Abstract
X-linked juvenile retinoschisis is a heritable condition of the retina in males caused by mutations in the RS1 gene. Still, the cellular function and retina-specific expression of RS1 are poorly understood. To address the latter issue, we characterized the minimal promoter driving expression of RS1 in the retina. Binding site prediction, site-directed mutagenesis, and reporter assays suggest an essential role of two nearby cone-rod homeobox (CRX)-responsive elements (CRE) in the proximal −177/+32 RS1 promoter. Chromatin immunoprecipitation associates the RS1 promoter in vivo with CRX, the coactivators CBP, P300, GCN5 and acetylated histone H3. Transgenic Xenopus laevis expressing a green fluorescent protein (GFP) reporter under the control of RS1 promoter sequences show that the −177/+32 fragment drives GFP expression in photoreceptors and bipolar cells. Mutating either of the two conserved CRX binding sites results in strongly decreased RS1 expression. Despite the presence of sequence motifs in the promoter, NRL and NR2E3 appear not to be essential for RS1 expression. Together, our in vitro and in vivo results indicate that two CRE sites in the minimal RS1 promoter region control retinal RS1 expression and establish CRX as a key factor driving this expression.
INTRODUCTION
X-linked juvenile retinoschisis (RS) is a degenerative disorder of the retina characterized by a splitting of the inner retinal layers which eventually leads to visual impairment (1). It is a common condition of juvenile macular degeneration in males caused by mutations in the retina-specific RS1 gene (2). The encoded protein, termed retinoschisin, is secreted as a disulfide-linked homo-oligomeric complex and is primarily localized on the outer surface of the inner segments of cones and rods as well as the outer nuclear and outer plexiform layers of the retina (3–7). A main feature of the protein is its highly conserved discoidin domain functionally implicating retinoschisin in cell–cell interactions (2,5,8). A recent study demonstrates that RS1 binding to the surface of photoreceptors and bipolar cells is mediated through its interaction with Na/K ATPase (9). To date, there is no medical treatment for the condition, although therapeutic gene delivery may be an option in the future (10–13).
High mRNA levels of RS1 are present in the retina as shown by northern blot hybridization to a number of human (2) and mouse (14) tissues. Furthermore, in situ hybridization experiments revealed RS1 transcripts in rod and cone photoreceptor inner segments (3,15) and also in other cell bodies of the retinal layers, namely in bipolar cells, amacrine cells, and retinal ganglion cells (16). In postnatal eye development of the mouse, measurable levels of RS1 expression appear around postnatal day 1 (P1) and reach a maximum between P5 and P7. This level of expression is then maintained throughout adult life, indicating that continued de novo synthesis of RS1 is required and is essential for the maintenance of retinal integrity. The pineal gland is the only site of RS1 expression outside the eye (17). However, RS1−/Y mice lacking retinoschisin reveal no evidence of morphological changes in the pineal gland, indicating that RS1 might have different functions in the pineal gland and the retina (17).
The molecular basis underlying retina-specific expression of RS1 is unknown so far, although knowledge about regulatory sequences at the RS1 locus might lead to designing novel tools for gene therapy ensuring targeted and efficient expression of the protein. Furthermore, the functional importance of promoter variants in patients lacking classical RS1 mutations or patients with highly variable RS phenotypes could be evaluated on the basis of such data.
Preliminary evidence for a potential role of the transcription factors cone-rod homeobox (CRX) and neural retina leucine zipper protein (NRL) in retinal expression of RS1 comes from its differential expression in mice deficient for CRX (18) and NRL (19). Recent genome-wide expression profiling and chromatin immunoprecipitation (ChIP) approaches have revealed complex retinal regulatory networks balanced by CRX and NRL and the orphan nuclear receptor NR2E3 (18,20–23). CRX is a nuclear protein critical for general photoreceptor maturation in both rods and cones (24,25), while NR2E3 and NRL have specific roles in rod photoreceptor maturation and suppression of cone proliferation (19,26,27). Mutations of any one of the three transcription factors or the DNA binding sites in their respective promoters lead to retinal pathology, most notably cone-rod dystrophy (28), enhanced S-cone syndrome (29) and autosomal dominant retinitis pigmentosa (30).
To determine whether CRX, NRL and NR2E2 have a direct regulatory effect on human RS1 gene expression, we characterized the cis-elements of the RS1 promoter with special emphasis on these three factors. Using a computational approach to predict regulatory sequences, we identified multiple putative cis-elements in the RS1 proximal promoter, including three evolutionarily conserved sites for CRX. We defined the exact binding sequences for CRX and demonstrated CRX-dependent regulation of the −177/+32 promoter region driving RS1 expression in vitro and in vivo.
MATERIALS AND METHODS
Bioinformatic analyses
RS1 promoter sequences from different species were retrieved with Gene2Promoter (Genomatix GmbH, Munich, Germany) and DataBase of Transcriptional Start Sites (dbtss; http://dbtss.hgc.jp). The upstream regulatory regions were analyzed for putative transcription factor binding sites by Matinspector (Genomatix GmbH) with the matrices V$CRX.01 (CRX), V$NRL.01 (NRL) and V$PNR.01 (NR2E3). Only matrices predicted with a core similarity of 1.0 and a matrix similarity >0.75 were included in the analysis. The computer algorithm NUBIScan (31) and a self-defined matrix based on known NRL and NR2E3 target genes was used to predict DNA recognition sites for both nuclear receptors. Transcription start sites of the human and mouse RS1 genes have been determined previously (14).
Promoter and expression constructs
RS1 promoter fragments were cloned into the KpnI/HindIII site of the luciferase reporter vector pGL4.10 (Promega, Madison, WI, USA). Three fragments, starting from −703, −419 and −177 and ending at +32, respectively, relative to the transcriptional start site were generated by PCR from human genomic DNA with KpnI- and HindIII-flanked oligonucleotide primers (Supplementary Table 1). Mutant and deletion promoter constructs were based on the wild-type KpnI−177/HindIII+32 promoter fragment: mutated promoter fragments, termed mCRE1, mCRE3 and mCRE13, were cloned by site-directed mutagenesis (QuikChange Multi Site-Directed Mutagenesis Kit, Stratagene, La Jolla, CA, USA) with primers replacing the central conserved nucleotides of the CRX binding site (Supplementary Table 2). The mCRE2 mutant was generated by cloning two PCR fragments overlapping the CRX-responsive element (CRE) motif with an ApaI restriction site. Similarly, double mutants mCRE12, mCRE23 and the triple mutant mCRE123 were generated with mutated plasmid templates for the PCR (Supplementary Table 3). Sequences of cloned fragments are listed in Supplementary Table 4.
