Abstract
In humans a single species of the RNAseIII enzyme Dicer processes both microRNA precursors into miRNAs and long double-stranded RNAs into small interfering RNAs (siRNAs). An interesting but poorly understood domain of the mammalian Dicer protein is the N-terminal helicase-like domain that possesses a signature DExH motif. Cummins et al. created a human Dicer mutant cell line by inserting an AAV targeting cassette into the helicase domain of both Dicer alleles in HCT116 cells generating an in-frame 43-amino-acid insertion immediately adjacent to the DExH box. This insertion creates a Dicer mutant protein with defects in the processing of most, but not all, endogenous pre-miRNAs into mature miRNA. Using both biochemical and computational approaches, we provide evidence that the Dicer helicase mutant is sensitive to the thermodynamic properties of the stems in microRNAs and short-hairpin RNAs, with thermodynamically unstable stems resulting in poor processing and a reduction in the levels of functional mi/siRNAs. Paradoxically, this mutant exhibits enhanced processing efficiency and concomitant RNA interference when thermodynamically stable, long-hairpin RNAs are used. These results suggest an important function for the Dicer helicase domain in the processing of thermodynamically unstable hairpin structures.
INTRODUCTION
Eukaryotic ribonuclease III (RNase III) enzymes exhibit specificity for double-stranded RNA (dsRNA) and are divided into three structural classes. The first class of RNase III enzymes are the simplest, consisting of a single catalytic domain (RIII), and are represented by Escherichia coli RNase III that promotes ribosomal RNA (rRNA), tRNA and mRNA maturation through cleavage of hairpin-like stem–loop structures (1). Members of the second RNase III class, which include Drosha and its homologs, contain two catalytic RIII domains, a dsRNA binding domain (dsRBD), and an N-terminal region that mediates interactions with protein partners such as DGCR8 (2). The third class of RNase III enzymes comprised of Dicer and Dicer-like proteins that play essential roles in several developmental processes through the processing of endogenous dsRNA substrates. For the most part, Dicer enzymes contain an N-terminal DExH-box RNA helicase-like domain, a domain of unknown function (DUF283), a PAZ domain, two RIII domains (RIIIa and RIIIb), and a dsRBD (3). The PAZ domain is highly conserved and is also found in proteins from the Argonaute superfamily that are important mediators of RNA interference (4).
Mammalian genomes harbor one Dicer gene encoding a ∼220-KDa protein that has the multidomain structure typical of class 3 RNase III enzymes (5,6). In vitro experiments using wild-type and various mutant versions of the human Dicer enzyme (hDicer) reveal that hDicer can process various forms of dsRNA, including long linear dsRNA molecules that contain 5′ PO4 and 3′ OH groups, as well as short hairpin RNAs (shRNA) that mimic Drosha-produced pre-miRNAs (5,7,8). For long dsRNA, cleavage by hDicer into 21-mer siRNAs can occur from both ends of the open, linear substrate in an ATP-independent manner (7–10). In contrast, processing of pre-miRNAs into mature miRNAs occurs through recognition of the 2-nt 3′ overhang at one end of the hairpin duplex (11). Analysis of dsRNA processing by hDicer enzymes engineered with specific RIII domain mutations reveal that human Dicer contains a single dsRNA processing center formed through intramolecular dimerization of the two RIII domains that function independently to cleave phosphodiester bonds on opposite strands of the dsRNA substrate (12,12). Biochemical analysis of hDicer, along with crystal structure data of the hDicer C-terminus, indicates that divalent cations (e.g. Mg2+) are present at the active site and are required for catalysis (7,12,13). At present, the crystal structure of the hDicer N-terminus has not been reported.
Biochemical fractionation of human cells indicates that hDicer does not exist alone, but rather associates in vivo with other dsRNA-binding partners such as members of the Ago family (14,15), HIV-1 TAR RNA-binding protein (TRBP) (16,17) and a related protein, protein activator of PKR (PACT) (18,19), RNA helicase A (RHA) (20) and possibly others. RNAi depletion of PACT or TRBP has a detrimental effect on miRNA-mediated and siRNA-mediated gene silencing, respectively, with loss of PACT expression resulting in a decrease in mature miRNA levels (19). Dicer is also found in a complex with Ago 2, the slicer component of RISC (21), suggesting that Dicer is involved in the hand-off of processed trigger molecules for use by RISC (17,22). Therefore, the processing of hairpin RNAs by hDicer is likely to occur in multiple steps that involve different domains of the enzyme. The contributions of other proteins such as Ago 2 and TRBP, which have been identified in a putative miRNA loading complex are likely to also be important for efficient hairpin processing (14,15).
Despite biochemical and crystallographic investigation of the hDicer C-terminal domain, the role of the N-terminal helicase domain in hDicer activity is still poorly understood. Recently a Dicer mutant cell line was created via AAV insertional mutagenesis of the hDicer helicase domain (23). This cell line has reduced levels of many, but not all, miRNAs relative to its wild-type parental cell line. We have further characterized the nature of the insertion, which is a homozygous in-frame 43-amino-acid insertion close to the helicase DExH motif. Our findings demonstrate that this mutation has detrimental effects on the processing of most pre-miRNAs, but paradoxically has enhanced processing relative to wild-type hDicer of long-hairpin transcripts. These studies suggest an important role for the helicase domain in the recognition and processing of imperfect Watson–Crick helices, and the absence of a functional helicase domain results in enhanced processivity in the cleavage of long hairpins. Our studies lead to a model in which the hDicer helicase domain is required for efficient processing of hairpin RNAs with thermodynamically unstable stem regions. Most likely this characteristic of hDicer reflects upon the requirement that Dicer processes a variety of stem–loop substrates, pointing to an important role for the helicase domain in expanding the function of human Dicer beyond pre-miRNA processing. Interestingly, perturbation of this domain results in enhanced processivity by the enzyme on long, thermodynamically stable hairpin RNAs.
