Abstract
O6-Methylguanine (O6-meG) is a major mutagenic, carcinogenic and cytotoxic DNA adduct produced by various endogenous and exogenous methylating agents. We report the results of transcription past a site-specifically modified O6-meG DNA template by bacteriophage T7 RNA polymerase and human RNA polymerase II. These data show that O6-meG partially blocks T7 RNA polymerase and human RNA polymerase II elongation. In both cases, the sequences of the truncated transcripts indicate that both polymerases stop precisely at the damaged site without nucleotide incorporation opposite the lesion, while extensive misincorporation of uracil is observed in the full-length RNA. For both polymerases, computer models suggest that bypass occurs only when O6-meG adopts an anti conformation around its glycosidic bond, with the methyl group in the proximal orientation; in contrast, blockage requires the methyl group to adopt a distal conformation. Furthermore, the selection of cytosine and uracil partners opposite O6-meG is rationalized with modeled hydrogen-bonding patterns that agree with experimentally observed O6-meG:C and O6-meG:U pairing schemes. Thus, in vitro, O6-meG contributes substantially to transcriptional mutagenesis. In addition, the partial blockage of RNA polymerase II suggests that transcription-coupled DNA repair could play an auxiliary role in the clearance of this lesion.
Methylating agents are among the array of chemicals that damage the genome. They react with nucleophilic sites in DNA, adding methyl groups to the purines, pyrimidines and the phosphodiester backbone, resulting in modifications that compromise the fidelity and efficiency of replication and transcription. These agents include exogenous compounds like N-methyl-N-nitrosourea, methyl methane sulfonate and N-methyl-N′-nitro-nitrosoguanidine (1), along with endogenous biochemical agents like S-adenosylmethionine (2) and metabolically nitrosated amines and amides such as betaine and choline (3). Nitrates and nitrites found in food and beverages can also form methylating agents in cells (4). Hence, aberrant DNA methylation is an unavoidable consequence of metabolism and exposure to the surrounding environment (5,6). Approximately 12 different methylated DNA lesions have been identified, with the relative amount of each depending largely on the chemical nature of the reactive alkylating species. The most frequent lesions occur at the N7- or O6-positions of guanine and the N3 position of adenine (6,7).
O6-Methylguanine (O6-meG) (Figure 1) is a stable DNA lesion formed by the reaction of methylating agents, including the chemotherapeutic agents procarbazine and temozolomide, with guanine (8,9). O6-MeG is highly mutagenic since it often instructs DNA polymerases to incorporate thymine instead of cytosine, resulting in GC to AT transitions (10–12). The highly mutagenic nature of O6-meG offers an explanation as to why methylating agents that produce it are carcinogenic (13). But O6-meG can also block replication (14), inducing chromosomal aberrations, sister chromatid exchange and intra-chromosomal homologous recombination (15–17).
Figure 1.
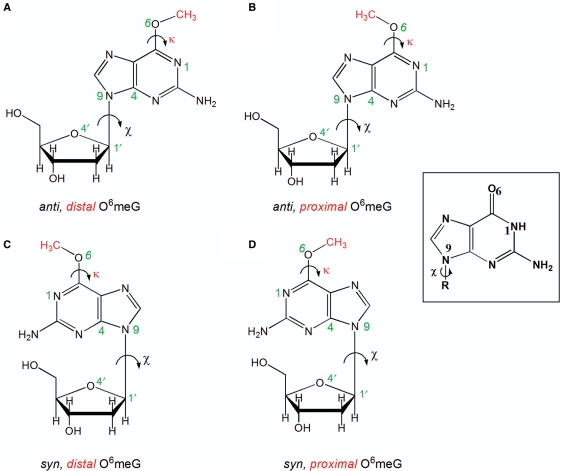
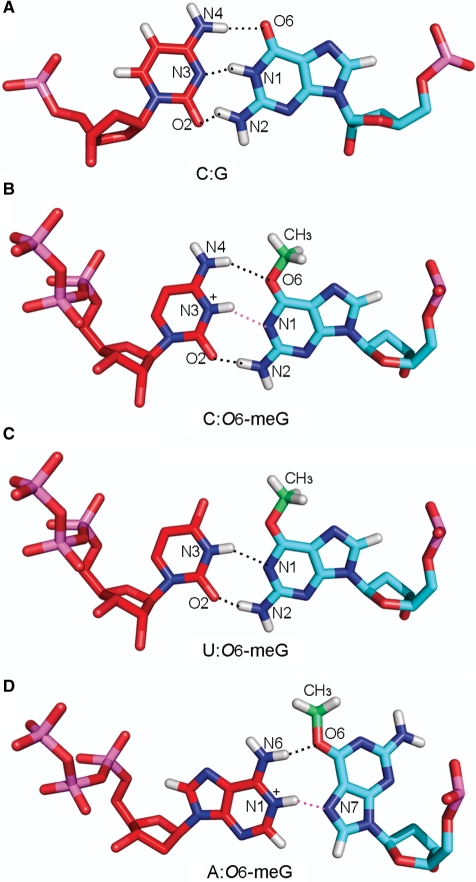
Structures of the four possible conformations of O6-meG. The glycosidic torsion angle (χ) defined by the atoms O4′–C1′–N9–C4 is shown in black and the torsion angle (κ) defined by the atoms N1–C6–O6–C is displayed in red. The angle χ is shown in both the anti (the six-membered ring points away from the sugar) or syn (the six-membered ring is over the sugar) conformations, while the torsion angle (κ) is in the distal (facing N1–C6) or the proximal (facing away from N1–C6) orientation. These torsional possibilities give rise to four conformational combinations for the lesion (A) anti,distal (B) anti,proximal (C) syn,distal and (D) syn,proximal O6-meG. Guanine is shown as an inset to highlight the different hydrogen-bond donor/acceptor properties.
O6-MeG is repaired in a non-enzymatic, stoichiometric fashion by the direct reversal protein O6-methylguanine–DNA methyltransferase (MGMT) (18). MGMT transfers the methyl group from the lesion to a cysteine residue within itself, rendering it incapable of continued repair; hence, the number of O6-meG lesions that can be cleared from a cell's genome is limited by the available MGMT molecules, unless more of the protein is made. In fact, variations in MGMT levels among cell types result in a differential ability of organs and tissues to repair O6-meG via direct reversal, leading to the accumulation of O6-meG after MGMT is depleted (19,20).
Replication past O6-meG by several DNA polymerases has been well studied (7,10,14,21–24); however, little is known about its effect on transcription elongation. In Escherichia coli, RNA polymerase bypasses O6-meG with high efficiency, often misincorporating uracil opposite the lesion. The resulting alterations to the transcripts have been termed transcriptional mutagenesis (TM) (25). Here, we examined transcriptional processing of O6-meG by bacteriophage T7 RNA polymerase (T7 RNAP) and human RNA polymerase II (hRNAPII). These two polymerases differ significantly in the architecture of their active sites and behave differently when facing DNA lesions (26–28). It is shown here for the first time that O6-meG poses a minimal hurdle to transcription by T7 RNAP, but it does impede hRNAPII transcription to a significant extent. Consistent with the previous study with E. coli RNA polymerase (25), bypass of O6-meG causes significant sequence alterations in the RNA transcripts, with predominant misincorporation of uracil by both polymerases. Computer modeling was utilized to provide a structural explanation of the experimental results. These models showed that the altered Watson–Crick hydrogen-bonding edge of O6-meG (Figure 1) underlies the observed selectivity for uracil as well as cytosine incorporation during transcription past the lesion, with hydrogen-bonding possibilities between O6-meG and uracil or cytosine that have been inferred in NMR solution studies (29–32) and observed in X-ray crystal structures of O6-meG (22,33–36). In addition, the more crowded active site region of hRNAPII explained the greater blocking effect of the lesion in this polymerase.
MATERIALS AND METHODS
Materials
Unless stated otherwise, all reagents, enzymes and biologicals were purchased from Promega, Sigma-Aldrich, New England Biolabs, Ambion or BioRad. DNA oligomers were obtained from Sigma-Aldrich. Radioactive materials were purchased from Perkin Elmer. All DNA sequencing was done using the GeneAmp PCR System 9700 (BioRad) and 3100 Capillary Sequencer (Applied Biosystems).