The NR2E3 binding site deletion fragment was produced by cloning two PCR fragments with EcoRI sequences at each of the 3′-ends (Supplementary Table 5). The NRE deletion fragment was generated in two steps. An EcoRI restriction site was cloned to replace the last six base pairs of the NRE fragment at nucleotide −80, so that the NRE site was flanked by an artificial EcoRI and an endogenous HpaI restriction site, through which NRE was removed (Supplementary Table 6).
Full-length NR2E3 and OTX2 were amplified from human retinal cDNA and cloned into the mammalian expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). The construction of CRX and NRL expression vectors has been reported previously (26). For Xenopus laevis transgene vectors, promoter fragments were cloned into Xop0.8-eGFP-N1 to replace the X. laevis opsin promoter. For verification, all cloned constructs were directly sequenced by capillary sequencing (Applied Biosystems, Darmstadt, Germany).
Cell culture and luciferase assays
Y79 retinoblastoma cells were maintained in DMEM containing 10%FCS and 100 U/ml penicillin/streptomycin. HEK293 cells were grown in DMEM medium as stated above, and supplemented with 500 µg/ml G418. BV-2 microglia cells were cultured as described previously (32). All cells were grown in a 37°C incubator with a 5% CO2 environment. For transfections, 1 million Y79 or 500 000 HEK cells were seeded overnight into 6-well plates. Y79 cells were then transfected with FugeneHD (Roche Applied Science, Mannheim, Germany), following the manufacturer's instructions. HEK293 cells were transfected using the standard calcium–phosphate method. Briefly, cells were transfected at 80% confluence with up to 7 µg of DNA in 218 µl of CaCl2 and 2× BBS buffer (50 mM BES, 280 mM NaCl, 1.5 mM Na2HPO4). After 48 h, cells were harvested and lyzed in Lysis Buffer (Promega). For luciferase assay, 20 µl of cytosolic extract and 100 µl of assay reagent were used. Light emission was measured with a FluoStar Optima (BMG LABTECH GmbH, Offenburg, Germany). Luciferase activity was normalized to the protein content of each well by dividing the measured relative light units by the protein concentration measured by Bradford assay. Each experiment was performed at least three times in duplicate wells.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) experiments were performed with nuclear extracts from Y79 cells, mouse retina, BV-2 cells and in vitro translated proteins. Oligonucleotides were designed based on the human RS1 promoter sequence. Nuclear extracts were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL, USA) and in vitro translated proteins were prepared with the TnT Quick-Coupled in vitro Transcription/Translation System (Promega, Mannheim, Germany). Double-stranded oligonucleotides were labeled with 32P-ATP and T4 polynucleotide kinase (Roche Biochemicals, Mannheim, Germany). Radiolabeled double-stranded oligonucleotides were incubated with 5 µg of nuclear extract or 4 µl of translated proteins and poly(dI-dC) in GSA binding buffer (Promega) for 20 min at room temperature. In competition experiments, unlabeled wild-type, mutant or consensus sequences as well as anti-CRX antibody were included in the binding reaction. Samples were loaded on a nondenaturing acrylamide gel in 0.5× TBE buffer and electrophoretically separated for 1.5 h at 250 V. Subsequently, gels were dried and audioradiographed.
ChIP assay
ChIP assays were performed in human retinoblastoma Y79 cells or Y79 cells transfected with CRX, NR2E3 and NRL as described previously (33). Briefly, 10 million cells were treated with 1% formaldehyde for 15 min and lyzed with SDS, Empigen and NP-40 (supplemented with 1 mM PMSF, 1 µg/ml aprotinin and 1 µg/ml pepstatin A). The nuclear pellet was homogenized by sonication twice at 30% amplitude for 10 s. Immunoprecipitation was performed on the lysate with 2.5 µg of anti-CRX antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-NR2E3 antibody (Santa Cruz Biotechnology), anti-NRL antibody (Santa Cruz Biotechnology), anti-di-acetylated (K9 and K14) histone H3 (Upstate Biotechnology, Lake Placid, NY, USA), anti-p300 clone RW128 (Upstate Biotechnology), anti-CBP (Upstate Biotechnology), anti-Gcn5 (Santa Cruz Biotechnology) or anti-IgG antibody (Santa Cruz Biotechnology). After washing and elution steps, cross-links were reversed at 65°C overnight. The immunoprecipitated DNA was purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and analyzed by PCR using the specific forward primer 5′-CTT CCT CGC GAA CTG AAT CT-3′ and reverse primer 5′-TGC AAT GAA TGT CAA TGG TT-3′ for the RS1 promoter fragment −177/+130 and the forward primer 5′-TGC ACA TGA TGC CTA GTT GC-3′ and reverse primer 5′-GAA GCA ATG GAG GGA GAG AAC-3′ for the RS1 promoter region −703/−530. Primers for the red-cone opsin promoter were used as positive controls and have been described previously (34).
Real-time quantitative RT–PCR
RNA was extracted from retinae of both wild-type C57/BL mice and Nrl−/− mice with RNeasy Plus Mini kit (Qiagen, Hilden, Germany). A total of 2.5 µg RNA was reverse transcribed into cDNA. Real-time quantitative RT–PCR was performed in triplicate with an Icycler (Biorad, Munich, Germany) in the presence of SYBR green dye. The signal from a pair of primers (5′-TCC AGA ATG CCC ATA TCA CA-3′ and 5′-GCA CAC CCA AAA CCT TGA CT-3′) amplifying a RS1 cDNA fragment was normalized by the HPRT signal from primers (5′-CAA ACT TTG CTT TCC CTG GT-3′ and 5′-CAA GGG CAT ATC CAA CAA CA -3′).