MATERIALS AND METHODS
Plasmid construction
The plasmids expressing sh- and lhRNA that target the HIV genome are driven by the human U6 promoter and are described in detail elsewhere (24). Luciferase reporter plasmids were constructed by inserting the annealed oligonucleotides that contain the relevant target site were inserted into XhoI and NotI sites of the psiCheck-2 plasmid (Promega) and were previously described in detail (24,25). The plasmid expressing wild-type Dicer was a generous gift of Dr. Patrick Provost (7).
Cell culture
Wild-type HCT116 and Dicer mHelicase cells, an HCT116-based cell line that contains a targeted insertion into exon 5 of both Dicer alleles was generously provided by Dr Bert Vogelstein (23). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 20 U/ml penicillin/streptomycin and 2 mM L-Gln. Cells were grown in a humidified, 5% CO2 incubator at 37°C and were passaged by standard means.
Mutant Dicer characterization
Genomic DNA was collected from wild-type or mHelicase cells by standard methods and subjected to PCR with primers DSF (5′ GTTCTCATTATGACTTGCTATGTCGC) and DSR (5′ CTCGATAGGGGTGGTCTAGGATT) under the following cycling conditions: 95°C, 5 min, one cycle; 94°C, 30 s, 55°C, 30 s, 72°C, 30 s, 30 cycles; 72°C, 10 min, one cycle. PCR products were examined by electrophoresis and cloned into a T/A cloning vector followed by sequencing with vector-specific primers. The in-frame mutation in the helicase domain was identified and used in BLAST queries to determine its origin.
For quantitative RT-PCR, total RNA from wild-type and Dicer mHelicase cells was collected with RNA STAT-60 (Tel-Test) and converted into cDNA using random hexamers and M-MLV reverse transcriptase. Control reactions omitting M-MLV were also included to rule-out genomic DNA contamination. Prepared cDNA was subjected to separate real-time PCR reactions using the iQ SYBR green master mix (Bio-Rad) and primers specific for human Dicer or the internal control gene, human acidic ribosomal phosphor-protein P0 (RPLP0). The PCR and fluorescence measurement was performed in a Bio-Rad iCycler using the following cycling conditions: 95°C for 9 min, followed by 40 cycles to 3-step PCR with 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Dicer expression was normalized to levels of the RPLP0 internal control gene. All data points were performed in triplicate and the experiment was performed twice. PCR primer sequences for the RT-PCR analysis are available from the authors on request.
For immunoblot analysis, whole-cell extracts were collected from wild-type and Dicer mHelicase cells and HEK293 cells (CRL-1573) using the MPer reagent according to the manufacturer's instructions (Pierce) and the protein concentration determined by Bradford analysis. Thirty micrograms of protein was separated on a 10% resolving SDS/PAGE gel, transferred to PVDF, blocked in 5% non-fat dried milk in TBST, and incubated with the following Dicer antibodies: N-terminus (Abcam, clone 13D6, 1:300 dilution) or C-terminus (Santa Cruz Biotechnologies, clone H-212, 1:200 dilution), followed by an incubation with HRP-labeled secondary antibodies and detection by ECL chemiluminescence. Separate parts of the membrane were incubated with primary antibodies specific to beta-actin and detected with secondary antibodies and HRP staining as described above for Dicer.
Homology modeling of the helicase insertion was carried out using the HEF helicase from Pyrococcus furiosus (PDB file 1WP9) as a template for visualizing the helicase domain of human Dicer since this domain of Dicer has not been crystallized (26). The sequences were aligned using Figure 1 in Nishino et al. as a guide (26). The program MODELLER 9.0 (27) running within Discovery Studio 2.0 (Accelrys, San Diego, CA) was used to build a model of HEF helicase with a 43-residue loop corresponding to the in-frame insertion mutation spliced into motif II.
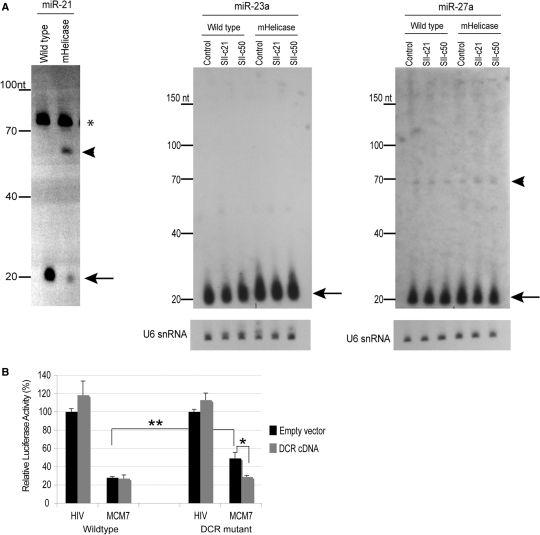
Figure 1.
