Abstract
The crystal structure of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) bound to an RNA/DNA hybrid reveals an extensive network of contacts with the phosphate backbone of the DNA strand ∼4–9 bp downstream from the ribonuclease H (RNase H) catalytic center. Collectively designated as ‘the RNase H primer grip’, this motif contains a phosphate binding pocket analogous to the human and Bacillus halodurans RNases H. The notion that the RNase H primer grip mediates the trajectory of RNA/DNA hybrids accessing the RNase H active site suggests that locally neutralizing the phosphate backbone may be exploited to manipulate nucleic acid flexibility. To examine this, we introduced single and tandem methylphosphonate substitutions through the region of the DNA primer contacted by the RNase H primer grip and into the RNase H catalytic center. The ability of mutant hybrids to support RNase H and DNA polymerase activity was thereafter examined. In addition, site-specific chemical footprinting was used to evaluate movement of the DNA polymerase and RNase H domains. We show here that minor alteration to the RNase H primer can have a dramatic effect on enzyme positioning, and discuss these findings in light of recent crystallography of human RNase H containing an RNA/DNA hybrid.
INTRODUCTION
The interaction of p66/p51 human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) with its duplex and hybrid nucleic acid substrates is mediated by an extensive network of contacts between protein subdomains and primarily the sugar–phosphate backbone of the latter (1–5). With respect to the DNA polymerase catalytic center, the β12–β13 hairpin of the thumb subdomain is designated the ‘primer grip’, while α-helix H and α-helix I of the same subdomain contact the backbone of the primer and template strands, respectively (1,2,5). Additional contacts with the template strand are mediated by a ‘template grip’, comprising β-strand 4 and α-helix B of the p66 fingers subdomain and the β8–αE connecting loop and β-strand 5a of the palm. Detailed biochemical studies have demonstrated the importance of these protein/nucleic acid contacts in substrate binding, catalysis and translocation (6–12).
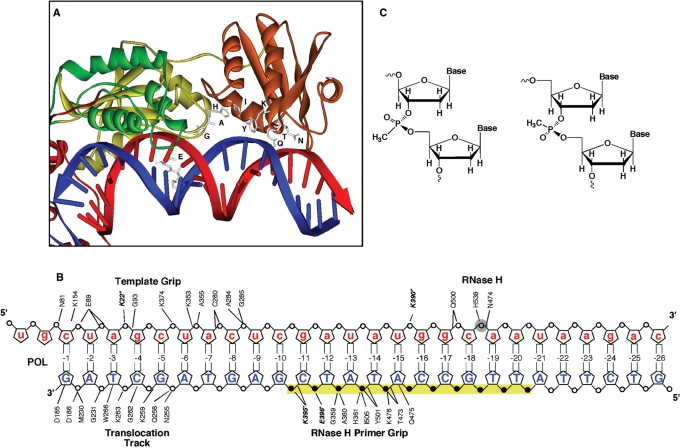
A second network of contacts, involving the sugar–phosphate backbone of the primer is located 4–9 bp from the ribonuclease H (RNase H) catalytic center. This region comprises p51 connection residues Lys395 and Glu396, p66 connection residues Gly359, Ala360, His361 and p66 RNase H domain residues Thr473, Gln475, Lys476, Tyr501 and Ile505 (Figure 1A). Based on the crystal structure of HIV-1 RT containing an RNA/DNA hybrid, Sarafianos et al. (5) designated this motif the RNase H primer grip, proposing that by contacting the DNA primer, it imposed the appropriate trajectory of the RNA template strand accessing the catalytic center for optimal catalysis. Although the significance of the RNase H primer grip has been documented via biochemical evaluation of mutant enzymes (13) and analysis of viral replication kinetics (14), the manner whereby it contributes to nucleic acid trajectory remains to be established. One clue may come from the recent high resolution crystal structures of Bacillus halodurans and human RNasesH containing their RNA/DNA substrates. Nowotny and coworkers (15–17) have shown that the bacterial and human enzymes contain a phosphate binding pocket 2–3 bp from the active site, around which backbone distortion and minor groove narrowing was proposed as a means of anchoring the DNA strand. Only Thr473 and Gln475 of HIV-1 RT were analogous to their bacterial counterparts (Thr43 and Asn45, respectively), suggesting that the extended motif of the retroviral enzyme may contribute to differential recognition of duplex DNA, duplex RNA and DNA/RNA hybrids, all of which are utilized by RT during the reverse transcription cycle.
Figure 1.
(A) A model depicting the RNase H primer grip residues and their proposed contacts with a DNA/RNA hybrid, based on the structure of Sarafianos et al. (5). Amino acids are presented in single letter format and the DNA and RNA strands are in blue and red, respectively. (B) Experimental strategy for methylphosphonate substitution. In the model DNA/RNA hybrid, the analog is substituted at positions −10 through −20 either as singlet, doublet or triplets in an overlapping manner. Filled circles represent the DNA phosphates proposed to interact with the RNase H primer grip residues. The open shaded RNA phosphate is the scissile bond. For reference, protein contacts from the polymerase domain (template grip and translocation track), and RNase H active center are also shown. (C) Chemical structure methylphosphonate Rp and Sp stereoisomers, where a methyl group replaces one oxygen of the phosphate.