Generation of transgenic X. laevis, immunolabeling and confocal microscopy
Transgenic X. laevis were generated using the method described by Kroll and Amaya (35) with modifications (36). For each transgenesis, one sperm donor and three egg donors were used. Embryos were housed in 4 l tanks in an 18°C incubator on a 12 h light/dark cycle. After 24 h, embryos were exposed to 18 µg/ml G418 for 120 h (37). Only developmentally normal animals were used in our analysis; animals that were abnormally small or had other developmental abnormalities were not used. Tadpoles were initially screened for GFP expression and localization using an epi-fluorescence microscope. Eyes from representative tadpoles were cryosectioned and examined by confocal microscopy 14 days postfertilization. Sections were stained with wheat germ agglutinin, which labeled the glycosylate membrane, and Hoechst 33342, which stained nuclei.
RESULTS
Identification and analysis of putative binding sites in the RS1 promoter
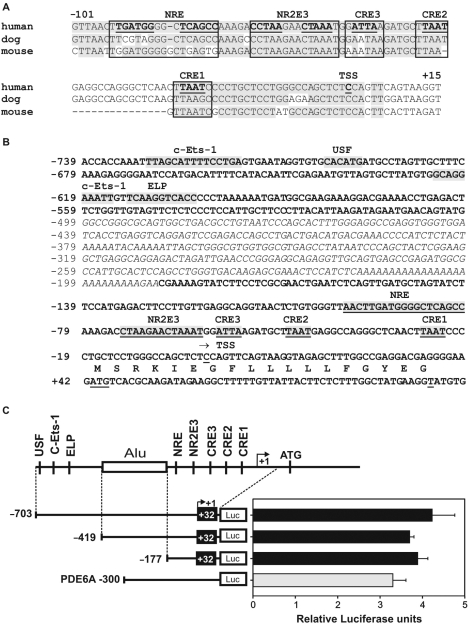
We have previously characterized the human and mouse RS1 genomic regions and experimentally defined the transcription start sites (14). Based on DNA-microarray expression data (18,19) and an in silico transcriptional network (38), we hypothesized that RS1 might be a direct target for transcription factors CRX, NRL and NR2E3. We therefore used a bioinformatic approach to predict evolutionarily conserved binding motifs for the three transcription factors in the putative human RS1 promoter sequences. Within the first 100-bp upstream of the major transcription start site in human, dog and mouse, comparative genomics locates three putative CRE at −26/−23, −47/−44, and −58/−55, as well as a putative NR2E3 binding motif (−76/−62) and an NRL response element (NRE, −99/−81) (Figure 1A). Further analysis revealed an ALU-repeat spanning the region −188/−499 and binding sites for c-Ets-1 (−729/−714; −624/−615), USF (−700/−695) and ELP (−612/−603) (Figure 1B).
Figure 1.
Analysis of conserved transcription factor binding sites in the RS1 promoter and delineation of the proximal region. (A) Sequence alignment of the putative RS1 promoter regions from human, dog and mouse. Conserved sequences are highlighted in gray and canonical binding sites for CRX (CRE1 to 3), NR2E3 and NRL (NRE) are boxed. (B) DNA sequence of the putative human RS1 promoter. Potential cis-elements are indicated. An interspersed ALU repeat is italicized. (C) Basal activity of three RS1 promoter–luciferase constructs (−703/+32, −419/+32, −177/+32) and of a PDE6A positive control (−300) transfected into Y79 retinoblastoma cells. RS1 promoter activities are comparable to the PDE6A promoter, which is known to be active in Y79 cells. A schematic of the RS1 promoter is shown with transcription factor binding sites and the ALU repeat (box). Each transfection was repeated three times with duplicate wells analyzed. Error bars represent the standard deviation of the mean from protein normalized luciferase activities.
Since repetitive sequences are rarely located in regulatory regions, we speculated that the RS1 core promoter is located downstream from the ALU-repeat. To further delineate this region, we transfected three different RS1 luciferase reporter constructs (−703/+32, −419/+32 and −177/+32) into human Y79 retinoblastoma cells which express endogenous RS1 transcripts. As a positive control, we used the pGL3-300 PDE6A (cyclic-GMP specific phosphodiesterase 6A α subunit) promoter region, which was shown to be transcriptionally active in Y79 cells (39). The longest RS1 promoter construct (−703/+32) showed a basal level of luciferase expression similar to the PDE6A promoter. When shortening the promoter to −419/+32 and further to −177/+32 the promoter activity was retained, indicating that critical core promoter sequences reside within the −177/+32 region (Figure 1C).
CRX strongly induces the RS1 proximal promoter
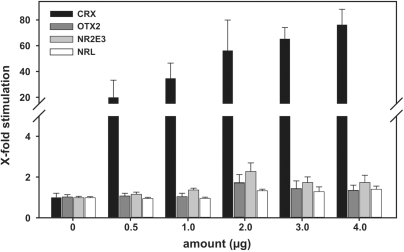
Having identified conserved transcription factor binding sites in the −177/+32 sequence, we evaluated the individual effects of transcription factor candidates potentially regulating the RS1 promoter via these sites. We analyzed the transactivation potential of CRX, its related family member OTX2, NRL and NR2E3 on the promoter region. Transcription factor expression vectors for the four nuclear proteins were cotransfected with the shortest RS1 promoter construct −177/+32 into HEK293 cells. These cells do not endogenously express retinal transcription factors and are therefore well suited to identify the specific transactivation potential of single heterologously expressed retinal DNA-binding proteins. As shown in Figure 2, CRX overexpression strongly transactivates the RS1 proximal promoter construct in a dose-dependent manner, resulting in an ∼80-fold induction with the highest CRX dose. In contrast, OTX2, NR2E3 and NRL reveal no significant effects on the RS1 promoter activity at all titrations analyzed (Figure 2).
Figure 2.
RS1 promoter induction levels by CRX, OTX2, NR2E3 and NRL. HEK293 cells were cotransfected with the −177/+32 RS1 promoter–reporter construct and transcription factor expression vectors. X-fold stimulation values were calculated by normalizing CRX, OTX2, NRL and NR2E3 cotransfected cells to mock-vector transfections. CRX (black bars) specifically induces the RS1 promoter in a dose-dependent manner. OTX2 (dark gray bars), NR2E3 (light gray bars) and NRL (white bars) do not induce or suppress the RS1 promoter. Each transfection was repeated three times with duplicate wells analyzed. Error bars represent the standard deviation of the mean from protein normalized luciferase activities.