Processing and efficacy of microRNAs in vivo. (A) Northern blot analyses of miRNA expression in wild-type and mHelicase cells. For miR-21, small RNAs were harvested from wild-type and Dicer mHelicase cells. The unprocessed pre-miR-21 (arrowhead) and mature miR-21 species (arrow) are indicated. The asterisk denotes a nonspecific signal that runs slightly >70 nt, which likely represents cross hybridization of the 2′-O-methyl-modified oligonucleotide to an abundant tRNA, serves as an internal loading control. The illustrated size marker is based on the migration of the Ambion Decade ladder. For miR-23a and miR-27a, total RNA was harvested from wild-type and mHelicase cells transfected with either the control U6 vector (Control) or vectors expressing U6-driven shRNA (SII-c21) or lhRNA (SII-c50). Arrows denote the migration of the mature miR-23a and miR-27a. For miR-27a, the ∼63 nt pre-miR-27a is indicated by the arrowhead. The illustrated size marker is based on the migration of the Ambion Decade ladder. U6 snRNA expression was also detected as a loading standard. (B) Inhibition of miRNA gene expression by an ectopic miRNA expression construct. Twenty-five nanograms of the psiCHECK-miR-93 reporter vector and 100 ng of the MCM7 miRNA expression vector were co-transfected with an empty vector or a vector expressing the wild-type Dicer cDNA into wild-type or Dicer mHelicase cells. Luciferase values were determined 24 h later and the Renilla target was normalized to Firefly luciferase, which is contained on the same reporter construct but is not subject to downregulation by the miRNA. Luciferase values in response to the irrelevant HIV miRNA expression vector (HIV) were set at 100%. The mean and standard deviations from three experiments are presented. *P < 0.02, **P < 0.05.
Luciferase assays
For miRNA assays, wild-type and Dicer mHelicase cells were transfected in 24-well dishes using Lipofectamine 2000 (LF2000) according to the manufacturer's directions. Each well received the following: 12.5 ng of psi-Check-93 target plasmid, 137.5 ng of miRNA expression plasmid (either MCM7 or the irrelevant control HIV) and 150 ng of pcDNA3.1 vector (either empty vector or wild-type Dicer cDNA), and 0.75 μl of LF2000. Twenty-four hours post-transfection, luciferase activity was determined using the Dual Luciferase system (Promega).
For sh- and lhRNA assays, wild-type and Dicer mHelicase mutant cells were transfected in 24-well dishes using LF2000. Each well received the following: 100 ng of the psiCheck target plasmid and 1–100 ng of the sh- or lhRNA expression plasmid. In experiments where 1 ng of sh- or lhRNA expression vector was used, the total amount of plasmid DNA was increased to 200 ng by adding the U6 empty vector. In experiments targeting the HIV genome, each well received 100 ng of pNL4-3.Luc target plasmid, 100 ng of sh or lhRNA expression and 1 ng of a Renilla luciferase expression plasmid (Promega). Twenty-four hours post-transfection, luciferase activity was analyzed using the Dual Luciferase system (Promega).
Northern blot analysis
For miRNA expression analysis, either total RNA or small RNAs were collected from wild-type or Dicer mHelicase cells using RNA STAT-60 (Tel-Test) (total RNA) or the miRVANA miRNA isolation kit according to the manufacturer's instructions (Ambion). Ten micrograms of total RNA or 2 μg of miRVANA RNA were separated in a 15%/8 M urea denaturing polyacrylamide gel, electro-transferred to a Hybond-N+ membrane (Amersham Bioscience), and probed with a 32P-labeled oligonucleotide that is complementary to the mature miR-21 and consists of 2′-O-methyl-modified RNA bases. For the detection of miR-23a and miR-27a, 32P-labeled DNA oligonucleotides that are complementary to the mature miRNA were used as probes for hybridization. Following successive washes of increasing stringency, the blot was analyzed by autoradiography.
For sh- and lhRNA expression analyses, total RNA was extracted and purified with RNA STAT-60 (Tel-Test) according to the manufacturer's instructions. Twenty micrograms of total RNA were separated in an 8% polyacrylamide denaturing gel, electro-transferred to a Hybond-N membrane and probed with 32P-labeled synthetic oligonucleotides complementary to either sense or antisense sequences of the specific siRNA. Following successive washes of increasing stringency, the blot was analyzed by autoradiography. The processing efficiency for each hairpin RNA (Probe 1, Probe 2, Probe 3) was determined by autoradiography and dosimetry as follows: Processing efficiency = matureIDV/(matureIDV + precursorIDV). IDV, Integrated Density Value.
In vitro processing analysis
Preparation of cell extracts and in vitro processing were performed as described elsewhere, with some modifications (28). Briefly, ∼1 × 107 cells (wild type and mHelicase) were washed twice with ice-cold PBS, resuspended in the following buffer: 20 mM HEPES-KOH pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 5% glycerol and protease inhibitor (Complete Mini, Roche), and lysed by sonication. Cell extracts were cleared by centrifugation at 13 000 g for 15 min at 4°C and then quickly frozen in an ethanol:dry ice bath. Protein concentrations of the cell extracts were determined by the Bradford assay.
For in vitro analysis of pre-miRNA, a synthetic RNA with the sequence (5′-UAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUC-3′) was 5′-end labeled with 32P-γ-ATP and the full-length RNA was gel-purified and precipitated with ethanol by standard methods after labeling. The labeled RNA was heated to 95°C, 3 min, followed by 75°C, 3 min, and then removed from the heat to cool to room temperature to allow the RNA to fold into the proper hairpin structure. For in vitro reactions using recombinant human Dicer (rDicer), 5 × 104 c.p.m. of labeled pre-miRNA was incubated with 1 μl of rDicer (Ambion) according the manufacturer's instructions. For in vitro reactions using cell extracts from wild-type or Dicer mHelicase cells, 5 × 104—1 × 105 c.p.m. of labeled pre-miRNA was incubated with 25 μg of cell extract, 3.2 mM MgCl2 and 0.5 mM ATP at 37°C for 1–3 h. Reactions were stopped by the addition of EDTA and immediately run on 10–15%/8 M Urea denaturing polyacrylamide gels. Following electrophoresis, separated RNAs were analyzed by exposing the gel to a phosphorscreen and analyzed using a Storm phosphorimager. The processing efficiency was determined as follows: processing efficiency = matureIDV/(matureIDV + precursorIDV) IDV, Integrated Density Value.