In order to examine the extent to which phosphate backbone contacts contribute to RNase H primer grip function, we examined the hydrolysis patterns of RNA/DNA hybrids whose DNA contained single or tandem neutralizing methylphosphonate linkages corresponding to the region contacted by RT between the RNase H primer grip and RNase H catalytic center. An equivalent strategy has been used to study the interaction of T7 DNA polymerase (18,19), T7 endonuclease I (20) and topoisomerase IB with their nucleic acid substrates (21). A second consequence of charge neutralization is local bending of the duplex, which has been proposed to promote specific protein–DNA interactions (22–24). Based on the proposal of Sarafianos et al. (5), locally altering nucleic acid geometry might be predicted to affect positioning of the scissile phosphodiester bond in the RNase H catalytic center. Data of this communication indicate that single methylphosphonate substitutions as far as 7 bp from the RNase H active site have a profound affect on RNase H cleavage specificity, the severity of which increases with the extent of substitution. Interestingly, RNase H activity is rarely inhibited, but rather re-directed to another position of the hybrid. In addition, when substituted DNA duplexes are assessed for their ability to support DNA polymerase activity, our results indicate that neutralizing phosphates which interact with RNase H primer grip stall the polymerization complex.
MATERIALS AND METHODS
Design, synthesis and purification of oligodeoxyribonucleotides
A 30-nt DNA oligonucleotide (3′ GATCGATGAGCTATACCGTTATTCTGAGGT 5′) was designed with minimal secondary structure as determined by mFOLD (25). Bases in bold and underlined indicate the positions of single, double or triple substitutions of nucleoside analogs (Figure 1C). Phosphoramidites of methylphosphonates were purchased from Glen Research, Sterling, VA, USA. Analog-substituted oligonucleotides were synthesized at 1-μmol scale on a PE Biosystems Expedite 8909 nucleic acid synthesizer by standard phosphoramidite chemistry. Stepwise coupling yields for incorporation of the analogs were >98% as determined by trityl cation monitoring. Deprotection and cleavage of oligonucleotides was carried out according to the manufacturer's protocol. Oligonucleotides were purified by preparative polyacylamide gel electrophoresis and quantified spectrophotometrically (260 nm) assuming a molar extinction coefficient equal to the sum of the constituent deoxynucleotides. The complementary RNA oligonucleotide (5′ UCAUGCCCUGCUAGCUACUCGAUAUGGCAAUAAGACUCCA 3′) with 10-nt 5′ overhang was purchased from Dharmacon Research (Boulder, CO, USA) and deprotected according to manufacturer's protocol. The sequence of the corresponding DNA oligonucleotide is 5′ TCATGCCCTGCTAGCTACTCGATATGGCAATAAGACTCCA 3′.
Purification of HIV-1 RT and RNase H assays
p66/p51 HIV-1 RT was purified according to Le Grice et al. (26). RNase H cleavage was evaluated as previously described (27). Briefly, substrates were generated by 5′-end labeling the 40-nt RNA template with γ-32ATP and annealing it to each of the methylphosphonate-substituted DNAs, as well as to an unsubstituted DNA. Hydrolysis was initiated by adding wild-type p66/p51 HIV-1 RT to DNA/RNA hybrids in 10 mM Tris–HCl (pH 8.0), 80 mM NaCl, 5 mM DTT and 6 mM MgCl2, with enzyme and substrate concentrations of 10 nM and 50 nM, respectively, at 37°C. Reactions were terminated after 10 min by adding an equal volume of a formamide-based gel-loading buffer [95% (v/v) formamide containing 0.1% (w/v) bromophenol blue and xylene cyanol]. Hydrolysis products were fractionated by high voltage electrophoresis through 15% (w/v) polyacrylamide gels containing 7 M urea. The gels were dried and visualized by autoradiography and/or phosphor-imaging using Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA, USA).
RNA- and DNA-dependent DNA polymerase activity
RNA- and DNA-dependent DNA synthesis was measured on a 40-mer RNA template (5′ UCAUGCCCUGCUAGCUACUCGAUAUGGCAAUAAGACUCCA 3′) and equivalent 40-nt DNA template, respectively. Substrates were generated by annealing either the RNA or DNA template to the methylphosphonate-substituted DNA primers. Polymerization was initiated by adding 10 nM enzyme to a mixture containing 50 nM template/primer, 200 μM dNTPs, with 50 μM [32P]-α-dCTP, 25 mM Tris–HCl (pH 7.8), 80 mM NaCl, 6 mM DTT and 9 mM MgCl2, at 37°C and terminated after 5 min by adding an equal volume of a formamide-based gel-loading buffer. Polymerization products were fractionated by denaturing 15% (w/v) polyacrylamide gels containing 7 M urea in 89 mM Tris–borate, pH 8.3, 2 mM EDTA.
Site-directed hydroxyl radical cleavage
Site-specific footprints with Potassium peroxynitrite (KOONO) and Fe2+ were monitored on DNA duplexes whose template strand was 5′-end labeled. Hybridization of the template (2.7 pmol) with the complementary primers containing the methylphosphonate analogs (8.1 pmol) was conducted in a buffer containing 20 mM sodium cacodylate, pH 7 and 20 mM NaCl. The duplex was incubated with HIV-1 RT (16.2 pmol) in a buffer containing 120 mM sodium cacodylate, pH 7, 20 mM NaCl, 6 mM MgCl2. Prior to the treatment with KOONO or Fe2+, complexes were preincubated for 20 min and the treatment with Fe2+ and KOONO, respectively, was performed essentially as described (28,29).