Since NRL and NR2E3 are known to function in association with CRX on opsin gene promoters (40), we tested the regulatory activity of both factors in combinatory transient transfections with CRX. No significant transcriptional activity of NRL and NR2E3 could be detected in any combination of NRL and NR2E3 individually or combined with CRX in either HEK293 or Y79 cells (data not shown).
CRX binds to the RS1 promoter, recruits CBP/P300/GCN5 and triggers histone H3 acetylation
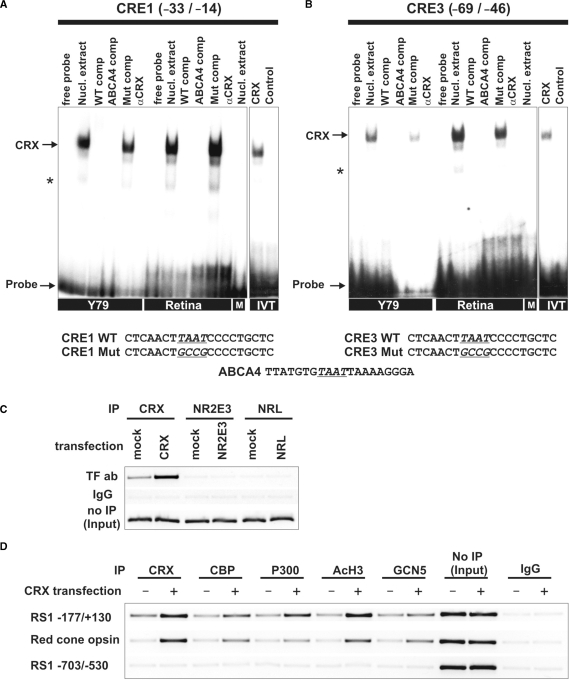
To assess in vitro promoter associations with the putative CRE/NR2E3/NRL binding sites, EMSAs were carried out with nuclear extracts from Y79 retinoblastoma cells and mouse retinal tissue in the presence of radiolabeled oligonucleotide probes representative of the binding motifs identified in the RS1 promoter (Figure 3A and B). Both Y79 and murine retina express CRX, NR2E3 and NRL transcripts as identified by real-time qRT–PCR (data not shown). BV-2 microglia cells were used as specificity control since these cells are of myeloid origin lacking endogenous CRX, NR2E3 and NRL expression (data not shown). The three putative binding sites for CRX (CRE1/2/3) were tested, but only probes homologous to CRE1 and CRE3 sequences showed specific binding (Figure 3A and B). In contrast to CRE1 and CRE3, only weak retardation bands were detectable with the CRE2 probe (data not shown). The mobility shifts from CRE1 and CRE3 could be blocked by both an excess amount of wild-type unlabeled oligonucleotides and an ABCA4 consensus sequence known to bind CRX (38). As expected, mutant CRE competitors had no effect on the specific CRX binding. Addition of a polyclonal anti-CRX antibody did not produce a supershift, but resulted in a specific displacement of CRX from its target oligonucleotides. In addition to analyzing the labeled probes with nuclear extracts, we tested the ability of CRX to bind the promoter by using in vitro translated CRX protein. As shown in Figure 3A and B, the in vitro translated CRX shifted the radiolabeled CRE1 and CRE3 probes to the same positions as with the nuclear extract. This result further confirms the specificity of the CRX binding and also suggests that CRX directly contacts these consensus DNA elements in the RS1 promoter.
Figure 3.
CRX, HATs and AcH3 are associated with the RS1 promoter region. (A and B) EMSAs with Y79, mouse retina, BV-2 microglia nuclear extracts or in vitro translated transcription factors. The sequences used for EMSA analysis are shown below with the mutated nucleotides underlined and conserved motifs printed in bold letters. Each gel was loaded with 5 µg of nuclear extracts or 4 µl of in vitro translated proteins, as indicated. Arrows represent specific binding identifiable by shifted bands that were inhibited by excess unlabeled weight or consensus oligonucleotides, but not by their mutant counterparts. Incubation with BV-2 microglia cells nuclear extract (M) did not result in a specific band shift. Addition of 1 µl anti-CRX antibody did not produce a supershift, but specifically inhibited CRX binding to the oligonucleotide. Asterisks indicate unspecific bands. (A) Interaction of RS1 CRE1 and (B) CRE3 with proteins in Y79 cells, mouse retina, BV-2 cells (M) and in vitro translated CRX (IVT). (C) ChIP assays using wild-type Y79 cells or Y79 cells transfected with CRX, NR2E3 or NRL. Immunoprecipitation was carried out with antibodies against CRX, NR2E3 or NRL. Input DNA served as positive control and IP with rabbit IgG antibody served as negative control. The RS1 promoter fragment −177/+130 was analyzed for in vivo CRX, NR2E3 and NRL binding by ChIP–PCR. (D) ChIP assays using wild-type or CRX-transfected Y79 cells with antibodies against CRX, CBP, P300, AcH3 and GCN5. Input DNA served as positive control and IP with rabbit IgG antibody served as negative control. DNA fragments were analyzed by PCR for two different RS1 promoter fragments and the red-cone opsin promoter. The RS1 promoter fragment −177/+130 and the red-cone opsin control promoter displayed ChIP-PCR positive signals whereas the RS1 promoter region −703/−530 lacking CRX sites did not show specific CRX binding.
In accordance with the lack of transactivational or repressional activities of NR2E3 and NRL in cotransfection experiments, probes spanning the two putative binding regions in the RS1 promoter showed no specific binding to Y79 and mouse retinal nuclear extracts or in vitro translated NR2E3 and NRL proteins (data not shown).
To further analyze whether CRX, NR2E3 and NRL bind to the proximal RS1 regulatory region in the natural chromatin context, ChIP assays were performed with wild-type or transcription factor-transfected Y79 cells. For PCR analysis of IP samples, a specific primer set was designed to amplify the −177/+130 RS1 promoter region containing the consensus CRE/NR2E3/NRL sites. CRX already associated with the proximal RS1 promoter in untransfected Y79 cells and its in vivo binding activity markedly increased after overexpression (Figure 3C). In contrast, neither NR2E3 nor NRL showed a specific interaction with the RS1 promoter region in the untransfected or transfected state (Figure 3C). This suggests that CRX binds to the minimal RS1 promoter in vivo and that the in silico predicted binding sites for NRL and NR2E3 may not be functional.