Transcription template construction
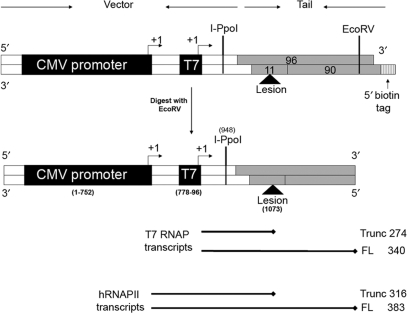
DNA templates suitable for transcription by T7 RNAP and hRNAPII were prepared as described in (26,27,37) (Figure 2). In brief, the restriction endonuclease BbsI was used to linearize the plasmid pCI-neo-G-less-T7, which contained the two promoters needed for both RNAPs (Supplementary Figure S1). A double-strand tail was assembled by annealing three synthetic single-strand DNA oligomers—an 11-mer and a 96-mer, to a complementary 90-mer containing a 5′-biotin tag. The 11-mer was either unmodified or modified with a site-specific O6-meG lesion, depending on whether a control or adduct-modified template was being assembled, respectively. DNA oligomers are shown in Table 1. T4 DNA ligase was used to ligate the tail to the digested vector for 16 h at 16°C. The product was precipitated using streptavidin-linked beads, gel purified and tested for nicks as described elsewhere (26,27,37).
Figure 2.
Site specifically modified DNA templates prepared for transcriptional assays. The vector displayed in white is comprised of the cloned T7 promoter and the CMV promoter/enhancer (black boxes). The tail region is formed by annealing a 96-mer, a 90-mer and an 11-mer (grey boxes). The position of the O6-meG lesion is shown with a triangle. The size of the template is 1140 bp after digestion with EcoRV, the last step during the purification of the template which removes the biotin tag from the tail end. The arrowheads indicate the +1 transcription start site. The expected sizes of the RNA products obtained during transcription by T7 RNAP and hRNAPII are shown below the template.
Table 1.
Sequence of DNA oligomers and primers used
| Oligomer name | DNA Sequence |
|---|---|
| 96-mera | 5′-TTGCGGTAGCGATGGGTGCTGTACTCAGGTGTGG AATCAACCCACAGCTGACAGGGCAGGTCTTGGCCAG TTGGGATATCCAAAACATCTTGTTGA-3′ |
| 90-mera | 5′-TTTTTTTTTTCAACAAGATGTTTTGGATATCCCAA CTGGCCAAGACCTGCCCTGTCAGCTGTGGGTTGATTCC ACACCTGAGTACAGCAC-3′ |
| 11-mera | 5′-CCATCGCTACC-3′ |
| 49Lb | 5′-GCTTCCTCATCCTTCTCCATCTCTCTGTGCAGCTG TGGGTTGATTCACG |
| 24Fc | 5′-AACGCAGTCAGTGCTTCTGACACA-3′ |
| 25Rc | 5′-GAGAGATGGAGAAGGATGAGGAAGC -3′ |
| 22R trunc | 5′-TACAGTGCTCGCATACGCTCAA-3′ |
| 24 chimera | 5′-UUUUGAGCGTATGCGAGCACTGTAidt-3′ |
aThe 96-mer, 90-mer and the 11-mer were annealed to make the tail of the DNA templates. The bold underscored G in the 11-mer represents either the undamaged guanine or the damaged O6-meG.
b49L is the primer used in the reverse transcription of the full-length RNA obtained from transcription of T7 RNAP past the DNA templates.
c24F and 25R are the forward and reverse PCR primers.
22R: The reverse transcription primer used in the ligation mediated reverse transcription. It is complementary to the chimera, which was ligated to the RNA prior to reverse transcription.
Transcription reactions with bacteriophage T7 RNA polymerase
Transcription using T7 RNAP was performed as recommended by the manufacturer Promega and as described elsewhere (26,27). Briefly, the transcription reactions contained 20 units of T7 RNAP (a unit is defined as the amount of enzyme required to catalyze the incorporation of 5 nmol of CTP into acid-insoluble product in 60 min at 37°C in a total volume of 100 μl), 25 ng (0.033 pmols) of DNA template, 2.5 mM each of ATP, GTP and UTP, 200 μM [α-32P]CTP (50 Ci/mmol), 100 mM DTT and 10 units RNasin, 40 mM Tris–HCl (pH 7.9 at 25°C), 10 mM NaCl, 6 mM MgCl2 and 2 mM spermidine in a final volume of 20 μl. The molar concentrations provided are the stock solution concentrations. The reaction was incubated at 37°C for 1 h, the unincorporated nucleotides were removed, and an equal volume of loading dye (80% formamide, 5 mM Tris–boric acid, 1 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol) was added to the product, which was then resolved using denaturing PAGE.
To test the efficiency of lesion bypass, a transcription time course was performed. The reaction was initiated as described earlier, and a radioactively labeled 108-base DNA oligomer was added to the master mix to act as a standard during PAGE. Aliquots were removed from the reaction mix at time points 0, 1, 10, 20, 30, 40, 50 and 60 min. Each aliquot was quenched by adding loading dye and heating to 65°C for 15 min to ensure polymerase inactivation. ImageQuant software (Molecular Dynamics) was used to visualize and quantify the radiolabeled transcripts, and for each time point, the RNA band intensity was compared to the DNA standard intensity.
Transcription reactions with human RNA polymerase II
Multiple rounds of transcription reactions using hRNAPII were performed as recommended by the manufacturer Promega and as described in (26,27,37,38). HeLa nuclear extract served as the source of the hRNAPII machinery, and the presence of the CMV enhancer/promoter on the DNA templates, which is specific for promoting transcription by RNAPII, ensured that only hRNAPII complexes formed that could carry out RNA elongation past the lesion (Promega and references herein). Briefly, transcription was carried out using 8 units of HeLa nuclear extract (a unit is defined as the amount of extract required for the incorporation of 50 fmol of nucleotides into a 363-nt runoff transcript ‘generated from the CMV immediate early promoter fragment’ per hour at 30 °C), 100 ng (0.15 pmols) DNA template, 10 units of RNase inhibitor, 3 mM MgCl2, 10 mM each of ATP, GTP and UTP, 0.4 μM CTP, 1 μl [α-32P]CTP (25 Ci/mmol), 20 mM HEPES (pH 7.9 at 25°C), 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol in a final volume of 25 μl (Molar concentrations provided are the stock solution concentrations). The reaction was incubated at 30°C for 60 min, and then quenched using a stop solution [0.3 M Tris–HCl (pH 7.4 at 25°C), 0.3 M sodium acetate, 0.5% SDS, 2 mM EDTA and 3 μg/ml tRNA] provided by the manufacturer (Promega). The resulting RNA was purified by phenol:chloroform extraction and resolved using denaturing 7% PAGE.
For a single round of transcription, GTP was omitted from the NTP mix, and the polymerase complex was allowed to transcribe up to the point at which the first GTP was required. After 10 min, heparin was added to bind all free hRNAPII. Finally, the addition of 100 μM GTP allowed transcription to continue, and after an additional 40 min, the reaction was quenched. The resulting RNA was purified by phenol:chloroform extraction and resolved using denaturing 7% PAGE.
To test the efficiency of lesion bypass, a transcription time course was performed. In order to prevent repair of O6-meG, HeLa nuclear extracts were pre-incubated with 0.5 μM O6-benzylguanine (BZG), a potent inhibitor of the MGMT that repairs O6-meG (39). A loading standard 175 bases in length was added to the reaction mix, and the transcription reaction was initiated by the addition of the DNA template as described earlier. Aliquots were removed from the reaction mix, and quenched in stop solution at time points 0, 1, 5, 10, 20, 30, 40 and 60 min. The resulting RNA was purified and resolved on 7% PAGE. ImageQuant software (Molecular Dynamics) was used to visualize and quantify the radiolabeled transcripts. At each time point, RNA band intensity was used to determine the percentage of each RNA species present.