RESULTS
Experimental design
Our experimental strategy is outlined schematically in Figure 1. The nucleic acid substrate was a 40-nt RNA/30-nt DNA hybrid whose DNA 3′-terminus was recessed to ensure its sequestration by the DNA polymerase domain. The position of methylphosphonate insertion is defined relative to the DNA primer 3′-terminus, i.e. position −10 represents the phosphodiester backbone between primer nucleotides −10 and −11. Backbone contacts with the RNase H primer grip extend from primer nucleotides −10 to −14 (Figure 1A and B), each of which was modified by introducing a single, dual or triple methylphosphonate linkage. In addition, backbone modifications were extended to include phosphates between positions −15 and −20 to investigate the effect of modifying nucleic acid geometry within the RNase H active site. In a similar manner to studies of Xu et al. (30) and Cannistraro et al. (18), methylphosphonate phosphoramidites were used as the mixture of Rp and Sp stereoisomers (Figure 1C). We cannot, therefore, exclude the possibility of one stereoisomer being selectively recognized by the enzyme.
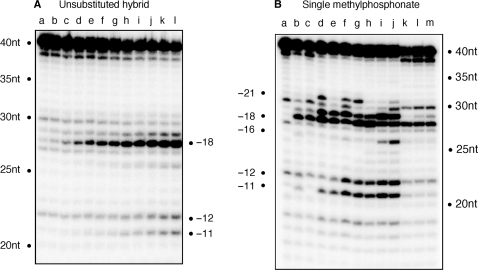
Hydrolysis of the unmodified RNA/DNA hybrid favors −18 cleavage
For reference, the hydrolysis profile of the unmodified RNA/DNA hybrid is shown in Figure 2A. Throughout this time course, the major 28-nt hydrolysis product represents ‘polymerization-dependent’ cleavage at position −18, with a minor −19 product evident at later time points. In contrast to our previous work with RNA/DNA hybrids of alternative sequence, we observed minimal cleavage at position −12/−11, indicative of ‘polymerization-independent’ hydrolysis under single hit conditions (31). Although subsequent data of this communication show that polymerization-independent cleavage can be induced by analog substitution, the inability of HIV-1 RT to cleave at −11/−12 on the wild-type RNA/DNA hybrid may indicate deviation from the preferred nucleotide cleavage sequence postulated by Champoux and co-workers (32).
Figure 2.
(A) Time course of RNase H hydrolysis profile with the unmodified RNA/DNA hybrid. Time points are: 0 s (lane a); 30 s (lane b); 1 min (lane c); 2 min (lane d); 3 min (lane e); 4 min, (lane f); 5 min (lane g); 6 min (lane h); 7 min (lane i); 8 min (lane j); 9 min (lane k) and 10 min (lane l). (B) RNase H cleavage for RNA/DNA hybrids containing single methylphosphonate substitution in the DNA primer. Lane a represents wild-type substrate without enzyme, and lane b wild-type substrate with enzyme. lane c, −10; lane d, −11; lane e, −12; lane f, −13; lane g, −14; lane h, −15; lane i, −16; lane j, −17; lane k, −18; lane l, −19 and lane m, −20. The time point for the hydrolysis of these substrates is 10 min.
Single methylphosphonate substitutions of the RNase H primer grip
A surprisingly complex series of hydrolysis profiles were obtained with RNA/DNA hybrids containing single methylphosphonate substitutions between primer phosphates −10 and −20 (Figure 2B). Since a non-specific breakdown product is present at position −20, we have excluded this from our analysis. Significant differences were evident for hybrids substituted between −11 and −17 (Figure 2B, lanes d–j, respectively), of which positions −11 to −14 represent backbone contacts with the RNase H primer grip (5). The hydrolysis profiles of hybrids bearing backbone modification at positions −10 (Figure 2B, lane c) and between −18 and −20 (Figure 2B, lanes k–m) were unaffected, strengthening the importance of the RNase H primer grip.
Although p51 residue Lys395 contacts primer phosphate −10, its neutralization has no effect on RNase H cleavage (Figure 2B, lane c). However, a second p51 contact, mediated through Glu396 to phosphate −11, is significantly affected by neutralization, resulting in additional cleavage at positions −19 and −21 (Figure 2B, lane d). Neutralizing phosphate −12, which is contacted by Gly359 and Ala360 of p66, also induces enhanced cleavage at position −19 (Figure 2B, lane e). While the altered hydrolysis profiles point to the importance of maintaining the correct phosphate backbone contacts, we also recognize that asymmetric phosphate neutralization can induce bending of duplex DNA towards the neutralized face (22,23). Since the RNase H primer grip is implicated in positioning the scissile bond at the active site, loss of contact with the primer backbone may combine with altered hybrid geometry to permit an adjacent phosphodiester bond to occupy the catalytic center, a situation which Figure 2A shows is evident, albeit to a far lesser extent, with the unsubstituted hybrid.