For in vitro analysis of sh- and lhRNAs, PCR products that include the T7 phage promoter were generated for SII-rev 21-, 50- and 80-bp hairpin sequences. The primers for producing the T7 transcription units were: upper (5′-TAATACGACTCACTATAGCCTGTGTCTCTTCAGTTACT-3′) and lower (5′-AAGCCTGTGCCTCTTCAGCTACC). Individually sized sh- or lhRNA RNAs were transcribed using the MAXIscript T7 kit (Ambion) in the presence of 32P-α-UTP according to the manufacturer's instructions and gel purified to obtain the full-length RNA. The labeled RNA was heated to 95°C, 3 min, followed by 75°C, 3 min, and then removed from the heat to cool to room temperature to allow the RNA to fold into the proper hairpin structure. For in vitro reactions using rDicer, 5 × 104 c.p.m. of labeled sh- or lhRNA was incubated with 1 U of rDicer (Genlantis) according the manufacturer's instructions. For in vitro reactions using cell extracts from wild-type or Dicer mHelicase cells, 5 × 104—1 × 105 c.p.m.of labeled sh- or lhRNA was incubated with 25 μg of cell extract, 3.2 mM MgCl2 and 0.5 mM ATP at 37°C for 1–3 h. Reactions were stopped by the addition of EDTA and immediately run in 10–15%/8 M Urea denaturing polyacrylamide gels. Following electrophoresis, separated RNAs were analyzed by exposing the gel to a phosphorscreen and analyzed using a Storm phosphorimager. The processing efficiency for each hairpin RNA was determined as follows: processing efficiency = matureIDV/(matureIDV + precursorIDV) IDV, Integrated Density Value.
RESULTS
Characterization of the Dicer helicase mutant HCT116 cells
A frozen aliquot of a subclone of HCT116 cells that contains a targeted insertion in exon 5 of both alleles of Dicer, herein referred to as the Dicer helicase mutant (or mHelicase) cells, along with the parental wild-type HCT116 cells, were kindly provided by Dr Bert Vogelstein (23). PCR amplification and sequence analysis of genomic DNA from wild-type and Dicer helicase mutant cells using primers that flank exon 5 of the human Dicer gene indicate that the Dicer helicase mutant cells contain a 129-bp insertion of an ectopic sequence derived from the targeting vector that is in-frame with the predicted amino acid sequence of Dicer (Supplementary Figure 1A). The insertion mutation resides two amino acids from the DExH motif DECH, an important motif in RNA helicases (29). Whereas the insertion mutation had no apparent effect on the level of hDicer mRNA in Dicer mHelicase cells (Supplementary Figure 1C), the level of hDicer protein is reduced in Dicer mHelicase cells compared with wild-type HCT116 cells (Supplementary Figure 1D). Because of the absence of structural data for the hDicer N-terminus, we cannot accurately determine the structural perturbations in the helicase induced by the 43-amino-acid insertion, nor can we rule out the possibility that other Dicer domains are affected by the insertion mutation. Based on empirical evidence presented below, showing the efficient processing of some endogenous pre-miRNAs and ectopic shRNAs in the mHelicase cells, we anticipate that the insertion into the helicase domain does not affect other functional domains of hDicer, such as the PAZ and C-terminal cleavage domains. By homology modeling, using the crystal structure of the Hef helicase from P. furiosus (26) that has sequence similarity with the Dicer helicase domain, we predict that the 43-amino-acid insertion disrupts the Hef DECH box and other motifs involved in ATP hydrolysis by the Hef helicase (Supplementary Figure 1E) (26,29).
Processing and efficacy of miRNAs in wild-type and the Dicer helicase mutant cells
While the expression of many miRNAs is reduced in mHelicase cells, as shown by the marked reduction in mature miR-21 (Figure 1A), a closer examination of the miRNA expression data generated by Cummins et al. (23) indicate that ∼25% of the 100 most highly expressed miRNAs in the wild-type and mHelicase cells did not exhibit significant changes in miRNA expression (data not shown). Our own northern blot analyses for two of these unaffected miRNAs (miR-23a and miR-27a) confirm that the processing and expression of these miRNA is the same in both wild-type and mHelicase cells (Figure 1A). In addition, transient expression of ectopic perfectly complementary shRNA (SII-c21) or lhRNA (SII-c50) targeting the human immunodeficiency virus type 1 (HIV-1) regulatory gene, rev (site II or SII) in wild-type and mHelicase cells had no effect on the expression of miR-23a and miR-27a (Figure 1A). The observation that multiple miRNAs escape downregulation in the mHelicase background suggests that the reduced level of Dicer is not limiting for pre-miRNA cleavage in these cells. Moreover, the fact that the expression of miR-23a and miR-27a remains unchanged in both cell lines that express ectopic shRNA or lhRNA indicates that transient transfection does not have a major impact on miRNA expression or the viability of the mHelicase cells. The expression of miR-21, whose steady-state level is reduced in mHelicase cells, also shows reduced expression following transient transfection of different shRNA or lhRNA vectors (SII-21 and SII-50; Figure 2). To determine whether the reduced production of some miRNAs in mutant cells has a detrimental effect on miRNA target knockdown, we cloned a polycistronic miRNA (miR-106b-93-25) from the MCM7 gene downstream of the CMV promoter and constructed a separate reporter vector that contains a target site perfectly complementary to miR-93 in the 3′ UTR of the Renilla luciferase gene (25). All three miRNAs that originate from this MCM7 cluster are reduced in the Dicer mHelicase cells (23). In wild-type cells, the MCM7 vector reduced miR-93 target gene expression by almost 70% compared with an irrelevant control vector (Figure 1B). In contrast, the efficacy of target knockdown was reduced ∼2-fold in the Dicer helicase mutant cells (Figure 1B). Upon co-transfection of Dicer helicase mutant cells with a plasmid expressing wild-type Dicer cDNA (7), target knockdown was restored to near wild-type levels, showing that the mutation is not dominant (Figure 1B).