RNA sequencing
To sequence full-length transcripts, transcription reactions were performed without radioactive CTP but with all four non-radioactive nucleotides, each at a concentration of 500 μM for T7 RNAP and 400 μM for hRNAPII. Transcription reactions were followed by digestion with RNase-free DNase at 37°C for 30 min to destroy any residual DNA template. The resulting RNA was purified and sequenced using the reverse transcriptase polymerase chain reaction (RT–PCR). In brief, for the reverse transcriptase reaction, 20 pmol of primers were annealed to the RNA template, ImProm II (Promega) was used as a source of reverse transcriptase, and the reaction was performed following the manufacturer's protocol. Residual RNA was digested with RNase, and the RNase was subsequently heat inactivated. Excess primers and small RNA fragments were eliminated by using Microcon Centrifugal Filter Columns. Finally, PCR was performed with the forward and reverse primers 24F and 25R (Table 1).
To sequence the truncated transcripts, 1 μM of a chimeric RNA–DNA oligomer, 24 chimera (Table 1), was ligated to the 3′-ends of the transcripts using T4 RNA ligase and DMSO (40). The chimeric RNA–DNA oligomer contains four ribonucleotides at its 5′-end to facilitate ligation to the transcripts using RNA ligase; the other bases of the oligomer are deoxyribonucleotides that make the chimeric oligomer resistant to degradation. The reverse transcriptase primer 22RTrunc was annealed to the tail, and RT–PCR was subsequently performed. This approach selected all RNA, both the full-length and the truncated transcripts to which the chimeric oligomer was ligated.
For both full-length and truncated RNA, the RT–PCR products were cloned and sequenced. The RT–PCR products were purified using a 1.2% non-denaturing agarose matrix and subsequently inserted into the pDrive vector (Promega) specifically designed for cloning PCR products. The cloned PCR products were used to transform Qiagen EZ competent cells grown on agar plates containing kanamycin and IPTG. Only white colonies, indicating the insertion of the PCR product in the vector, were selected and grown overnight in Luria broth supplemented with ampicillin. Plasmids were extracted from the cells using the QIAprep Spin Miniprep Kit (Qiagen) and were then sequenced. For each polymerase, 100 clones were sequenced for full-length products and 50 clones were sequenced for the truncated products (26,27).
Molecular modeling of O6-meG in T7 RNAP
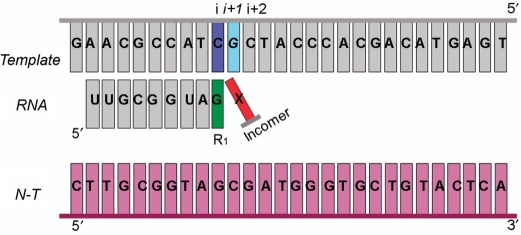
To model the O6-meG adduct in the active site of T7 RNAP, four available X-ray crystal structures of the protein were used. The crystal structures represent four phases of one cycle of nucleotide addition during the elongation phase of transcription: the pre-insertion substrate complex, the insertion substrate complex, the pre-translocation product complex and the post-translocation complex with PDB codes 1S0V (41), 1S76 (42), 1S77 (42) and 1MSW (43), respectively. These structures are shown in Supplementary Figure S2. In each phase, the protein was crystallized in a ternary complex with a DNA duplex of approximately 30 bases, a 9 base DNA:RNA hybrid, and an incoming ribonucleotide, except for the post-translocation phase. Sequences of the template/non-template DNA and the RNA transcript were re-modeled to match the sequences used in the study reported here (Figure 3).
Figure 3.
Sequences employed in the experimental and modeling studies in T7 RNAP and yRNAPII complexes. The insertion position i + 1 occupied by the template nucleotide to be transcribed, G or O6-meG is shown in cyan. The pre-insertion position i + 2 occupied by the next to be transcribed nucleotide is in grey. The ribonucleotide previously incorporated at the 3′-end of the RNA is designated R1 and shown in green. The rNTP to be inserted opposite the lesion is called Incomer and colored red and the non-template DNA strand is colored in purple.
The structure of the damaged O6-meG base was obtained from a crystal structure of the lesion paired with a cytosine [PDB code 1D24 (33)]. The unmodified guanine at position i+1 or i+2 (only in the post-translocation complex) of the template strand was replaced by this O6-meG structure (Figure 3). The subsequent search for optimal structures employed the following procedure. The glycosidic torsion angle of the lesion was retained in the original anti conformation present in the crystal (χ = 252°) (Figure 1A and B). The base of the crystal NTP was replaced by cytosine or uracil while retaining the original anti conformation of the glycosidic torsion angle (χ = 196°). The torsion angle (κ) (Figure 1) governing the methyl group conformation of the adduct was then rotated utilizing the INSIGHTII (Accelrys) utility bumpcheck in a search for structures with minimal collisions. This torsion angle has been observed in crystal structures to adopt two conformational ranges termed distal and proximal with (κ) values in the range of 0–76° and 159–208°, respectively (22,33–36,44). However, to accommodate the incoming pyrimidines, only the proximal conformation for (κ), ranging from 182° to 191°, provided structures without clashes. The structure with κ = 183° was used for further study, as this value fell in the middle of the range of proximal κ-values (159–208°) previously observed in crystal structures of O6-meG (22,34–36,44). Subsequently, the slight, close contacts remaining (<2 Å) were relieved by rotation of the side chain of Arg-632, namely around the Cα–Cβ, Cβ–Cγ and Cγ–Cδ bonds. Finally, additional adjustments were made to the torsion angle (χ) of the NTP (<15°) in order to locate structures with optimal hydrogen bonds between O6-meG and the incoming NTP.
In the case of an incoming ATP, the original anti conformation of O6-meG exhibited severe collisions with the ribonucleotide. To relieve the clashes observed in the model and explain the insertion of adenine in the growing RNA as identified by the biochemical results, the torsion angle (χ) of O6-meG was continuously rotated to find the range of minimal collision, which was in the syn domain. As with the incoming pyrimidines, slight close contacts (<2 Å) were relieved by rotation of the side chain of the same Arg-632, namely around the Cβ–Cγ, Cγ–Cδ and Cδ–Nε bonds. When a collision-free range was obtained, the torsion angle (χ) of the lesion was further rotated slightly (<3°) to locate a conformation with optimal hydrogen-bonding possibilities between the incoming ATP and O6-meG. No NTP was present opposite the base at i+2 in the post-translocation complex. The hydrogen-bonding criteria in the insertion complex were in the following observed ranges (22,45–50): (1) donor to acceptor atom distance of 2.5 Å < X–A< 3.9 Å; (2) hydrogen to acceptor atom distance of 1.5 Å < H–A < 2.9 Å; and (3) angle θ defined by the donor–hydrogen–acceptor atoms (X–H–A) >90°. Full details for the torsion angles and the resulting hydrogen bonds in all the modeled structures are provided in Tables S1 and S2, respectively.
Molecular modeling of O6-meG in yeast RNA polymerase II
Crystal structures for the hRNAPII are not available at this writing. However, crystal structures for yeast S. cerevisiae RNAPII (yRNAPII) shares 53% sequence identity with the human polymerase, and the conserved residues are distributed over the entire structure (51). In addition, cryo-electron microscopy with image reconstruction and variance analysis of the hRNAPII together with homology modeling of the human enzyme based on the yRNAPII show that the human and the yeast enzymes’ cores that contain the active sites are structurally indistinguishable (52). This makes the yeast polymerase a good model for the human enzyme. The X-ray crystal structure of S. cerevisiae RNAPII, PDB code 2NVZ (53), was in a ternary complex form with a 29-base DNA template strand, a 10-base RNA transcript and a 14-base non-template DNA strand complementary to the template strand downstream of the RNA. This structure was used as the starting model for yRNAPII containing the O6-meG lesion in its active site. The modeling procedures employed for yRNAPII were very similar to the procedures described earlier for T7 RNAP. First, the undamaged templating base present in the crystal was replaced by the lesion while retaining its original glycosidic torsion angle (χ = 187°), placing the damaged guanine in an anti conformation. Then, the torsion angle κ was continuously rotated to find a collision-free range for the methyl group (κ = 200–250°). The original crystal contained a uracil as an incoming ribonucleotide. Therefore, to explain the incorporation of uracil opposite the lesion as shown in the experimental results, the possibility of hydrogen bonding between O6-meG and uracil was evaluated. Hydrogen-bonding requirements followed the criteria specified earlier for the T7 RNAP models. Next, the incoming ribonucleotide was remodeled to a cytosine, to interpret its observed incorporation in the RNA; again the glycosidic torsion angle was retained as in the crystal (χ = 220°), and finally, hydrogen-bonding possibilities with O6-meG were investigated. All modeling was carried out with the Biopolymer module of INSIGHTII (Accelrys), and all images were generated using the PYMOL program.