While analog insertions −11 and −12 invoke polymerization-dependent RNase H cleavage around position −18 (we define polymerization-dependent cleavage as RNase H activity that results from the primer 3′-OH being located at the DNA polymerase active site), these substitutions do not affect polymerization-independent RNase H function (Figure 2B, lanes d and e). In contrast, neutralizing primer phosphate −13, which is contacted by p66 residues His351, Tyr501 and Ile505, causes both relaxed cleavage around position −18 and establishes efficient polymerization-independent cleavage at positions −12 and −11 (Figure 2B, lane f). Rather than re-alignment of the RNA/DNA hybrid within the nucleic acid binding cleft, we believe this reflects physical re-location of RT to the latter mode of hydrolysis. Although primer phosphate −13 lies directly upstream of the polymerization-independent cleavage site, the profiles of lanes k–m indicate that once the modified substrate is located at or upstream of the catalytic center, correct cleavage is restored. Between positions −14 and −17, this two-phase hydrolysis pattern continues, ceasing abruptly with analog insertions between positions −18 and −20. While this ‘phosphate scanning’ strategy highlights the importance of the RNase H primer grip, a surprising observation is that the primer backbone opposite the scissile bond, in our case −18 (Figure 2B, lane k), can be neutralized without affecting cleavage efficiency or specificity.
Triple methylphosphonate scanning—effects of a ‘neutral patch’
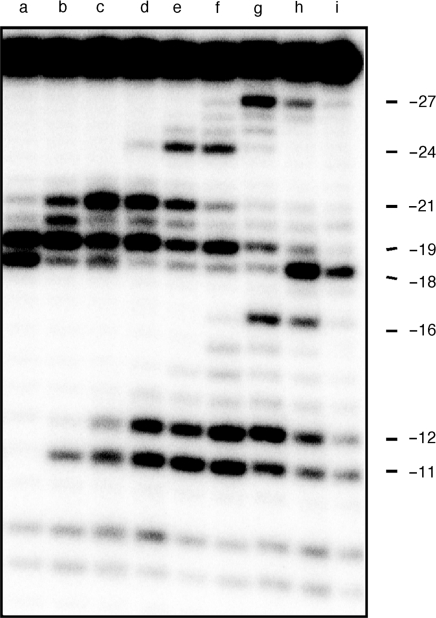
We next analyzed overlapping, triple methylphosphonate substitutions to examine the effects of scanning the RNase H primer grip and the RNase H catalytic center with a ‘neutral patch’. Figure 3 again reveals a complex set of hydrolysis profiles defining multiple RT binding sites. Substitution at positions −10/−11/−12 relaxes specificity, inducing equally efficient cleavage at positions −18 and −19 (Figure 3, lane a). Beyond this, and until the neutral patch centers on primer nucleotide −16 (Figure 3, lane f), −19 cleavage is favored, while both sites are unfavorable when analog substitution centers over primer phosphate −17 (Figure 3, lane g). Finally, and as might be predicted from data of Figure 2B, −18 cleavage predominates when the triple substitution centers on either primer phosphate −18 or −19 (Figure 3, lanes h and i). Thus, with respect to cleavage defined by the ∼17 bp spatial separation of the DNA polymerase and RNase H active sites of HIV-1 RT (1,2,5), extensively neutralizing the phosphate backbone of the RNase H primer grip has the consequence of realigning the RNase H catalytic center over the −19 phosphodiester bond.
Figure 3.
RNase H activity of wild-type HIV-1 RT on substrates containing triple methylphosphonate substitutions. Lane denominations are: lane a, −10/−11/−12; lane b, −11/−12/−13; lane c, −12/−13/−14; lane d, −13/−14/−15; lane e, −14/−15/−16; lane f, −15/−16/−17; lane g, −16/−17/−18; lane h, −17/−18/−19 and lane i, −18/−19/−20.
Similar to the observations of Figure 2B, analog substitutions centered on primer phosphates −14, −15, −16 and −17 result in efficient polymerization-independent hydrolysis at positions −11 and −12 (Figure 3, lanes d–g). As suggested earlier, we propose that extensive duplex bending towards the site of neutralization (22,24,33) distorts the RNA/DNA hybrid where it would normally contact the RNase H active site and as a consequence promotes enzyme binding further downstream in the polymerization-independent hydrolysis mode. When an enzyme adopts this binding mode, the modified portion of the hybrid is positioned upstream of the RNase H active site, where data of Figure 3, lanes h and i indicate that it is tolerated.
Lastly, while analog insertion might be predicted to affect the efficiency of polymerization-dependent and -independent RNase H activity, upstream cleavage as far as −27 (Figure 3, lanes g and h), and −24 (Figure 3, lanes e and f) was unexpected. Structures of HIV-1 RT containing nucleic acid (1,2,5) indicate a 45° bend in the duplex, centered ∼7-bp upstream of the polymerase active site and ∼10 bp from the RNase H catalytic center. We observe that a triple substitution centered over primer nucleotide −17 induces cleavage at position −27 (Figure 3, lane g), while a second, centered around position −15, induces cleavage at position −24 (Figure 3, lane e). Assuming that nucleic acid ‘bending’ in the vicinity of the polymerase active site aids correct enzyme positioning, the 9–10 bp spatial separation between the site of analog insertion and upstream cleavage suggests that the distortion induced by triple analog insertion may be recognized by the p66 thumb subdomain. This notion might also contribute to enhanced cleavage at position −21 induced by a triple analog substitution centered over template nucleotide −13 (Figure 3, lane b). While speculative, a precedent for such events has been established with RT from the Saccaromyces cerevisiae LTR retrotransposon Ty3, which can also be induced to cleave analog-modified RNA/DNA hybrids ∼12 bp from the site of insertion (34,35).