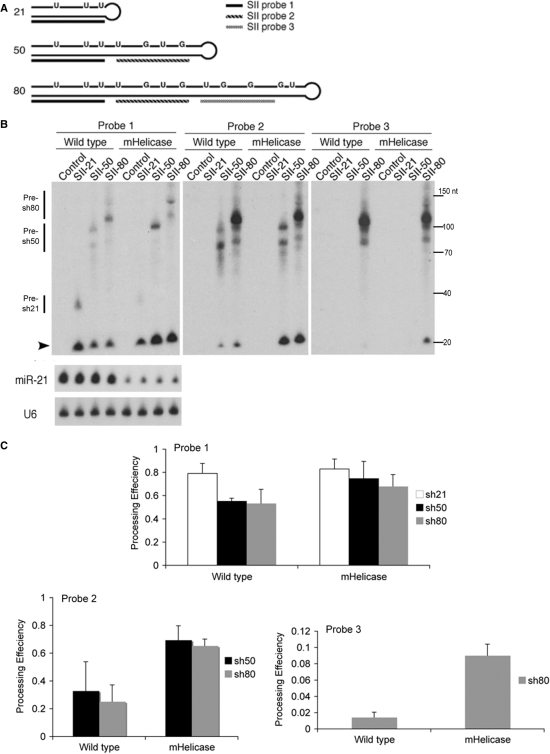
Figure 2.
Expression and efficacy of shRNA or lhRNAs in vivo. (A) Schematic representation of short and long hairpin constructs. Predicted structures of shRNA and lhRNAs, which target the HIV-1 rev exon (site II or SII) and the relative positioning of the oligonucleotide probes for detection of processed siRNAs are shown. The bases shown in the top strand indicate altered nucleotides in the passenger stand of the duplex, which create G:U wobble base parings within the sh- or lhRNA but do not affect the perfect complementarity of the guide strand with its target. (B) Northern analyses of siRNA production in wild-type or mHelicase cells transfected with each hairpin RNA expression vector. Total RNA extracted from cells transfected with the empty vector was used as a negative control for cross-hybridization of each probe. The mature siRNA is indicated with the arrowhead and the respective precursor molecules are indicated by the vertical bars. The membrane was subsequently stripped and probed for miR-21. U6 snRNA expression was also detected as a loading standard. The same membrane was also stripped and probed for siRNAs using Probe 2 and Probe 3. The illustrated size marker is based on the migration of the Ambion Decade ladder. (C) The processing efficiency for each hairpin RNA (Probe 1, Probe 2, Probe 3) was determined by autoradiography and dosimetry as follows: processing efficiency = matureIDV/(matureIDV + precursorIDV). IDV, Integrated Density Value. The mean and standard deviation of three experiments is represented.
Processing and efficacy of the shRNA or lhRNA in wild-type and the Dicer helicase mutant cells
To gain additional insight into the role of the hDicer helicase domain in hairpin processing, we used previously reported plasmids which express a 21-bp shRNA (SII-21), 50-bp lhRNA (SII-50) or 80-bp lhRNA (SII-80) targeted against the human immunodeficiency virus type 1 (HIV-1) regulatory gene, rev (site II or SII), (24). The SII-21 hairpin produces a single siRNA, whereas the SII-50 and SII-80 hairpins are capable of generating two and three individual siRNAs, respectively. These hairpin sequences contain G:U and U:G wobble pairings but do not alter the antisense guide strand of the processed siRNA (Figure 2A). The wobble pairing slightly distorts the stem helix of the hairpin RNA but was shown to facilitate cloning of these long hairpins in E. coli (24). To determine the extent of shRNA or lhRNA processing into ∼21-mer siRNAs, sh- and lhRNA expression plasmids were transfected into wild-type and Dicer helicase mutant cells followed by northern blot analyses using 5′-end labeled 21-nt oligonucleotides complementary to the antisense guide strand of each siRNA that is predicted to derive from Dicer-processing of the hairpins (Figure 2A and B). Reduced accumulation of siRNAs was observed for the SII-21 shRNA containing G:U wobble base-pairing in Dicer helicase mutant cells (Figure 2B), a result that was consistently observed upon repeated transfections, but which did not correlate with the apparent shRNA processing efficiency (Figure 2C, Probe 1). Northern analyses of the siRNAs produced from the SII-50 and SII-80 vectors showed somewhat increased amounts of the first siRNA processed from the lhRNAs in Dicer helicase mutant cells compared to lhRNA processing in the wild type Dicer background (Figure 2B and C, Probe 1). However, the processing efficiency of the second and third siRNAs from these lhRNA vectors was 3–5-fold greater in Dicer helicase mutant cells compared with wild-type cells (Figure 2B and C, Probes 2 and 3). Throughout our experiments we repeatedly observed the inverse accumulation of mature siRNA in wild-type (high-shRNA/low-lhRNA) and Dicer helicase mutant cells (low-shRNA/high-lhRNA) (Figure 2B).