RESULTS
DNA templates suitable for transcription were synthesized and confirmed to be free of nicks
DNA templates were constructed with a site-specific O6-meG on the transcribed strand. The lesion was positioned downstream from the CMV enhancer/promoter and the T7 RNAP promoter to allow for comparisons between the two RNAPs transcribing past O6-meG (Figure 2). Unmodified templates acted as control DNA. To test for the presence of nicks in the ligated products, the constructed templates were digested with I-PpoI, an endonuclease that cuts with high specificity at a 15-bp recognition sequence. The fragments were radiolabeled at the 5′-ends and analyzed on a denaturing PAGE. When ligation is complete, the resulting DNA fragments should be 187, 191 and 953 bases in length. In contrast, incomplete ligation that results in the presence of nicks in the template would result in fragments 73, 77, 110 or 118 bases in length. The absence of the shorter bands indicates that ligation was indeed efficient, and the templates for transcription were free of nicks (Supplementary Figure S3).
O6-MeG minimally blocks T7 RNA polymerase elongation
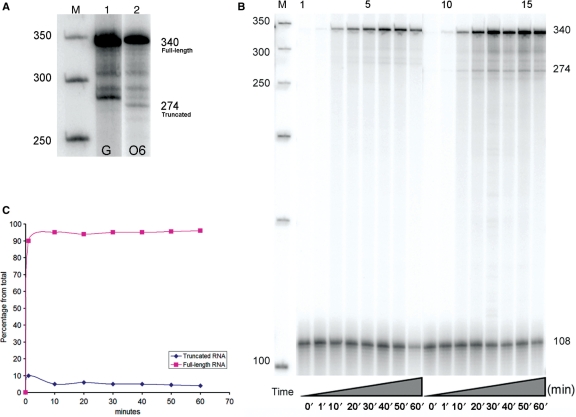
Transcription of the unmodified DNA with T7 RNAP produced only full-length RNA 340 bases in length. In contrast, transcription of the modified DNA template primarily produced full-length transcripts 340 bases in length, with some truncated RNA of approximately 274 bases (Figure 4A). The percentage of full-length RNA averaged from nine separate reactions was 95 ± 1%. The one percent value represents the standard deviation from the mean. The shorter transcripts coincided with the position of O6-meG on the template strand, indicating that this lesion impedes elongating T7 RNAP. The shorter RNA molecules observed between 274 and 340 bases were also obtained with the unmodified templates; hence, inherent pause sites on the DNA likely cause their formation, not the damaged base.
Figure 4.
Effect of O6-meG modified-DNA on transcription by T7 RNAP. (A) T7 RNAP transcription of the unmodified DNA resulted in full-length RNA (FL) approximately 340 bases in length (lane 1) while the modified template resulted in a mixture of two RNA species approximately 274 and 340 bases in length (lane 2). A 50 bp DNA step ladder is labeled M. (B) Time course of transcription with T7 RNAP past the undamaged DNA (lanes 1 through 8) and the O6-meG template (lanes 9 through 16). Aliquots were removed every 10 min to measure the extent of lesion blockage or bypass over time. Fragments were compared to a DNA standard of 108 bases in length used to minimize loading errors. The increased intensity across lanes indicates an increased amount of RNA over time. Transcription past the damaged templates shows that the lesion poses a minor obstruction to the rapid RNA synthesis. (C) The graph illustrates the percent of full-length RNA. It indicates that the amount of full-length RNA remains the same over time (95%) and that the lesion causes the production of ∼5% of truncated RNA at any time point.
Experiments were carried out to examine T7 RNAP lesion bypass efficiency over time. Aliquots were removed from the reaction mix at different times as described in the Materials and Methods section. For each time point, the RNA band intensity was compared to a DNA standard of 108 bases in length. The experiment was performed in triplicate, and in each case, the undamaged templates produced increasing amounts of full-length RNA over time (Figure 4B, lanes 1 through 8), while the O6-meG-damaged templates produced mainly full-length transcripts (∼95%), with a small amount of truncated RNA (∼5%) (Figure 4B, lanes 9 through 16). The results suggest that rapid RNA synthesis is mildly obstructed by O6-meG.
O6-MeG partially blocks human RNA polymerase II transcription
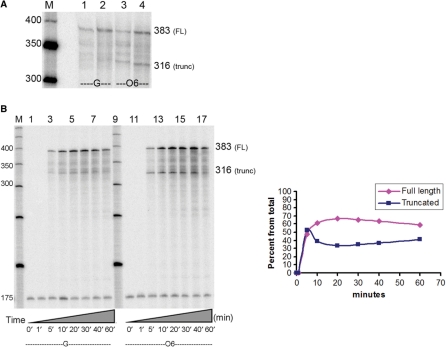
HeLa nuclear extract was used as the source of hRNAPII machinery. The presence of the CMV enhancer/promoter on the DNA templates supports hRNAPII transcription initiation alone, as described in the manufacturer's protocol (www.promega.com/tbs/tb123/tb123.html). Both single and multiple rounds of transcription were carried out on undamaged and damaged templates. For single rounds of transcription, a G-less cassette was utilized to synchronize the reaction so that a template could be transcribed only once by a single RNA polymerase (54). For multiple rounds of transcription, a template could be transcribed multiple times by several polymerase complexes. After 60 min, both single and multiple rounds of transcription using the unmodified control resulted in the formation of full-length transcripts 383 bases in length, with no truncated transcripts occurring in the vicinity of the O6-meG (Figure 5A, lane 1). However, transcription using the damaged template produced truncated and full-length RNA of 316 and 383 bases in length, respectively. Single round of transcription produced 64 ± 6% of full-length RNA, while multiple rounds of transcription produced 74 ± 2% of full-length RNA. The data obtained represent the average of 10 separate transcription experiments, with standard deviations from the mean given.
Figure 5.
Effect of O6-meG modified-DNA on transcription by hRNAPII. (A) Single and multiple rounds of transcription using HeLa nuclear extracts. Undamaged templates generated full-length (FL) transcripts of approximately 383 bases in length (lanes 1 and 2) while O6-meG-damaged templates produced two RNA species of 383 and 316 bases in length (lanes 3 and 4). A 50 bp DNA step ladder is shown (M). (B) Time course of transcription with hRNAPII past the undamaged DNA (lanes 1 through 8) and the O6-meG template (lanes 10 through 17). Aliquots were removed at the time points indicated on the figure to measure the extent of lesion blockage or bypass over time. Fragments were compared to a DNA standard of 175 bases in length used to minimize loading errors. Transcription past the damaged templates shows that the lesion partially obstructs RNA synthesis (only producing ∼60% of full-length RNA at time points 20 min through 60 min). The graph illustrates the % of full-length and truncated RNA over time (x-axis). It indicates that the amount of full-length and truncated RNA remain almost constant after the first ten minutes of the transcription reaction.
As with T7RNAP, time course experiments were carried out to examine hRNAPII lesion bypass efficiency over time. We carried out these transcription reactions in the presence of 0.5 µM BZG to ensure that repair of O6-meG does not occur. The time course shows that undamaged templates only produce full-length RNA over time (Figure 5B, lanes 1 through 8), while templates containing O6-meG produced a constant ratio of full-length (∼60%) and truncated RNA (∼40%) at each time point after the first 10 min (Figure 5B, lanes 10 through 17). These results suggest that RNA synthesis is partially obstructed by O6-meG and that repair of the damaged templates is unlikely to occur during the course of the performed transcription reaction as performed here.
Note that the addition of BZG to the HeLa nuclear extracts resulted in a decrease in the amount of full-length RNA, from 74% to ∼58% at 60 min, indicating that the extracts may have some MGMT activity which is thwarted in the presence of BZG, eliminating repair of the lesion. Note that nucleotide excision repair (NER) does not recognize O6-meG in human cell extracts (55), indicating that this pathway is not responsible for the repair of the lesion. Furthermore, mismatch repair does not remove O6-meG but replaces the mismatched thymine on the opposite strand (56); hence, this pathway is also not responsible for the clearance of this lesion in the extracts.