Temporal analysis of polymerization-dependent and -independent RNase H cleavages
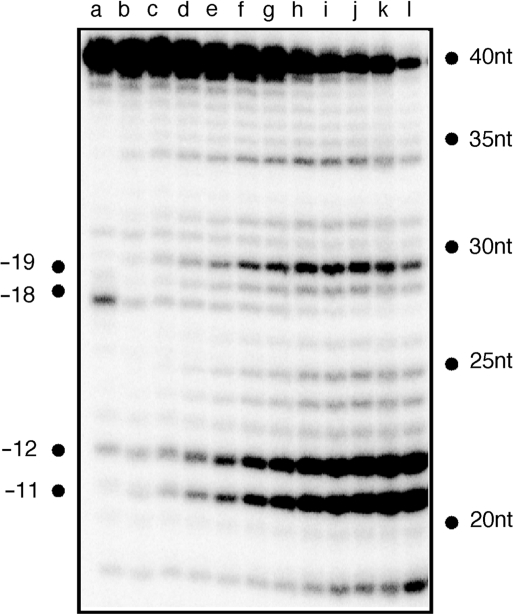
Previous studies demonstrated that polymerization-independent RNase H cleavage followed polymerization-dependent events (10). We therefore elected to determine whether analog substitution favoring polymerization-independent cleavage reflected: (i) acceleration of the former process or (ii) repositioning of RT to favor the latter. To address this, a time course was performed on substrate containing a −15/−16/−17 analog insertion, the results of which are presented in Figure 4. Throughout the entire analysis the polymerization-dependent (i.e. −18) cleavage product failed to accumulate, indicating that extensive charge neutralization affects hybrid structure in a manner that HIV-1 RT is ‘forced’ to occupy, and hydrolyze, its secondary binding site.
Figure 4.
Time-dependent RNase H activity of HIV-1 RT with the −15/−16/−17 triply substituted substrate. Sample time points are: 0 s (lane a); 30 s (lane b); 1 min (lane c); 2 min (lane d); 3 min (lane e); 4 min, (lane f); 5 min (lane g); 6 min (lane h); 7 min (lane i); 8 min (lane j); 9 min (lane k) and 10 min (lane l).
Site-specific footprinting of methylphosphonate-substituted DNA duplexes
To further investigate how charge neutralization affects positioning of the DNA polymerase and RNase H domains, we examined binding of RT to methylphosphonate-substituted DNA duplexes by site-specific chemical footprinting. KOONO provides a metal-free source of hydroxyl radicals generated during decomposition of the conjugate acid HOONO. Mediated through Cys280 of the p66 subunit, this approach induces cleavage of the template strand primarily at position −7/−8 (29). In contrast, replacing the catalytic Mg2+ of the RNase H domain with Fe2+ provides a metal-dependent source of hydroxyl radicals (28) capable of cleaving the template of duplex DNA at position −18.
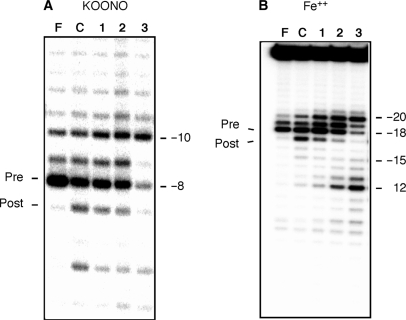
In Figure 5A, lane C, the −8 and −7 KOONO products represent cleavage of the unmodified substrate by HIV-1 RT positioned in the pre- and posttranslocation mode, respectively. In a single cycle of nucleotide incorporation, pyrophosphate is generated following the formation of a new phosphodiester bond. Binding of the next complementary nucleotide requires the release of pyrophosphate from this product complex. The enzyme must translocate a single template position further downstream to clear the nucleotide binding site (N-site). This motion relative to the nucleic acid substrate brings the 3′-end of the primer to the priming or product site (P-site) (5). The latter configuration is referred to as the posttranslocational state and the former pretranslocational state (29). As shown previously (36), the pyrophosphate analog Foscarnet (PFA) captures the pretranslocation complex (Figure 5A, lane F). When equivalent complexes are analyzed via Fe2+-mediated cleavage, the −18 and −17 products represent enzyme in the pre- and posttranslocation state, respectively (Figure 5B, lanes F and C). When a −13 or −13/−14 substitution (which collectively comprise the majority of primer grip contacts) is introduced, the DNA polymerase domain remains for the most part correctly positioned (Figure 5A, lanes 1 and 2), but increased −10 cleavage suggests that a fraction is relocated 2-bp upstream. This notion is supported when the two substituted duplexes are characterized by Fe2+ cleavage, where positions −19 and −20 are now accessible. The combined footprinting approaches on the −13 and −13/−14-substituted duplexes thus indicate that, while contacts to the DNA duplex at the DNA polymerase domain are preserved (Figure 1B), modifying the RNase H primer grip affects positioning of the RNase H catalytic center.
Figure 5.
Site-specific chemical footprinting of analog-substituted substrates using (A) KOONO and (B) Fe2+. Lane denominations are: lane F, PFA; lane C, wild-type substrate; lane 1, −13 substitution; lane 2. −13/−14 substitution; lane 3, −13/−14/−15 substitution. The notations ‘Pre’ and ‘Post’ refer to cleavage resulting from enzyme positioned in the pre- and posttranslocation state, respectively.