Using two different luciferase-based reporter systems, we measured a ∼2-fold increase in the efficacy of target knockdown for the lhRNA vectors in mHelicase cells compared with target knockdown in wild-type cells. In contrast, target knockdown in response to the SII-21 shRNA vector was reduced by about 2-fold in the mHelicase Dicer background, consistent with the reduced steady-state level of processed siRNA determined by northern analysis (Figure 2B and Supplementary Figure 2). The enhanced processing and greater target knockdown in the mHelicase Dicer background is not specific to the site II HIV lhRNA, as different lhRNA vectors targeting the HIV tat/rev common exon (site I or SI), yielded more siRNA and greater efficacy for each lhRNA in the Dicer helicase mutant cells compared to wild-type cells (Supplementary Figure 3). In addition to the increased steady-state levels of the siRNA guide strand from lhRNA vectors in Dicer mHelicase cells, northern analysis revealed stronger sense strand signals for the lhRNA-derived siRNAs in the mHelicase Dicer background relative to wild-type cells (Supplementary Figure 4). Target knockdown by the lhRNA-derived sense strand was also greater in the Dicer mHelicase cells indicating that lhRNA passenger strand siRNAs are also incorporated into RISC (Supplementary Figure 4). No significant differences in sense-strand accumulation or target knockdown from the SII-21 shRNA were observed between wild-type and Dicer mHelicase cells. Taken together, our results suggest that the Dicer mHelicase protein functions markedly more efficiently than the wild-type enzyme in the processing of lhRNAs, but not shRNAs containing G:U wobble pairings in the stem.
Effects of the Dicer helicase mutation on hairpin RNA processing in vitro
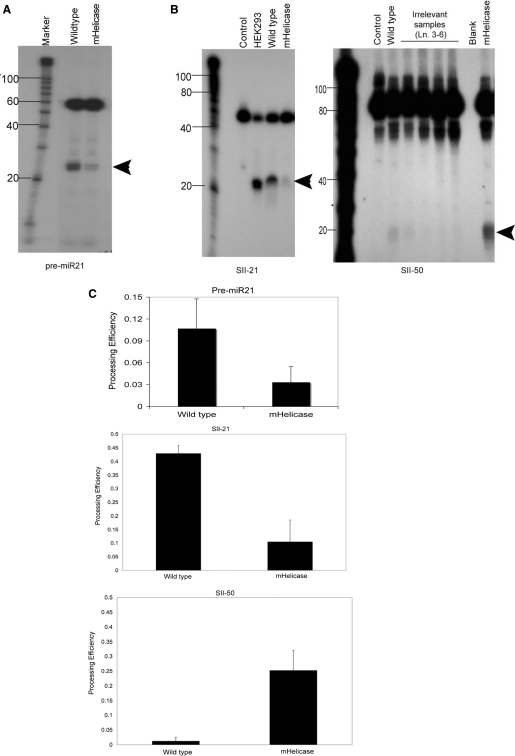
To more carefully investigate the role of the hDicer helicase domain on hairpin RNA processing, we assayed cell extracts prepared from wild-type and Dicer mHelicase cells for their ability to process various hairpin RNA structures in vitro, including a synthetic 60-nt RNA that mimics the pre-miR-21 hairpin RNA, and in vitro transcribed RNAs that derive from templates corresponding to the 21-bp shRNA (SII-21) and a 50-bp hairpin (SII-50). Recombinant hDicer was capable of processing all hairpin substrates into ∼21-nt siRNA products indicating that they are annealed into appropriate hairpin structures (Supplementary Figure 5). Upon incubation with a cell extract prepared from wild-type cells, a ∼21-nt RNA processed product was generated from both the pre-miRNA and SII-21 shRNA substrates (Figure 3A and B). Only a small amount of processed siRNA was observed when the SII-50 lhRNA was incubated with the wild-type cell extract (Figure 3B). A 3-fold decrease in the processing efficiency of the pre-miR 21 substrate was observed upon incubation with the Dicer mHelicase cell extract, a result consistent with the low levels of endogenous mature miR-21 determined by northern analysis (Figure 3C, Pre-miR21). The processing efficiency of the SII-21 shRNA containing G:U wobbles was also reduced by more than 4-fold in the Dicer mHelicase extract (Figure 3C, SII-21). In contrast, the processing efficiency of the SII-50 lhRNA was enhanced by more than 6-fold in the extract from the Dicer mHelicase cells relative to SII-50 processing in the wild-type Dicer extract (Figure 3B and C, SII-50).
Figure 3.
In vitro processing of hairpin RNA substrates. (A) In vitro processing assays were performed using cell extracts from wild-type or Dicer mHelicase HCT116 cells. Cell extracts (25 μg each) from either wild-type or mHelicase cells were incubated with 5′-end-labeled synthetic pre-miR-21 at 37°C for 1 h, run on a 15% denaturing polyacrylamide gel and visualized by autoradiography. The Ambion Decade marker and mature miR-21 (arrowhead) are illustrated. (B) Uniformly radiolabeled RNAs corresponding to the SII-21 shRNA and SII-50 lhRNA were incubated with either the wild-type (25 μg) or mHelicase (25 μg) cell extract at 37°C for 2 h and then electrophoresed in a 10% denaturing polyacrylamide gel and visualized by phosphorimager analysis. The control lane represents substrate RNA that was incubated with reaction buffer but no cell extract. The Ambion Decade marker is illustrated and the arrowheads mark the migration of the processed siRNAs. In the SII-50 gel, lanes 3–6 marked ‘Irrelevant samples’ were performed for a different experiment and are not included in our analysis. The blank lane has no RNA. (C) The processing efficiency for each radiolabeled substrate RNA was determined by phosphorimager analysis as follows: processing efficiency = matureIDV/(matureIDV + precursorIDV). IDV, Integrated Density Value. The mean and standard deviation of three experiments is represented.