T7 RNAP and hRNAPII generate extensive base misincorporation opposite O6-meG in full-length RNA
RT–PCR was used to sequence the full-length and truncated RNA following transcription of DNA templates that contain O6-meG. The resulting PCR products consisted of a heterogeneous population of DNA, necessitating subcloning to analyze individual RT-PCR products. For full-length transcripts resulting from T7 RNAP transcription past the O6-meG templates, the polymerase primarily incorporated both pyrimidines equally (47% each) and adenine to a very small extent (6%) (Table 2). Undamaged templates produced only full-length transcripts having the correct base sequence (data not shown). In the case of full-length RNA obtained for hRNAPII bypass of O6-meG, cytosine and uracil were exclusively incorporated opposite the lesion with a 3:1 ratio. Misincorporation of adenine was not observed in full-length RNA generated by hRNAPII (Table 2).
Table 2.
Bases incorporated opposite O6-meG in the full-length RNA during transcription with T7 RNAP and hRNAPII
| % rNTP | C | U | A |
|---|---|---|---|
| RNAP | |||
| T7 RNAP | 47 | 47 | 6 |
| hRNAPII | 75 | 25 | – |
For the truncated transcripts, T7 RNAP often stalled exactly at the site of the lesion and did not permit nucleotide incorporation opposite it. In some cases, T7 RNAP was able to misincorporate either one adenine or a cytosine followed by two non-template instructed adenines before terminating transcription (Table 3). In contrast, hRNAPII typically stopped at the site of O6-meG, but without nucleotide incorporation opposite the lesion. Additionally, hRNAPII misincorporated cytosine opposite the base immediately preceding O6-meG, which was cytosine, and then stopped (Table 3).
Table 3.
Bases incorporated opposite O6-meG in the truncated RNA during transcription with T7 RNAP and hRNAPIIa
| Sequence | RNA length (bases) | Percentage | |
|---|---|---|---|
| Expected RNA sequence | …UCUUGCGGUAGCGAUGG | ||
| Sequence obtained with T7 RNAP | …UCUUGCGGUAG | 272 | 25 |
| …UCUUGCGGUAGA | 273 | 25 | |
| …UCUUGCGGUAGCAA | 275 | 50 | |
| Sequence obtained with hRNAPII | …UCUUGCGGUAG | 272 | 80 |
| …UCUUGCGGUAC | 20 |
aBold, underscored bases represent misincorporated bases after the lesion.
Modeling suggests hydrogen-bonding possibilities that explain observed nucleotide selectivity
Bacteriophage T7 RNAP
Molecular modeling was employed to provide a structural rationale for the experimentally observed findings: full-length RNA exhibited significant base misincorporation events opposite the lesion with base selectivity C = U >> A, and shorter transcripts were truncated one residue before the lesion. Two glycosidic bond conformations (χ) for the O6-meG nucleoside were investigated, namely anti and syn (Figure 1). Our models showed that when the O6-meG nucleoside was in the anti conformation, it allowed the insertion of pyrimidine NTPs with no steric collisions, provided that the methyl group adopted the proximal conformation relative to the N1 atom (Figure 1B). This conformation avoided collision with incoming pyrimidines and allowed base pairing to occur. For CTP, two hydrogen-bonding interactions were identified, one between N4 of cytosine and O6 of O6-meG and another between O2 of cytosine and N2 of O6-meG. In case of an N3-protonated cytosine, an additional hydrogen bond could occur with the N1 of O6-meG. A protonated cytosine would allow a Watson–Crick-like pairing between the lesion and the incoming NTP similar to what was identified crystallographically (29,33,36) (Figure 6B). In the case of UTP, two hydrogen bonds were found possible, one between N3 of uridine and N1 of O6-meG, and the second between O2 of uridine and N2 of O6-meG. This O6-meG:U pair is in a ‘Watson–Crick isosteric’ conformation (36), also termed ‘pseudoWatson–Crick’ (22), and was observed crystallographically (33,34,36) (Figure 6C).
Figure 6.
Hydrogen-bonding interactions between (A) a G:C pair, O6-meG and (B) cytosine, (C) uridine and (D) adenine, respectively (see text). (A) G:C pair; G(O6):C(N4), G(N1):C(N3), and G(N2): C(O2), (B) anti,proximal O6-meG:C pair; O6-meG(O6):C(N4) and O6-meG(N2): C(O2), (C) anti,proximal O6-meG:U pair; O6-meG(N1): U(N3) and O6-meG(N2):U(O2) and (D) syn, distal O6-meG:A pair; O6-meG(O6): A(N6). Hydrogen bonds involving potential protonation sites, O6-meG(N1):C(N3+) and O6-meG(N7):A(N1+) are shown in pink. The methyl group carbon is colored in green. Color code is the same as in Figure 3. Details of the potential hydrogen-bonding partners are described in Supplementary Table S2.
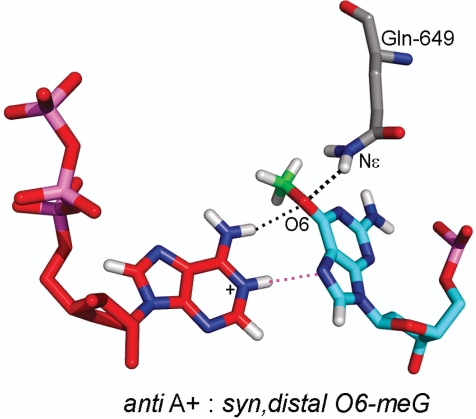
In order to accommodate an incoming adenine, the O6-meG nucleoside had to adopt the syn conformation around its glycosidic bond (χ) to avoid overlap with the six-membered ring of the adenine. In addition, the methyl group had to be oriented in the distal conformation (Figure 1C). In this conformation, the methyl could be housed, without collision, in the polymerase active site pocket on the major groove side of the damaged guanine, allowing the incorporation of adenine opposite it. Interestingly, this syn conformation was stabilized by one hydrogen bond between the O6 of O6-meG and a protein side chain of Gln-649 (Figure 7 and Supplementary Table S2B). The syn (χ) distal (κ) conformation of the lesion allowed for one hydrogen-bonding interaction between the N6 of the incoming ATP and the O6 of the O6-meG. For the case of an N1-protonated adenine, as observed in crystal structures (57,58), an additional hydrogen bond could form between the N1 hydrogen of the ATP with the N7 of the lesion (Figure 6D).
Figure 7.
H-bond interaction between O6-meG and the T7 RNAP residue, Gln-649. The O6-meG in the syn conformation is stabilized by one hydrogen bond between the O6 of O6-meG and the Nε of Gln-649. The hydrogen bond is shown as a dotted black line. Hydrogens of the backbone were removed for clarity. Color code is the same as in Figures 3 and 6.
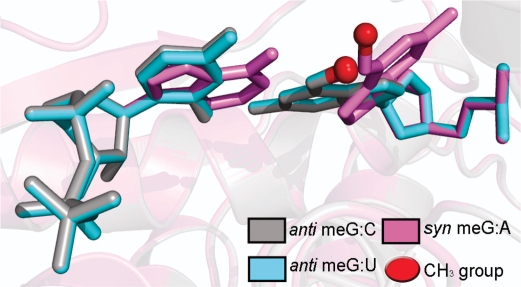
These hydrogen-bonding possibilities offer an explanation for the incorporation of cytosine and the misincorporation of uracil and adenine in the RNA opposite O6-meG during transcription elongation by T7 RNAP: equal insertion of cytosine and uracil is consistent with their equivalent hydrogen-bonding possibilities with O6-meG, while the much smaller selection for adenine is explained by the requirement for the abnormal and less-prevalent syn conformation of the templating lesion in order to allow hydrogen bonding with the incoming ATP (Figures 6 and 8). We have given detailed results for the substrate insertion complex, but the models were essentially identical for the pre-translocation product complex and quite similar in the post-translocation and the substrate pre-insertion complexes. Detailed results of the torsion angles and the hydrogen-bonding partners are given in Tables S1 and S2.
Figure 8.