In contrast, a triple analog substitution centered over primer phosphate −14 dramatically alters positioning of both the DNA polymerase and RNase H domains. KOONO footprinting fails to detect enzyme in the pretranslocation (i.e. −8 cleavage) mode (Figure 5A, lane 3), and the major products of Fe2+ footprinting indicate cleavage at positions −20 and −12 (Figure 5B, lane 3). Combining these observations suggests RT has redistributed into a population that binds 2-bp upstream and 10-bp downstream of the region it would normally be predicted to occupy, i.e. the 3′-OH of the nucleic acid substrate is displaced from the polymerase active site.
RNA- and DNA-dependent DNA synthesis using methylphosphonate-substituted DNA as primers
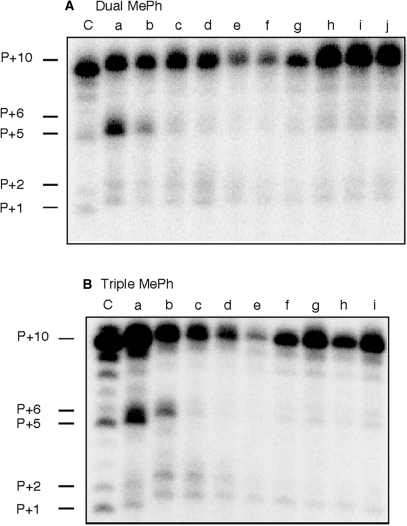
Although the RNase H primer grip is implicated in directing template orientation at the RNase H active site, it was important to understand how its alteration affected DNA synthesis. Consequently, RNA/DNA hybrids and DNA duplexes containing dually- and triply substituted primers were assayed for their ability to support DNA- and RNA-dependent DNA synthesis. In these experiments, extension products were visualized by incorporating [32P]-labeled dCTP as the first nucleotide. RNA-dependent DNA synthesis on modified RNA/DNA hybrids is presented in Figure 6. While double and triple methylphosphonate substitutions supported efficient DNA synthesis, we observed a prominent P + 5 product on hybrids containing a dual −10/−11 substitution (Figure 6A, lane a) and a triple −10/−11/−12 substitution (Figure 6B, lane a). Such stalling suggests that neutralizing the phosphate backbone in the vicinity of position −15 relative to an enzyme engaged in DNA synthesis may cause loss of contact with the RNase H primer grip and transient pausing. A prediction of this notion would be that a binary complex where methylphosphonate substitutions were introduced around position −15 would not initiate RNA-dependent DNA synthesis as efficiently. This prediction was borne out experimentally with hybrids containing dual −14/−15 and −15/−16 substitutions (Figure 6A, lanes e and f) and a triple −14/−15/−16 substitution (Figure 6B, lane e).
Figure 6.
RNA-dependent DNA polymerase activity of HIV-1 RT on dual-substituted RNA/DNA hybrids (A), Lane C, wild-type substrate; lane a, −10/−11; lane b, −11/−12; lane c, −12/−13; lane d, −13/−14; lane e, −14/−15; lane f, −15/−16; lane g, −16/−17; lane h, −17/−18; lane i, −18/−19 and lane j, −19/−20. (B), RNA-dependent DNA synthesis with triply-substituted RNA/DNA hybrids. Lane C, wild-type substrate; lane a, −10/−11/−12; lane b, −11/−12/−13; lane c, −12/−13/−14; lane d, −13/−14/−15; lane e, −14/−15/−16; lane f, −15/−16/−17; lane g, −16/−17/−18; lane h, −17/−18/−19 and lane i, −18/−19/−20.
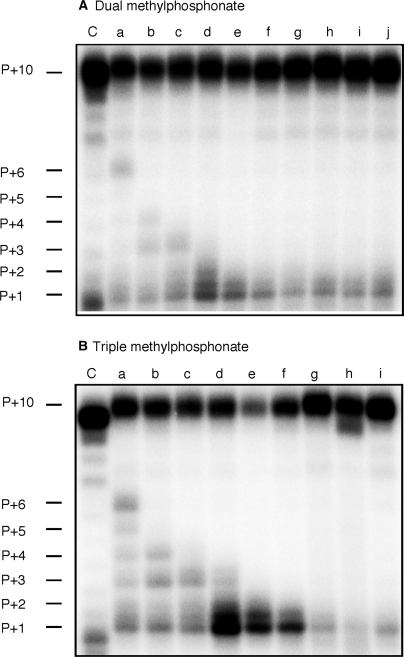
A similar pattern emerged when DNA-dependent DNA synthesis on substituted duplexes was examined, although in this case P + 1 and P + 2 products were more pronounced with −13/−14 and −14/−15 substitutions (Figure 7A, lanes d and e). The severity of this effect increased with triple methylphosphonate substitution, evidenced by significant accumulation of the P + 1 product on a −13/−14/−15-substituted duplex (Figure 7B, lane d). Since radiolabeled dCTP is incorporated as the first nucleotide of nascent DNA, the −13/−14/−15 substitution induces pausing directly after RT initiates DNA synthesis, when the methylphosphonate substitutions occupy positions −14/−15/−16. P + 1 pausing was also evident when duplex DNA containing the −14/−15/−16 substitution was examined, and accompanied by severely reduced full-length DNA synthesis (Figure 7B, lane e). This phenotype is partially restored with −16/−17/−18-substituted DNA, i.e. a slight diminution of P + 1 product and restoration of the full-length product. Assuming asymmetric backbone neutralization induces duplex bending, primer positions at which methylphosphonate substitution affects DNA synthesis implies that an irregular geometry of the duplex invokes a steric clash with structural elements of the RNase H active site that blocks further movement of the replication complex. Surprisingly, P + 1 pausing was considerably less pronounced during RNA-dependent DNA synthesis (Figure 6), which may reflect greater processivity of HIV-1 RT on RNA templates (37,38).