Thermodynamic properties of shRNAs determine the necessity of the Dicer helicase domain
To investigate whether structural properties of the shRNA or lhRNA affect processing by hDicer, we constructed SII vectors that express perfectly complementary shRNA (SII-c21) or lhRNAs (SII-c50) by restoring the G:U wobble pairs to their natural base-pairing partners (G:C, A:U) and introduced these vectors into the wild-type and Dicer mHelicase cells for northern blot analysis. As was observed for the G:U wobble-containing lhRNAs, the processing efficiency and target knockdown by both the first and the second siRNAs generated from the SII-c50 lhRNA was more pronounced in the Dicer mHelicase cells compared with wild-type cells, a result consistent with the increased steady-state levels of mature siRNA in the mHelicase Dicer background (Figure 4 and Supplementary Figure 6).
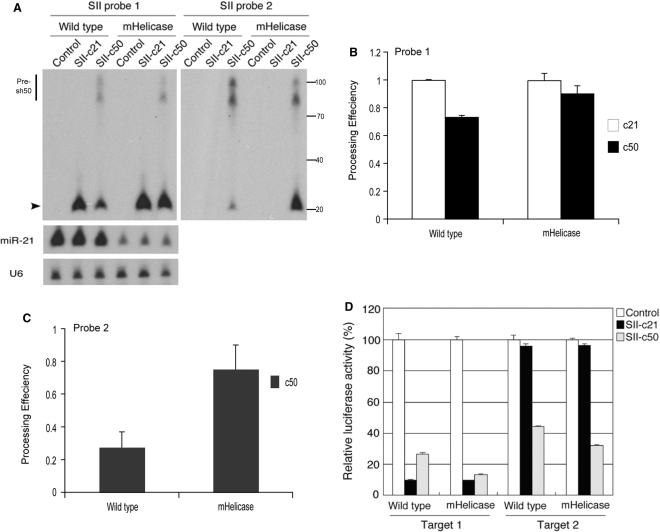
Figure 4.
Expression and efficacy of perfectly complementary shRNA and lhRNAs. (A) Northern analyses of siRNA production in wild-type or mHelicase cells following transfection with perfectly complementary hairpin RNA constructs. Total RNA extracted from cells transfected with the empty vector was used as a negative control for cross-hybridization of each probe. The mature siRNA is indicated with the arrowhead and the precursor molecules are indicated by the vertical bars. The membrane was subsequently stripped and probed for miR-21. U6 snRNA expression was also detected as a loading standard. The same membrane was also stripped and probed for siRNA using Probe 2. The illustrated size marker is based on the migration of the Ambion Decade ladder. (B, C) The processing efficiency for each siRNA [Probe 1 (B) and Probe 2 (C)] was determined by autoradiography and dosimetry as follows: processing efficiency = matureIDV/(matureIDV + precursorIDV) IDV, Integrated Density Value. The mean and standard deviation of three experiments is represented. (D) Inhibition of reporter gene expression by the shRNA or lhRNA constructs. The sequence corresponding to either target 1 or 2 was inserted into the cloning site in the 3′ UTR of the Renilla luciferase gene in the psiCHECK-2 plasmid. One hundred nanograms of each reporter construct and 1 ng of either SII-c21 or SII-50 hairpin RNA expression vector were co-transfected into wild-type or mHelicase cells. Luciferase values were determined 24 h later and the Renilla target was normalized to Firefly luciferase, which is contained on the same reporter construct but is not subject to downregulation by the shRNA. Luciferase values in response to the empty vector (control) were set at 100%. P-values compare mHelicase versus wild type: target 1: SII-c21:P < 0.6; SII-c50:P < 0.001; target 2: SII-c50: P < 0.001.
Similarities in the processing efficiency of the second siRNA from the perfectly paired lhRNA with lhRNAs containing G:U wobble pairs (compare Figure 2C, probe 2 and Figure 4C, probe 2) were observed suggesting that the G:U wobble pairs in the lhRNAs were not specific for the observed increase in processivity. Interestingly, we observed no appreciable differences in the processing efficiency or siRNA accumulation from the SII-c21 vector in wild-type and Dicer mHelicase cells (Figure 4A and B), in contrast to the data obtained with G:U-containing shRNA, which showed reduced siRNA accumulation in vivo in the mHelicase cells. Furthermore, no significant differences in target knockdown were observed in wild-type and Dicer mHelicase cells in response to the SII-c21 shRNA (Figure 4D, SII-c21, P < 0.6). These results are in contrast with the significant reduction in knockdown efficacy by the G:U wobble containing SII-21 shRNA in the Dicer mHelicase background, suggesting that shRNAs with thermodynamically stable stem regions are largely unaffected by the mHelicase insertion into hDicer.