View of the O6-meG lesion pairing with cytosine (gray), uracil (cyan) and adenine (violet) inside the active site of T7 RNAP. The backbone of the protein is rendered in transparent cartoons, superimposed on the three pairs and colored using the same code as the DNA:RNA pair shown in the figure. The methyl group of the lesion is shown as a red sphere. For lesion pairing with cytosine or uracil, the O6-meG:C and O6-meG:U pairs superimpose completely and mimic a normal Watson–Crick base pair. With the O6-meG:A pair, the O6-meG must adopt the less-prevalent syn conformation for collision-free placement of the incoming rATP.
Yeast RNAPII (yRNAPII)
As detailed in the Materials and Methods section, current evidence indicates that the active site regions of yeast and human RNA polymerase II are in fact nearly identical (51,52). This makes the yeast polymerase a suitable model for the human enzyme. Hence, molecular modeling was employed to explain the observed incorporation of C and to lesser extent U opposite the lesion in the full-length RNA using yRNAPII. No adenine incorporation was observed. In the case of the truncated transcripts, nucleotide incorporation did not occur opposite the O6-meG lesion. To explain the observed bypass events, the models had the glycosidic bond (χ) for the O6-meG nucleoside in the anti conformation and there was a range in κ around the O6–C6 bond which was collision free (κ = 200–250°), similar to the T7 RNAP bypass models. In this domain, the lesion was in an anti, proximal orientation (Figure 1B). We term this orientation the bypass conformation because it is compatible with nucleotide incorporation into the RNA. The anti, proximal orientation of the O6-meG allowed for the formation of two hydrogen bonds with each of the incoming pyrimidines. For CTP, the hydrogen bonds involved the N4 of cytosine with O6 of O6-meG, and the O2 of cytosine with N2 of O6-meG. Again, in the case of an N3-protonated cytosine, an additional hydrogen bond could form with the N1 of O6-meG. For UTP, the two hydrogen bonds involved the N3 of uridine with N1 of O6-meG and the O2 of uridine with N2 of O6-meG. These hydrogen-bonding schemes, which are detailed in Figure 6 and Supplementary Table S3, are similar to what was obtained for T7 RNAP, and explain the incorporation of uracil and cytosine in the growing RNA.
When the methyl group was outside the allowed κ range, it collided with either the incoming ribonucleotide or neighboring protein residues, namely Thr-827, Ala-828, Val-829 of the bridge helix. These collisions could not be alleviated by minimal rotation of the side chains of the helix, because slight rotations of the bridge helix residues cause severe distortions to the yRNAPII structure (53). This conformational range (κ ≠ 200–250°), termed the blocking structure, provides a model to explain the halting of the polymerase opposite O6-meG.
The modeling studies showed that ATP could not be accommodated with the O6-meG in the anti glycosidic bond conformation due to collisions between the lesion and the large purine. While, it was possible to obtain a collision-free structure containing ATP, this structure was otherwise unfavorable. The lesion could only be placed in a narrow region of the syn domain (χ = 238–243°) with κ in the distal conformation (κ = 172–238°) (Figure 1C). This structure had two hydrogen bonds between the Hoogsteen edge of the O6-meG and the A [N6(A):O6(meG) and N1+(A):N7(meG)], but it contained an unfavorable repulsive interaction between the O6 and an oxygen of Ala-828 of subunit 1 of yRNAPII, with close distance of ∼2.6 Å (Supplementary Figure S4). This repulsive interaction together with the small region of conformational space in χ that is permissible sterically would make this structure unlikely to occur. Thus, the observed failure to incorporate adenine is reasonable on the basis of these models.
DISCUSSION
Gene expression is a tightly regulated process that requires RNA polymerase to read a transcription unit when directed. To achieve this, an unperturbed, processive elongation complex centered around an RNA polymerase is essential. However, transcription is often obstructed by pausing sequences, protein roadblocks (59) or DNA damage (60). More specifically, unrepaired damage present on the transcribed strand of a gene can either block an elongating RNA polymerase or lead to bypass with nucleotide insertion opposite or near the site of the damage (60). While arrest of transcription is believed to trigger TCR, lesion bypass can induce TM (25).
Here we report that, in the current sequence context, O6-meG partially blocks transcription elongation of both T7 RNAP and hRNAPII, albeit to different extents (Figures 4 and 5); the lesion poses a more significant block to hRNAPII. Although, there have been contradictory reports on the preferential repair of O6-meG from the transcribed strand of a gene (61,62), our results indicate that partial polymerase blockage by O6-meG raises the possibility that this lesion might invoke TCR (63). Hence, the blocked polymerase could provide an additional means for cells to identify O6-meG, not recognized by the MGMT protein. This possibility needs to be investigated further.
In addition, we show the contribution of O6-meG to TM in vitro. T7 RNAP bypasses the lesion efficiently but with low fidelity, resulting in a high frequency of nucleotide misinsertion opposite the lesion with selectivity C = U = 47% >> A = 6%. Similarly, hRNAPII bypasses O6-meG with misincorporation of uracil in 25% of the full-length RNA. During gene expression and mRNA translation, each protein-encoding mRNA is translated about 40 times on average; therefore, an error in the primary code of the RNA, such as produced by O6-meG, can lead to a burst effect of flawed proteins (64). In this connection, it is interesting that the incorporation of uracil was evidenced during a transcriptional assay past O6-meG in human cells (Burns,J.A. Dreij,K. and Scicchitano,D.A. unpublished data). Hence, our work adds O6-meG to the pool of smaller DNA lesions that partially block transcription and may induce TM; others include the oxidative lesions 8-oxoguanine (65,66), thymine glycols (67) and cisplatin 1,2-intrastrand d(GpG) crosslinks (68).
Sequences of the truncated RNAs showed that when T7 RNAP and hRNAPII stopped at the lesion, nucleotide incorporation opposite O6-meG was not permitted (Table 3). Modeling indicated that in both polymerases transcription blockage would ensue if the lesion adopts an anti, distal orientation, shown in Figure 1A. In the distal orientation, the methyl group is in close proximity to the incoming ribonucleotide, producing steric collisions that thwart incorporation and elongation. Moreover, the greater blockage observed with hRNAPII could be attributed to the occurrence of a distal conformation of the methyl group which highly crowds the incoming NTP in the more confined hRNAPII. Interestingly, two additional truncated RNAs were obtained during transcription by T7 RNAP with either an additional adenine or a cytosine followed by two non-template instructed adenines (Table 3). These two truncated RNA species were also observed in other studies where T7 RNAP stalled at damaged guanines namely 1,N2-ethenogunaine (1,N2-εG) (26) and 5-guanidino-4-nitroimidazole (NI) (27). This behavior may therefore be inherent to the stalled T7 RNAP and independent of the lesion. It has been suggested that stalled RNA polymerases tend to add one or two bases at the 3′-end of transcripts (69) further supporting our observation.
The bypass events observed with O6-meG can be explained on the basis of the altered hydrogen-bonding possibilities of O6-meG compared to guanine (Figure 1). Specifically, the hydrogen-bond acceptor atom N1 of guanine becomes a hydrogen-bond donor in O6-meG. With the lesion in an anti-proximal conformation our models showed that, in both polymerases, there were no collisions with the incoming UTP or CTP. We readily modeled a pseudo Watson–Crick base pair with uracil (22) and a Watson–Crick-like pair with cytosine (36). Each pair allowed two hydrogen bonds between the lesion and the incoming nucleotide (Figure 6B and C). A protonated cytosine allowed three hydrogen bonds to form with an alignment that is very near Watson–Crick. These pairing schemes were feasible in both T7 RNAP and yRNAPII, and a number of crystal structures (22,33–36) and NMR solution studies (29–32) have revealed the base-pairing possibilities for O6-meG present in our models. While a wobble interaction with cytosine [N4(C):N1(O6-meG) and N3(C):N2(O6-meG)] was also seen in crystal structures (70) and inferred by NMR solution studies (32), this scheme requires rearrangement in the DNA backbone which could be energetically costly. The preference for cytosine over uracil incorporation opposite O6-meG in yRNAPII probably stems from the more confining active site region of yRNAPII, which likely selects more stringently for a hydrogen-bonding scheme closest to Watson–Crick. Our models show that adenine could only be accommodated in T7 RNAP when O6-meG was in the abnormal syn orientation around its glycosidic bond; explaining the low adenine misincorporation (Figure 7). No favorable models were possible for the yRNAPII, explaining why adenine incorporation was not seen in the full-length transcripts.