We were furthermore interested in whether CRX binding to the RS1 promoter recruits the histone acetyl transferase (HAT) coactivators CBP, P300 and GCN5 with subsequent histone H3 (K9/K14) acetylation. These coactivators have been recently shown to regulate CRX-dependent transcription of several photoreceptor-specific genes (34). ChIP assays were performed with wild-type or CRX-transfected Y79 cells to simulate different levels of nuclear CRX protein. Antibodies against CRX, each of the three coactivators, and AcH3 (K9/K14) were used for immunoprecipitation. The IP samples were analyzed by PCR amplification of the −177/+130 RS1 promoter region or the adjacent upstream promoter region −703/−530 lacking CRX-binding elements. The proximal red-cone opsin promoter was used as positive control (34). As shown in Figure 3D, CRX, the three HATs, and AcH3 were already associated with the proximal RS1 promoter region in Y79 cells. CRX overexpression caused further recruitment of CBP and P300 and increased AcH3 levels as indicated by strong ChIP signals. GCN5 was also bound to the RS1 promoter, albeit with lesser intensity. The control red-cone opsin promoter was positive for binding to CBP, P300, AcH3 and GCN5 especially in the CRX-transfected state (Figure 3D), whereas PCR analysis of IgG-precipitated DNA did not yield detectable PCR products. Furthermore, the RS1 upstream control region −703/−530 lacking CRX sites showed no in vivo DNA–protein interactions. These results suggest that CRX specifically binds to the RS1 promoter in vivo with high affinity, recruits CBP/P300/GCN5 and triggers histone H3 acetylation required for RS1 gene expression.
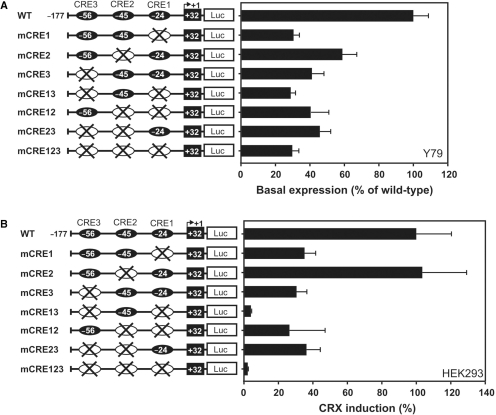
CRE1 and CRE3 are essential for RS1 expression
We next determined the individual contribution of the three CRE sites for RS1 expression. Four and six base pair substitutions were introduced in each CRE sequence motif eliminating the most conserved TAAT core sequence (Figure 1A). We also generated double and triple mutants based on the wild-type −177/+32 promoter construct. CRE-mutant promoter plasmids were transfected into two different cell systems, Y79 cells (Figure 4A) and CRX-overexpressing HEK293 cells (Figure 4B), and normalized luciferase signals were quantified. Base pair changes within either the first (CRE1) or the third (CRE3) site markedly diminished RS1 basal promoter activity (Figure 4A) and reduced CRX-dependent induction (Figure 4B) by more than 50%, although residual promoter activity was still present in these single mutant constructs. When mutating the second CRX-binding element (CRE2), constitutive RS1 promoter activity declined to 60% (Figure 4A), but this construct was still CRX inducible to the level of wild type (Figure 4B), suggesting that CRX-mediated RS1 expression is independent of CRX binding to CRE2. Double and triple mutants further emphasized the importance of either CRE1 or CRE3 in stimulating RS1 expression. Mutations at both CRE1 and CRE3 sites reduced the basal promoter activity below 30% and fully abolished luciferase induction as shown by the CRE13 double and CRE123 triple mutant (Figure 4A and B). In contrast, the constitutive RS1 promoter strength and induction levels obtained with the double mutants CRE12 or CRE23 were comparable to single mutants CRE1 or CRE3, respectively (Figure 4A and B). These results are in full agreement with the findings from our CRE-EMSA experiments, emphasizing CRE1 and CRE3 as the critical cis-elements in CRX-dependent RS1 promoter induction.
Figure 4.
Delineation of the critical sequences required for CRX binding and RS1 promoter activity. Luciferase assays were performed with the RS1 wild-type promoter construct −177/+32, and CRE single-, double- and triple mutant promoters in Y79 cells (A) and HEK293 cells cotransfected with CRX (B). The basal luciferase activity of the wild-type contruct −177/+32 (A) or the CRX-induction of the wild-type contruct −177/+32 was set to 100%. Each transfection was repeated three times with duplicate wells analyzed. Error bars represent the standard deviation of the mean from protein normalized luciferase activities.
In vivo analysis of functional cis-elements in the RS1 promoter
To assess the possible relevance of our findings in vivo, we examined the contribution of all three CRE motifs and the putative NR2E3/NRL-binding sites for RS1-driven gene expression in transgenic X. laevis. Transgenic tadpoles were generated which express GFP driven by either the X. laevis opsin promoter as control, the human RS1 promoter or mutated versions of the RS1 promoter. Tadpoles that survived G418 selection were screened for the presence of GFP in vivo. G418 selection effectively identifies transgenic animals, as demonstrated by the high incidence of GFP expression in G418-selected tadpoles resulting from injection of the opsin promoter plasmid (∼92%). Since the opsin promoter is very strong, assessing GFP positive tadpoles is relatively easy. However, in the RS1 promoter groups fewer GFP positive tadpoles were observed and the GFP signal was markedly lower (Table 1), indicating that the RS1 promoter is less strong relative to the opsin promoter. The percentage of GFP positives may be used as a crude measure of the strength of the promoter constructs. Consequently, our in vivo results suggest that mCRE1 and mCRE13 are significantly weaker promoters than WT, mCRE2, mNRE and mNR2E3 (Table 1).
Table 1.
Analysis of relative GFP expression of RS1 promoter constructs
| Promoter construct | Number of tadpoles (i.e. surviving G418 selection) | Percentage with detectable GFP at dpf14a (%) | Number examined by confocal microscopy |
|---|---|---|---|
| XOP | 13 | 92 | 3 |
| WT | 11 | 73 | 4 |
| mCRE1 | 23 | 9 | 4 |
| mCRE2 | 18 | 67 | 6 |
| mCRE13 | 19 | 5 | 5 |
| mNRE | 15 | 53 | 6 |
| mNR2E3 | 22 | 59 | 4 |
aAs determined by fluorescent screening of tadpoles eyes in vivo.