Figure 7.
DNA-dependent DNA synthesis activity of HIV-1 RT on dually (A) and triply (B) substituted DNA duplexes. Lane notations are as in the Figure 6 legend.
DISCUSSION
Crystal structures of HIV-1 RT containing duplex DNA (1,2) and an RNA/DNA hybrid (5) indicate multiple contacts to the phosphate backbone in the vicinity of the RNase H catalytic center. While the contribution of these contacts (designated the RNase H primer grip) to RNase H function remains to be fully established, complexes of B. halodurans and human RNases H containing an RNA/DNA hybrid revealed a ‘phosphate binding pocket’, albeit involving fewer contacts, in the equivalent region (15–17,39). The latter finding has led to the proposal that backbone distortion and minor groove narrowing might serve to anchor the DNA strand as the scissile phosphate of the RNA accesses the catalytic site. The notion that the RNase H primer grip of HIV-1 RT functions in an analogous manner is therefore not unreasonable, and prompted us to examine the consequences of neutralizing the DNA primer backbone where it contacts the RNase H primer grip.
Introducing a single −11 phosphodiester modification, where the primer contacted by the p51 subunit, significantly alters RNase H cleavage specificity (Figure 2B, lane d). A general structural alteration resulting from backbone neutralization seems unlikely here, since a −10 substitution has little effect on RNase H activity, despite being likewise contacted by the small RT subunit (Figure 2B, lane c). A more extensive triple substitution, centered around primer phosphate −16, virtually eliminates polymerization-dependent −18 cleavage in favor of the polymerization-independent site at −11/−12 (Figure 4). Finally, modifying the RNase primer grip ∼14 bp from the DNA polymerase active site efficiently stalls RT after initiation of DNA synthesis. Collectively, our data indicates that modifying critical contacts between RT and the RNA/DNA hybrid near the RNase H domain and altering hybrid geometry by charge neutralization affect the position of the RNase H active site on a static enzyme as well as the location of the entire enzyme on the hybrid. Although re-positioning of RT is suggested by the hydrolysis profile of a −15/−16/−17 triply modified hybrid (Figure 4), this can be ‘overridden’ by including dNTPs, which favor enzyme binding in a polymerization orientation with its polymerase active site over the primer 3′-terminus (Figure 6B, lane g).
Rausch et al. (13) have reported that alanine substitution of RNase H primer grip residues Thr473, Asn474, Glu475 and Tyr501 dramatically reduced polymerization-independent cleavage although considerable levels of polymerization-dependent RNase H activity was retained by these mutant enzymes. Our results reveal that neutralizing the DNA phosphates (−13 and −14) contacting these residues affect both polymerization-dependent and -independent cleavages. These observations suggest that methylphosphonate substitutions may affect both the contacts from the RNase H primer grip residues and the trajectory of the substrate. Closer examination of the phenotype displayed by a subset of substituted RNA/DNA hybrids suggests that altering the geometry of nucleic acid contacted by the RNase H primer grip may have consequences for the translocation status of RT. Figure 3, lanes d–f, indicates that triple methylphosphonate substitutions centered around positions −13, −14 and −15 virtually eliminate −18 cleavage, instead favoring hydrolysis at −19. According to Marchand et al. (29,36), polymerization-dependent RNase H cleavage at −18 is indicative of a posttranslocated complex, while cleavage at −19 represents a pretranslocation complex that would not support DNA synthesis. The data in Figure 6b, lanes d–f shows that DNA polymerase activity on duplex DNA substrates containing these substitutions.
In summary, this comprehensive analysis of the RNase H primer grip motif suggest that neutralizing phosphate charges of the DNA primer repositions HIV-1 RT on the RNA/DNA hybrid, thereby cleaving in both polymerization-dependent and -independent manner. Moreover, the footprinting analysis shows that the RNase H primer grip also appears to affect translocation of HIV-1 RT highlighting the importance of this motif in RT function.
FUNDING
Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (to C.D. and S.F.J.LeG.); Canadian Institutes of Health Research (to M.G.). Funding for open access charge: Intramural Research Program of the NIH, NCI and Center for Cancer Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government.