CONCLUSION
The present work extends our understanding of lesion processing by RNA polymerases and the possible attendant sequelae of either TM or TCR. Our work and that of others have provided studies of bulky lesions derived from polycyclic aromatic carcinogens [reviewed in (60,71)] with diverse shapes and stereochemical properties, as well as the rigid 1,N2-εG (26), the flexible NI (27) and here the smaller O6-meG. From these investigations, we deduce that the unique and specific hydrogen-bonding capabilities of the lesion with different incoming NTPs play an important role in TM. For the specific case of O6-meG, the altered hydrogen-bond donor/acceptor properties (Figure 1) allow for enhanced selectivity for uracil compared to guanine in both T7 RNAP and hRNAPII. In addition, lesion flexibility has been pinpointed as an important structural factor based on our studies of the conformationally heterogenous NI lesion (27). Our results show that for O6-meG, the possible variation in the torsion angle κ, which governs the rotation about the O6–C6 bond, and hence the placement of the bulky methyl group in the polymerase active site is key. Both distal and proximal conformations have been explicitly observed by NMR (29–32) and X-ray crystallography (33,35,36). The conformation of the methyl group is the determinant for whether bypass or blockage occurs. Polymerase blockage and the subsequent induction of TCR is connected to steric hindrance by the lesion in the polymerase active site. Blockage structures stem from a distal orientation of the methyl group which places this sterically challenging group in collision with the templating base. Our results are consistent with earlier modeling studies suggesting that multiple conformations of O6-meG account for the observed blockage and pyrimidine insertion during in vitro replication (72). Thus, the subtle structural distinctions between guanine and its O6-methylated derivative produce significant consequences, not only as a mutational damage during replication, but through its impact on transcription. Further research is needed to discern the impact of O6-meG on the production of faulty proteins in cells via TM and the potential impact of TCR on the lesion's clearance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (ES010581 to D.A.S., 2R01 CA75449 and 5R01 CA28038 for partial support of computational infrastructure and systems management) Funding for open access charge: ES010581 to D.A.S.
Conflict of interest statement. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Nicholas Geacintov for his critical reading of the manuscript and helpful discussions. We also thank the members of the Scicchitano Lab for reading and evaluation of the manuscript.
REFERENCES
- 1.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 2.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sedgwick B. Nitrosated peptides and polyamines as endogenous mutagens in O6-alkylguanine-DNA alkyltransferase deficient cells. Carcinogenesis. 1997;18:1561–1567. doi: 10.1093/carcin/18.8.1561. [DOI] [PubMed] [Google Scholar]
- 4.Liu JG, Li MH. Roussin red methyl ester, a tumor promoter isolated from pickled vegetables. Carcinogenesis. 1989;10:617–620. doi: 10.1093/carcin/10.3.617. [DOI] [PubMed] [Google Scholar]
- 5.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 6.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loechler EL. A violation of the Swain-Scott principle, and not SN1 versus SN2 reaction mechanisms, explains why carcinogenic alkylating agents can form different proportions of adducts at oxygen versus nitrogen in DNA. Chem. Res. Toxicol. 1994;7:277–280. doi: 10.1021/tx00039a001. [DOI] [PubMed] [Google Scholar]
- 8.Souliotis VL, Kaila S, Boussiotis VA, Pangalis GA, Kyrtopoulos SA. Accumulation of O6-methylguanine in human blood leukocyte DNA during exposure to procarbazine and its relationships with dose and repair. Cancer Res. 1990;50:2759–2764. [PubMed] [Google Scholar]
- 9.Taverna P, Catapano CV, Citti L, Bonfanti M, D’Incalci M. Influence of O6-methylguanine on DNA damage and cytotoxicity of temozolomide in L1210 mouse leukemia sensitive and resistant to chloroethylnitrosoureas. Anticancer Drugs. 1992;3:401–405. doi: 10.1097/00001813-199208000-00014. [DOI] [PubMed] [Google Scholar]
- 10.Toorchen D, Topal MD. Mechanisms of chemical mutagenesis and carcinogenesis: effects on DNA replication of methylation at the O6-guanine position of dGTP. Carcinogenesis. 1983;4:1591–1597. doi: 10.1093/carcin/4.12.1591. [DOI] [PubMed] [Google Scholar]
- 11.Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc. Natl Acad. Sci. USA. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer B, Dosanjh MK. Site-directed mutagenesis for quantitation of base-base interactions at defined sites. Mutat. Res. 1990;233:45–51. doi: 10.1016/0027-5107(90)90150-3. [DOI] [PubMed] [Google Scholar]
- 13.Marchesi F, Turriziani M, Tortorelli G, Avvisati G, Torino F, De Vecchis L. Triazene compounds: mechanism of action and related DNA repair systems. Pharmacol. Res. 2007;56:275–287. doi: 10.1016/j.phrs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Voigt JM, Topal MD. O6-methylguanine-induced replication blocks. Carcinogenesis. 1995;16:1775–1782. doi: 10.1093/carcin/16.8.1775. [DOI] [PubMed] [Google Scholar]
- 15.Galloway SM, Greenwood SK, Hill RB, Bradt CI, Bean CL. A role for mismatch repair in production of chromosome aberrations by methylating agents in human cells. Mutat. Res. 1995;346:231–245. doi: 10.1016/0165-7992(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 16.Kaina B, Fritz G, Coquerelle T. Contribution of O6-alkylguanine and N-alkylpurines to the formation of sister chromatid exchanges, chromosomal aberrations, and gene mutations: new insights gained from studies of genetically engineered mammalian cell lines. Environ. Mol. Mutagen. 1993;22:283–292. doi: 10.1002/em.2850220418. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Tsujimura T, Bhattacharyya NP, Maher VM, McCormick JJ. O6-methylguanine induces intrachromosomal homologous recombination in human cells. Carcinogenesis. 1996;17:2229–2235. doi: 10.1093/carcin/17.10.2229. [DOI] [PubMed] [Google Scholar]
- 18.Pegg AE, Dolan ME, Scicchitano D, Morimoto K. Studies of the repair of O6-alkylguanine and O4-alkylthymine in DNA by alkyltransferases from mammalian cells and bacteria. Environ. Health Perspect. 1985;62:109–114. doi: 10.1289/ehp.8562109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pegg AE, Fang Q, Loktionova NA. Human variants of O(6)-alkylguanine-DNA alkyltransferase. DNA Repair (Amst). 2007;6:1071–1078. doi: 10.1016/j.dnarep.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JM, Zhang YP, Wang C, Sun Y, Fujimoto J, Ikenaga M. O6-methylguanine-DNA methyltransferase activity in human tumors. Carcinogenesis. 1992;13:1503–1507. doi: 10.1093/carcin/13.9.1503. [DOI] [PubMed] [Google Scholar]
- 21.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 22.Eoff RL, Irimia A, Egli M, Guengerich FP. Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by “wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. J. Biol. Chem. 2007;282:1456–1467. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 23.Woodside AM, Guengerich FP. Misincorporation and stalling at O(6)-methylguanine and O(6)-benzylguanine: evidence for inactive polymerase complexes. Biochemistry. 2002;41:1039–1050. doi: 10.1021/bi011496f. [DOI] [PubMed] [Google Scholar]
- 24.Woodside AM, Guengerich FP. Effect of the O6 substituent on misincorporation kinetics catalyzed by DNA polymerases at O(6)-methylguanine and O(6)-benzylguanine. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- 25.Viswanathan A, Liu J, Doetsch PW. E. coli RNA polymerase bypass of DNA base damage. Mutagenesis at the level of transcription. Ann. NY Acad. Sci. 1999;870:386–388. doi: 10.1111/j.1749-6632.1999.tb08909.x. [DOI] [PubMed] [Google Scholar]