In order to confirm these findings, confocal microscopy of 14-day-old tadpole retinae was carried out. We first assessed GFP expression of the opsin positive promoter compared with nontransgenic control retina (Figure 5A and B) and a strong GFP signal was detected exclusively in photoreceptors (Figures 5B and 6A). The wild-type RS1 promoter drives GFP expression most strongly in the photoreceptor layer (Figure 5C). This is consistent with previous results in which retinoschisin mRNA was detected in photoreceptors by in situ hybridization (6) and suggests that the human RS1 promoter exhibits normal activity in the X. laevis retina. GFP was also present in the inner nuclear layer (likely bipolar cells) albeit at lower levels (Figures 5C and 6B, asterisks). RS1 was previously detected on the exterior of bipolar cells (4) but it was not clear whether the protein originated from bipolar cells or was secreted by photoreceptors. The present results provide the first direct evidence that RS1 is expressed by bipolar cells.
Figure 5.
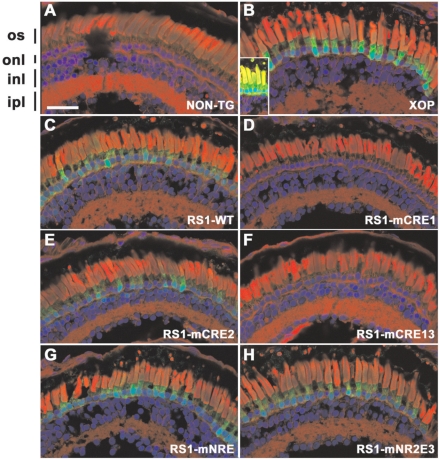
In vivo analysis of RS1 promoter–GFP constructs. Confocal micrographs of transgenic X. laevis retinae expressing GFP (green) under the control of various promoters. Cryosections were counterstained with wheat germ agglutinin (red) and Hoechst nuclear stain (blue). (A) Nontransgenic control retina (NON-TG); (B) Xenopus opsin promoter (XOP); (C) RS1 promoter wild type (RS1-WT); (D) RS1 promoter with mutated CRE1 (mCRE1); (E) RS1 promoter with mutated CRE2 (mCRE2); (F) RS1 promoter with mutated CRE1 and CRE3 (mCRE13); (G) RS1 promoter with mutated NRE (mNRE); (H) RS1 promoter with mutated NR2E3 (mNR2E3). GFP was not expressed in the nontransgenic retina but was highly expressed in photoreceptors in the XOP retina. The wild-type RS1 promoter (NON-TG) drives GFP expression in photoreceptors albeit at lower levels. Abolishing NRL and NR2E3 binding did not significantly alter GFP expression (G and H). Mutating the CRE1 site (D and F) reduced GFP expression to below detectable limits, but no obvious effect resulted from mutating the CRE2 site (E). The GFP signals in panels A and C-H are directly comparable as the same laser intensity and amplifier setting were used, and identical image processing was applied. In panel B, the amplifier gain was reduced due to the intensity of the GFP signal. The inset shows an image acquired with settings equivalent to the other panels. Photoreceptor outer segments (os), outer nuclear layer (onl), inner nuclear layer (inl) and inner plexiform layer (ipl). Scale bar, 50 µm.
Figure 6.
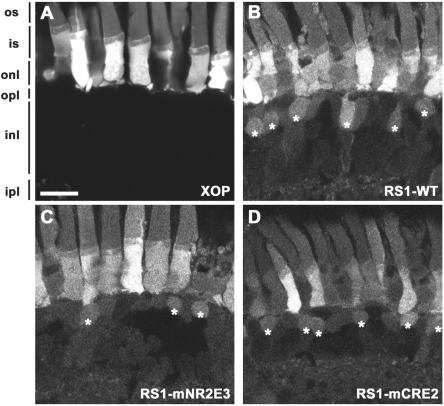
RS1 promoter activity in bipolar cells. (A–D) Confocal micrographs of transgenic retinas expressing GFP. (A) Within the retina, the X. laevis opsin promoter drives expression exclusively in rod photoreceptors. In contrast, the RS1 promoter (B) drives expression in both photoreceptors and bipolar cells (asterisks). Mutant RS1 promoters lacking NR2E3 (C) or CRE2 (D) sequences were also capable of driving expression in bipolar cells. Photoreceptor outer segments (os), inner segments (is), outer nuclear layer (onl), outer plexiform layer (opl), inner nuclear layer (inl) and inner plexiform layer (ipl). Scale bar, 20 µm.
The vast majority of mCRE1 and mCRE13 retinae exhibited no detectable GFP in any retinal cell layer (Figure 5D and F). mNRE drives significant photoreceptor expression but less bipolar expression (Figure 5G). In contrast, mCRE2 and mNR2E3 promoters revealed similar results when compared with the wild-type RS1 promoter in the retina (Figure 5E and H) and also drive GFP expression in bipolar cells (Figure 6C and D).
DISCUSSION
In this study, we have identified cis-regulatory elements required for in vitro and in vivo promoter activity of the RS1 gene. This defines a core promoter downstream of an ALU repeat that is sufficient to drive gene expression in the retina. Most importantly, we identified CRX as an important regulator of RS1 promoter activity and showed that CRX acts via two evolutionarily conserved binding sites, CRE1 and CRE3, which strongly stimulate RS1 transcription. CRX binds to these sites with high affinity even in the absence of additional nuclear proteins. Cooperative binding of CRX to both CRE1 and CRE3 seems to be required for full promoter induction, since substitution of conserved nucleotides in either of the two binding motifs causes a more than 50% reduction in promoter activity and CRX-dependent transactivation. Despite a strong binding prediction score and high interspecies sequence conservation, the CRE2 motif appears dispensable for CRX binding and only moderately affects full promoter activity.