REFERENCES
- 1.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 2.Jacobo-Molina A, Ding J, Nanni RG, Clark A.D., Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl Acad. Sci. USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarafianos SG, Das K, Tantillo C, Clark A.D., Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curr K, Tripathi S, Lennerstrand J, Larder BA, Prasad VR. Influence of naturally occurring insertions in the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and mutation frequencies in vitro. J. Gen. Virol. 2006;87:419–428. doi: 10.1099/vir.0.81458-0. [DOI] [PubMed] [Google Scholar]
- 7.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J. Virol. 2002;76:9143–9151. doi: 10.1128/JVI.76.18.9143-9151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer PL, Ferris AL, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes SH. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 (HIV-1) reverse transcriptase. J. Mol. Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 9.Wohrl BM, Krebs R, Thrall SH, Le Grice SF, Scheidig AJ, Goody RS. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J. Biol. Chem. 1997;272:17581–17587. doi: 10.1074/jbc.272.28.17581. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix JL, Le Grice SF. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 11.Jacques PS, Wohrl BM, Ottmann M, Darlix JL, Le Grice SF. Mutating the ‘primer grip’ of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem. 1994;269:26472–26478. [PubMed] [Google Scholar]
- 12.Chao SF, Chan VL, Juranka P, Kaplan AH, Swanstrom R, Hutchison C.A., 3rd Mutational sensitivity patterns define critical residues in the palm subdomain of the reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1995;23:803–810. doi: 10.1093/nar/23.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice SF. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 14.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J. Virol. 2003;77:8548–8554. doi: 10.1128/JVI.77.15.8548-8554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannistraro VJ, Taylor JS. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J. Biol. Chem. 2004;279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- 19.Cannistraro VJ, Taylor JS. Ability of polymerase eta and T7 DNA polymerase to bypass bulge structures. J. Biol. Chem. 2007;282:11188–11196. doi: 10.1074/jbc.M608478200. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Declais AC, Lilley DM. Mechanistic aspects of the DNA junction-resolving enzyme T7 endonuclease I. Biochemistry. 2006;45:3934–3942. doi: 10.1021/bi0523254. [DOI] [PubMed] [Google Scholar]
- 21.Tian L, Claeboe CD, Hecht SM, Shuman S. Remote phosphate contacts trigger assembly of the active site of DNA topoisomerase IB. Structure. 2004;12:31–40. doi: 10.1016/j.str.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Hardwidge PR, Zimmerman JM, Maher L.J., 3rd Charge neutralization and DNA bending by the Escherichia coli catabolite activator protein. Nucleic Acids Res. 2002;30:1879–1885. doi: 10.1093/nar/30.9.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosikov KM, Gorin AA, Lu XJ, Olson WK, Manning GS. Bending of DNA by asymmetric charge neutralization: all-atom energy simulations. J. Am. Chem. Soc. 2002;124:4838–4847. doi: 10.1021/ja0125218. [DOI] [PubMed] [Google Scholar]
- 24.Okonogi TM, Alley SC, Harwood EA, Hopkins PB, Robinson BH. Phosphate backbone neutralization increases duplex DNA flexibility: a model for protein binding. Proc. Natl Acad. Sci. USA. 2002;99:4156–4160. doi: 10.1073/pnas.072067799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Grice SF, Cameron CE, Benkovic SJ. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 27.Dash C, Rausch JW, Le Grice SFJ. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 2004;32:1539–1547. doi: 10.1093/nar/gkh307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotte M, Maier G, Gross HJ, Heumann H. Localization of the active site of HIV-1 reverse transcriptase-associated RNase H domain on a DNA template using site-specific generated hydroxyl radicals. J. Biol. Chem. 1998;273:10139–10146. doi: 10.1074/jbc.273.17.10139. [DOI] [PubMed] [Google Scholar]
- 29.Marchand B, Gotte M. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J. Biol. Chem. 2003;278:35362–35372. doi: 10.1074/jbc.M304262200. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. Contacts between the 5′ nuclease of DNA polymerase I and its DNA substrate. J. Biol. Chem. 2001;276:30167–30177. doi: 10.1074/jbc.M100985200. [DOI] [PubMed] [Google Scholar]
- 31.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 32.Schultz SJ, Zhang M, Champoux JJ. Sequence, distance, and accessibility are determinants of 5′-end-directed cleavages by retroviral RNases H. J. Biol. Chem. 2006;281:1943–1955. doi: 10.1074/jbc.M510504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss-Soukup JK, Rodrigues PD, Maher L.J., 3rd Effect of base composition on DNA bending by phosphate neutralization. Biophys. Chem. 1998;72:297–306. doi: 10.1016/s0301-4622(98)00112-4. [DOI] [PubMed] [Google Scholar]
- 34.Lener D, Kvaratskhelia M, Le Grice SF. Nonpolar thymine isosteres in the Ty3 polypurine tract DNA template modulate processing and provide a model for its recognition by Ty3 reverse transcriptase. J. Biol. Chem. 2003;278:26526–26532. doi: 10.1074/jbc.M302374200. [DOI] [PubMed] [Google Scholar]
- 35.Yi-Brunozzi HY, Brinson RG, Brabazon DM, Lener D, Le Grice SF, Marino JP. High-resolution NMR analysis of the conformations of native and base analog substituted retroviral and LTR-retrotransposon PPT primers. Chem. Biol. 2008;15:254–262. doi: 10.1016/j.chembiol.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchand B, Tchesnokov EP, Gotte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J. Biol. Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 37.Huber HE, McCoy JM, Seehra JS, Richardson CC. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J. Biol. Chem. 1989;264:4669–4678. [PubMed] [Google Scholar]
- 38.Reardon JE. Human immunodeficiency virus reverse transcriptase. A kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J. Biol. Chem. 1993;268:8743–8751. [PubMed] [Google Scholar]
- 39.Nowotny M, Cerritelli SM, Ghirlando R, Gaidamakov SA, Crouch RJ, Yang W. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008;27:1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]