaeger D, Devoto M, Jackson LG, Spinner NB, Krantz ID. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. A. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, et al. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum. Mol. Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- 33.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the Cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;E259:1325–1339. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, et al. Association of Cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]