Over the years, a complex regulatory network involving CRX and associated factors like OTX2 has been described (41). In this context, the presence of three CRE motifs in the RS1 promoter is in full agreement with data from the rhodopsin gene and its prototypic retina-specific promoter. The rhodopsin proximal regulatory region harbors at least three adjacent CRX sites required for strong expression (24). Also, in contrast to the RS1 core promoter, CRX seems to only moderately activate a rhodopsin luciferase reporter in the absence of NRL or NR2E3 binding to the basal promoter region (42). One explanation for these obvious differences could be that Y79 retinoblastoma cells are not fully differentiated retinal cells, such as photoreceptor cells or bipolar cells. Thus, Y79 may only serve as a model system for basal constitutive expression of RS1. Moreover, HEK293 cells transfected with expression plasmids for individual retina-specific transcription factors may be more suitable for transactivation experiments with CRX than with NRL or NR2E3. Therefore, we cannot rule out that the signaling-dependent expression or the developmental regulation of RS1 still requires a synergistic activity of CRX with other retina-specific or general transcription factors.
The CRX-related homeobox gene OTX2 binds to similar motifs, since OTX2 and its expression precedes CRX early in neuronal development (43). We therefore analyzed the ability of OTX2 to regulate the RS1 promoter in cotransfection assays. In contrast to CRX, however, OTX2 was not able to transactivate the RS1 gene, indicating that OTX2 is not a major factor in regulating the RS1 CRE motifs in vitro.
Our ChIP results in untransfected and CRX-transfected Y79 cells showed that CRX binds to the proximal RS1 promoter in its native chromatin configuration. The coactivators CBP, P300 and GCN5 were subsequently recruited and histone H3 acetylation was detected in this promoter region, indicative of active transcription. CBP, P300 and GCB5 are critical components of retinal development and are required for maintenance of photoreceptor gene expression (34,40). Low AcH3 levels are correlated with retinal diseases and transcriptional suppression of opsin genes (44). Therefore, the low expression levels of RS1 mRNA in the Crx−/− retina (18) may be at least partially related to compressed chromatin and prevention of the basal transcription machinery to access the RS1 promoter.
Further evidence for in vivo regulation of RS1 expression by CRX comes from our transgenic X. laevis experiments. The wild-type −177/+32 regulatory region consistently drives GFP expression in the tadpole retina. The low GFP signal found in CRE1 single- and CRE13 double mutant transgenic X. laevis correlates well with the 60% reduced promoter activity found in the in vitro transfection experiments, providing additional support that binding at both sites, CRE1 and CRE3, is crucial for RS1 promoter activity. Furthermore, localization of RS1 minimal promoter-driven GFP in X. laevis is consistent with findings of endogenous RS1 expression in human and mice (4). In transgenic X. laevis, prominent RS1 expression is found in the photoreceptor layer and a cell-type reminiscent of bipolar cells. In contrast, GFP expression was not seen in other cells types of the frog retina including ganglion cells or glial cells, which have previously been suggested to express RS1 (16). In summary, we conclude from our own findings as well as data in CRX-deficient mice, which reveal strongly reduced RS1 transcript levels in retinal tissue (18,23) that CRX regulates RS1 transcription in vivo.
Reduced RS1 mRNA levels in retinae from NRL-deficient mice (19,22), in addition to bioinformatic binding site predictions, has led us to address the contribution of NRL and NR2E3 in regulating RS1 promoter activity. However, no significant influence of NRL and NR2E3 on the level of RS1 transcription in retinal cells could be detected in vitro or in vivo. Since NRL and NR2E3 are known to interact with a multitude of other transcription factors (41), those partners might be absent in our transfection assays. Nevertheless, we can exclude a direct interaction of NR2E3 and NRL with the RS1 promoter in vivo or with CRX as analyzed by our combination cotransfection studies in the luciferase reporter assay. Double cotransfections of these three transcription factors were previously shown to synergistically activate several rod phototransduction genes (26). In transgenic X. laevis, mutant NRE and NR2E3 promoter constructs could drive GFP expression to a level similar to wild type. This implies that both binding sites are dispensable for basal induction of the RS1 gene in the retina.
Overall, our data establish CRX as a crucial transcriptional regulator of RS1 expression in the retina. Additional in vivo models, such as transgenic mice will help to further elucidate the CRX transcriptional network driving gene expression from retinal promoters.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) (WE1259/12-3 to BHFW); (LA1203/6-1 to TL). Funding for open access charge: DFG (WE1259/12-3).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWEDGEMENTS
We thank A. Swaroop for providing CRX and NRL expression vectors.
REFERENCES
- 1.Roesch MT, Ewing CC, Gibson AE, Weber BH. The natural history of X-linked retinoschisis. Can. J. Ophthalmol. 1998;33:149–158. [PubMed] [Google Scholar]
- 2.Sauer CG, Gehrig A, Warneke-Wittstock R, Marquardt A, Ewing CC, Gibson A, Lorenz B, Jurklies B, Weber BH. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat. Genet. 1997;17:164–170. doi: 10.1038/ng1097-164. [DOI] [PubMed] [Google Scholar]
- 3.Grayson C, Reid SN, Ellis JA, Rutherford A, Sowden JC, Yates JR, Farber DB, Trump D. Retinoschisin, the X-linked retinoschisis protein, is a secreted photoreceptor protein, and is expressed and released by Weri-Rb1 cells. Hum. Mol. Genet. 2000;9:1873–1879. doi: 10.1093/hmg/9.12.1873. [DOI] [PubMed] [Google Scholar]
- 4.Molday LL, Hicks D, Sauer CG, Weber BH, Molday RS. Expression of X-linked retinoschisis protein RS1 in photoreceptor and bipolar cells. Invest Ophthalmol. Vis. Sci. 2001;42:816–825. [PubMed] [Google Scholar]
- 5.Wu WW, Molday RS. Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis. J. Biol. Chem. 2003;278:28139–28146. doi: 10.1074/jbc.M302464200. [DOI] [PubMed] [Google Scholar]