- 26.Dimitri A, Goodenough AK, Guengerich FP, Broyde S, Scicchitano DA. Transcription processing at 1,N2-ethenoguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. J. Mol. Biol. 2008;375:353–366. doi: 10.1016/j.jmb.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimitri A, Jia L, Shafirovich V, Geacintov NE, Broyde S, Scicchitano DA. Transcription of DNA containing the 5-guanidino-4-nitroimidazole lesion by human RNA polymerase II and bacteriophage T7 RNA polymerase. DNA Repair (Amst) 2008;7:1276–1288. doi: 10.1016/j.dnarep.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perlow RA, Kolbanovskii A, Hingerty BE, Geacintov NE, Broyde S, Scicchitano DA. DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner. J. Mol. Biol. 2002;321:29–47. doi: 10.1016/s0022-2836(02)00593-4. [DOI] [PubMed] [Google Scholar]
- 29.Williams LD, Shaw BR. Protonated base pairs explain the ambiguous pairing properties of O6-methylguanine. Proc. Natl Acad. Sci. USA. 1987;84:1779–1783. doi: 10.1073/pnas.84.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel DJ, Shapiro L, Kozlowski SA, Gaffney BL, Jones RA. Structural studies of the O6meG.C interaction in the d(C-G-C-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986;25:1027–1036. doi: 10.1021/bi00353a012. [DOI] [PubMed] [Google Scholar]
- 31.Patel DJ, Shapiro L, Kozlowski SA, Gaffney BL, Jones RA. Structural studies of the O6meG.T interaction in the d(C-G-T-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986;25:1036–1042. doi: 10.1021/bi00353a013. [DOI] [PubMed] [Google Scholar]
- 32.Gaffney BL, Goswani B, Jones RA. Nitrogen-15-labeled oligodeoxynucleotides. Use of 15H NMR to probe H-bonding in an O6MeG.C base pair. J. Am. Chem. Soc. 1993;115:12607–12608. [Google Scholar]
- 33.Ginell SL, Kuzmich S, Jones RA, Berman HM. Crystal and molecular structure of a DNA duplex containing the carcinogenic lesion O6-methylguanine. Biochemistry. 1990;29:10461–10465. doi: 10.1021/bi00498a005. [DOI] [PubMed] [Google Scholar]
- 34.Ginell SL, Vojtechovsky J, Gaffney B, Jones R, Berman HM. Crystal structure of a mispaired dodecamer, d(CGAGAATTC(O6Me)GCG)2, containing a carcinogenic O6-methylguanine. Biochemistry. 1994;33:3487–3493. doi: 10.1021/bi00178a004. [DOI] [PubMed] [Google Scholar]
- 35.Vojtechovsky J, Eaton MD, Gaffney B, Jones R, Berman HM. Structure of a new crystal form of a DNA dodecamer containing T.(O6Me)G base pairs. Biochemistry. 1995;34:16632–16640. doi: 10.1021/bi00051a011. [DOI] [PubMed] [Google Scholar]
- 36.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O(6)-methyl-guanine lesions. Proc. Natl Acad. Sci. USA. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perlow RA, Schinecker TM, Kim SJ, Geacintov NE, Scicchitano DA. Construction and purification of site-specifically modified DNA templates for transcription assays. Nucleic Acids Res. 2003;31:e40. doi: 10.1093/nar/gng040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schinecker TM, Perlow RA, Broyde S, Geacintov NE, Scicchitano DA. Human RNA polymerase II is partially blocked by DNA adducts derived from tumorigenic benzo[c]phenanthrene diol epoxides: relating biological consequences to conformational preferences. Nucleic Acids Res. 2003;31:6004–6015. doi: 10.1093/nar/gkg771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc. Natl Acad. Sci. USA. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 42.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 43.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 44.Yamagata Y, Kohda K, Tomita K. Structural studies of O6-methyldeoxyguanosine and related compounds: a promutagenic DNA lesion by methylating carcinogens. Nucleic Acids Res. 1988;16:9307–9321. doi: 10.1093/nar/16.19.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabernero L, Bella J, Aleman C. Hydrogen bond geometry in DNA-minor groove binding drug complexes. Nucleic Acids Res. 1996;24:3458–3466. doi: 10.1093/nar/24.17.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panigrahi SK, Desiraju GR. Strong and weak hydrogen bonds in the protein-ligand interface. Proteins. 2007;67:128–141. doi: 10.1002/prot.21253. [DOI] [PubMed] [Google Scholar]
- 47.Torshin IY, Weber IT, Harrison RW. Geometric criteria of hydrogen bonds in proteins and identification of “bifurcated” hydrogen bonds. Protein Eng. 2002;15:359–363. doi: 10.1093/protein/15.5.359. [DOI] [PubMed] [Google Scholar]
- 48.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 49.Fabiola F, Bertram R, Korostelev A, Chapman MS. An improved hydrogen bond potential: impact on medium resolution protein structures. Protein Sci. 2002;11:1415–1423. doi: 10.1110/ps.4890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spratt TE, Levy DE. Structure of the hydrogen bonding complex of O6-methylguanine with cytosine and thymine during DNA replication. Nucleic Acids Res. 1997;25:3354–3361. doi: 10.1093/nar/25.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 52.Kostek SA, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14:1691–1700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viswanathan A, Doetsch PW. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem. 1998;273:21276–21281. doi: 10.1074/jbc.273.33.21276. [DOI] [PubMed] [Google Scholar]
- 55.Kusumoto R, Masutani C, Sugasawa K, Iwai S, Araki M, Uchida A, Mizukoshi T, Hanaoka F. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat. Res. 2001;485:219–227. doi: 10.1016/s0921-8777(00)00082-3. [DOI] [PubMed] [Google Scholar]
- 56.Karran P, Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 57.Brown T, Leonard GA, Booth ED, Chambers J. Crystal structure and stability of a DNA duplex containing A(anti).G(syn) base-pairs. J. Mol. Biol. 1989;207:455–457. doi: 10.1016/0022-2836(89)90268-4. [DOI] [PubMed] [Google Scholar]
- 58.Pan B, Mitra SN, Sundaralingam M. Crystal structure of an RNA 16-mer duplex R(GCAGAGUUAAAUCUGC)2 with nonadjacent G(syn).A+(anti) mispairs. Biochemistry. 1999;38:2826–2831. doi: 10.1021/bi982122y. [DOI] [PubMed] [Google Scholar]
- 59.Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- 60.Scicchitano DA, Olesnicky EC, Dimitri A. Transcription and DNA adducts: what happens when the message gets cut off? DNA Repair (Amst) 2004;3:1537–1548. doi: 10.1016/j.dnarep.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Souliotis VL, Sfikakis PP, Anderson LM, Kyrtopoulos SA. Intra- and intercellular variations in the repair efficiency of O6-methylguanine, and their contribution to kinetic complexity. Mutat. Res. 2004;568:155–170. doi: 10.1016/j.mrfmmm.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Pegg AE, Scicchitano D, Dolan ME. Comparison of the rates of repair of O6-alkylguanines in DNA by rat liver and bacterial O6-alkylguanine-DNA alkyltransferase. Cancer Res. 1984;44:3806–3811. [PubMed] [Google Scholar]
- 63.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 64.Libby RT, Gallant JA. The role of RNA polymerase in transcriptional fidelity. Mol. Microbiol. 1991;5:999–1004. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 65.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 66.Kuraoka I, Suzuki K, Ito S, Hayashida M, Kwei JS, Ikegami T, Handa H, Nakabeppu Y, Tanaka K. RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair (Amst) 2007;6:841–851. doi: 10.1016/j.dnarep.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 67.Tornaletti S, Maeda LS, Lloyd DR, Reines D, Hanawalt PC. Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2001;276:45367–45371. doi: 10.1074/jbc.M105282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cullinane C, Mazur SJ, Essigmann JM, Phillips DR, Bohr VA. Inhibition of RNA polymerase II transcription in human cell extracts by cisplatin DNA damage. Biochemistry. 1999;38:6204–6212. doi: 10.1021/bi982685+. [DOI] [PubMed] [Google Scholar]
- 69.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sriram M, van der Marel GA, Roelen HL, van Boom JH, Wang AH. Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex. Biochemistry. 1992;31:11823–11834. doi: 10.1021/bi00162a022. [DOI] [PubMed] [Google Scholar]
- 71.Scicchitano DA. Transcription past DNA adducts derived from polycyclic aromatic hydrocarbons. Mutat. Res. 2005;577:146–154. doi: 10.1016/j.mrfmmm.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 72.Dosanjh MK, Loechler EL, Singer B. Evidence from in vitro replication that O6-methylguanine can adopt multiple conformations. Proc. Natl Acad. Sci. USA. 1993;90:3983–3987. doi: 10.1073/pnas.90.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.