- 6.Reid SN, Yamashita C, Farber DB. Retinoschisin, a photoreceptor-secreted protein, and its interaction with bipolar and muller cells. J. Neurosci. 2003;23:6030–6040. doi: 10.1523/JNEUROSCI.23-14-06030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu WW, Wong JP, Kast J, Molday RS. RS1, a discoidin domain-containing retinal cell adhesion protein associated with X-linked retinoschisis, exists as a novel disulfide-linked octamer. J. Biol. Chem. 2005;280:10721–10730. doi: 10.1074/jbc.M413117200. [DOI] [PubMed] [Google Scholar]
- 8.Vogel WF, Abdulhussein R, Ford CE. Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal. 2006;18:1108–1116. doi: 10.1016/j.cellsig.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Molday LL, Wu WW, Molday RS. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J. Biol. Chem. 2007;282:32792–32801. doi: 10.1074/jbc.M706321200. [DOI] [PubMed] [Google Scholar]
- 10.Min SH, Molday LL, Seeliger MW, Dinculescu A, Timmers AM, Janssen A, Tonagel F, Tanimoto N, Weber BH, Molday RS, et al. Prolonged recovery of retinal structure/function after gene therapy in an Rs1h-deficient mouse model of x-linked juvenile retinoschisis. Mol. Ther. 2005;12:644–651. doi: 10.1016/j.ymthe.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y, Takada Y, Kjellstrom S, Hiriyanna K, Tanikawa A, Wawrousek E, Smaoui N, Caruso R, Bush RA, Sieving PA. RS-1 gene delivery to an adult Rs1h knockout mouse model restores ERG b-Wave with reversal of the electronegative waveform of X-linked retinoschisis. Invest Ophthalmol. Vis. Sci. 2004;45:3279–3285. doi: 10.1167/iovs.04-0576. [DOI] [PubMed] [Google Scholar]
- 12.Janssen A, Min SH, Molday LL, Tanimoto N, Seeliger MW, Hauswirth WW, Molday RS, Weber BH. Effect of late-stage therapy on disease progression in AAV-mediated rescue of photoreceptor cells in the retinoschisin-deficient mouse. Mol. Ther. 2008;16:1010–1017. doi: 10.1038/mt.2008.57. [DOI] [PubMed] [Google Scholar]
- 13.Takada Y, Vijayasarathy C, Zeng Y, Kjellstrom S, Bush RA, Sieving PA. Synaptic pathology in retinoschisis knockout (Rs1-/y) mouse retina and modification by rAAV-Rs1 gene delivery. Invest Ophthalmol. Vis. Sci. 2008;49:3677–3686. doi: 10.1167/iovs.07-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehrig AE, Warneke-Wittstock R, Sauer CG, Weber BH. Isolation and characterization of the murine X-linked juvenile retinoschisis (Rs1h) gene. Mamm. Genome. 1999;10:303–307. doi: 10.1007/s003359900991. [DOI] [PubMed] [Google Scholar]
- 15.Reid SN, Akhmedov NB, Piriev NI, Kozak CA, Danciger M, Farber DB. The mouse X-linked juvenile retinoschisis cDNA: expression in photoreceptors. Gene. 1999;227:257–266. doi: 10.1016/s0378-1119(98)00578-2. [DOI] [PubMed] [Google Scholar]
- 16.Takada Y, Fariss RN, Tanikawa A, Zeng Y, Carper D, Bush R, Sieving PA. A retinal neuronal developmental wave of retinoschisin expression begins in ganglion cells during layer formation. Invest Ophthalmol. Vis. Sci. 2004;45:3302–3312. doi: 10.1167/iovs.04-0156. [DOI] [PubMed] [Google Scholar]
- 17.Takada Y, Fariss RN, Muller M, Bush RA, Rushing EJ, Sieving PA. Retinoschisin expression and localization in rodent and human pineal and consequences of mouse RS1 gene knockout. Mol. Vis. 2006;12:1108–1116. [PubMed] [Google Scholar]
- 18.Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr. Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 19.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 20.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, et al. Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 22.Corbo JC, Myers CA, Lawrence KA, Jadhav AP, Cepko CL. A typology of photoreceptor gene expression patterns in the mouse. Proc. Natl Acad. Sci. USA. 2007;104:12069–12074. doi: 10.1073/pnas.0705465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 26.Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 27.Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum. Mol. Genet. 2001;10:1619–1626. doi: 10.1093/hmg/10.16.1619. [DOI] [PubMed] [Google Scholar]
- 28.Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 29.Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 30.Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl Acad. Sci. USA. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podvinec M, Kaufmann MR, Handschin C, Meyer UA. NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol. Endocrinol. 2002;16:1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- 32.Ebert S, Schoeberl T, Walczak Y, Stoecker K, Stempfl T, Moehle C, Weber BH, Langmann T. Chondroitin sulfate disaccharide stimulates microglia to adopt a novel regulatory phenotype. J. Leukoc. Biol. 2008;84:736–740. doi: 10.1189/jlb.0208138. [DOI] [PubMed] [Google Scholar]
- 33.Weigelt K, Ernst W, Walczak Y, Ebert S, Loenhardt T, Klug M, Rehli M, Weber BH, Langmann T. Dap12 expression in activated microglia from retinoschisin-deficient retina and its PU.1-dependent promoter regulation. J. Leukoc. Biol. 2007;82:1564–1574. doi: 10.1189/jlb.0707447. [DOI] [PubMed] [Google Scholar]
- 34.Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis. Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- 35.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 36.Moritz OL, Tam BM, Knox BE, Papermaster DS. Fluorescent photoreceptors of transgenic Xenopus laevis imaged in vivo by two microscopy techniques. Invest Ophthalmol. Vis. Sci. 1999;40:3276–3280. [PubMed] [Google Scholar]
- 37.Moritz OL, Biddle KE, Tam BM. Selection of transgenic Xenopus laevis using antibiotic resistance. Transgenic Res. 2002;11:315–319. doi: 10.1023/a:1015612022976. [DOI] [PubMed] [Google Scholar]
- 38.Qian J, Esumi N, Chen Y, Wang Q, Chowers I, Zack DJ. Identification of regulatory targets of tissue-specific transcription factors: application to retina-specific gene regulation. Nucleic Acids Res. 2005;33:3479–3491. doi: 10.1093/nar/gki658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittler SJ, Zhang Y, Chen S, Mears AJ, Zack DJ, Ren Z, Swain PK, Yao S, Swaroop A, White JB. Functional analysis of the rod photoreceptor cGMP phosphodiesterase alpha-subunit gene promoter: Nrl and Crx are required for full transcriptional activity. J. Biol. Chem. 2004;279:19800–19807. doi: 10.1074/jbc.M401864200. [DOI] [PubMed] [Google Scholar]
- 40.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 41.Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 43.Rath MF, Morin F, Shi Q, Klein DC, Moller M. Ontogenetic expression of the Otx2 and Crx homeobox genes in the retina of the rat. Exp. Eye Res. 2007;85:65–73. doi: 10.1016/j.exer.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 44.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl Acad. Sci. USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.