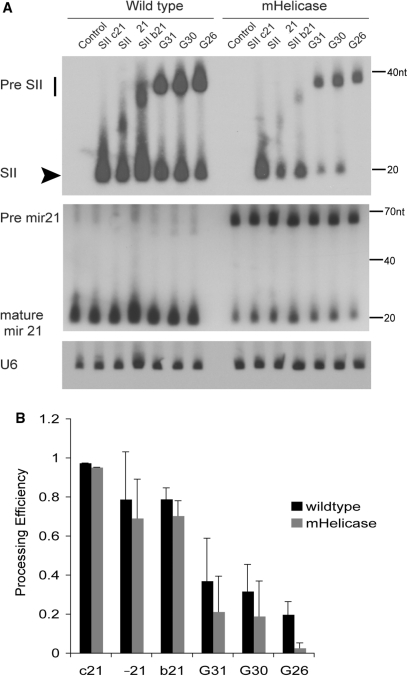
These results suggest that the thermodynamic stability of helical stem region of hairpin RNAs is an important feature for the Dicer helicase mutant phenotype and affects the processing efficiency and siRNA accumulation in the Dicer mHelicase cells. Supporting this hypothesis is our observation that miRNAs with unpaired nucleotides at the end of the stem (positions 1–3) or around the Dicer processing site (positions 18–22) were cloned less frequently in the Dicer helicase mutant cells compared with wild-type cells (Supplementary Figure 7 and Figure 1A) (23). In light of these observations, we sought to carry out a systematic approach to delineate the role of the hDicer helicase domain in processing shRNAs. To do this, we constructed several additional shRNA variants by inserting G:U wobble pairings or creating bulges within the stem of the Sh-c21 shRNA vector and performed northern blot analysis for processed siRNA. Since each shRNA contains different G:U wobble pairs and/or bulges within the stem, the resultant shRNAs have different calculated thermodynamic stabilities (Supplementary Figure 8A). In both cell lines, the processing of SII-21 shRNA variants with lower thermodynamic stabilities was reduced compared with the parental SII-c21 vector (Figure 5). In particular, the processing efficiency of SII-G26, which has a similar predicted thermodynamic stability to the pre-miR-21 hairpin (ΔG = –24.9 kcal/mol), was most affected by the Dicer helicase mutation. The steady-state ratios of the endogenous pre-miR21/mature miR-21 are also increased in the Dicer mHelicase cells (Figure 5A). Even though the processing efficiency of most SII-21 shRNA variants did not differ significantly between wild-type and Dicer mHelicase cells (Figure 5B), the efficacy of target knockdown was significantly reduced in the mHelicase Dicer background for most shRNAs with lower internal stabilities (Supplementary Figure 8B), consistent with the impaired production of siRNAs and suggestive of a defect in guide strand handoff to RISC by the Dicer mHelicase mutant.
Figure 5.
Structural properties of short hairpin precursors affect their processing in Dicer mHelicase cells. (A) Northern blot detection of siRNAs derived from various shRNAs in the wild type or mHelicase cells. Northern blotting analyses were performed as described in Figure 3. The mature siRNA is indicated with the arrowhead and the precursor molecules are indicated by the vertical bar. The membrane was subsequently stripped and probed for miR-21. U6 snRNA expression was also detected as a loading standard. The illustrated size marker is based on the migration of the Ambion Decade ladder. (B) The processing efficiency for each shRNA was determined by autoradiography and dosimetry as follows: processing efficiency = matureIDV/(matureIDV + precursorIDV). IDV, Integrated Density Value. The mean and standard deviation of three experiments is represented.
DISCUSSION
Human Dicer possesses several domains that are required for functional processing of pre-miRNAs as well as longer double-stranded RNAs. Prior to this study and a recent publication by Ma et al. (30) the role of the N-terminal helicase domain in Dicer function was unstudied. We have taken advantage of a human cell line that expresses a mutant form of hDicer that has an in-frame, 43-amino-acid insertion immediately adjacent to the DExH motif in the N-terminal helicase domain. This mutant Dicer enzyme exhibits poor processing of pre-miRNAs and shRNA with thermodynamically unstable stem regions, particularly when normal Watson–Crick base pair interactions are disrupted at the base of the stem (positions 1–3) and close to the Dicer cleavage site (i.e. base-paired between nucleotides 18–22) (Figure 1A and Supplementary Figure 7). By restoring Watson–Crick base pairing in the stem region of an shRNA, the processing efficiency and target knockdown in the mHelicase Dicer background could be restored to levels achieved in wild-type cells. A somewhat surprising characteristic of the Dicer mHelicase is the enhanced processing of lhRNA with stem regions ≥ 50-nt. The processing of endogenous lhRNAs by Dicer in mammals has gained importance with the discovery of long RNAs in mouse oocytes that appear to regulate gene expression through Dicer- and Ago 2-dependent processes (31,32). Taken together, our results provide strong evidence that the helicase domain is required for efficient processing and possibly handoff of the guide strand to RISC when the substrates are thermodynamically unstable duplexes.
Eukaryotes possessing only one Dicer gene that lack the helicase domain, such as Giardia and Schizosaccharomyces Pombe, encounter only perfectly matched duplex or hairpin RNAs and do not encode known miRNAs (31). Given the varied sequence and secondary structures of human endogenous pre-miRNAs and the newly discovered oocyte-restricted lhRNAs (32), it is possible that the N-terminal domain of human Dicer evolved to broaden the array of structured hairpin RNAs that this enzyme can efficiently cleave. Biochemical and crystallographic data for Dicer and other RNAse III enzymes that lack a helicase domain have produced models using linear dsRNA molecules that do not take into account the unidirectional entry of Dicer on substrates containing a loop at one end (1,8,33–36). What is clear from these earlier reports, however, is that significant conformational changes in the protein and RNA occur during cleavage (8,33,35). Therefore, hairpins with slightly different structures due to their sequence composition and thermodynamic stability are likely to require different conformational changes in order for processing to be completed. Our own results, as well as those of other labs, have shown a polarity in the processing of long hairpins in cells resulting in reduced amounts and efficacies of distally produced siRNAs (24,37,38). Our current analysis of long hairpin processing in cells harboring either wild-type or mutant helicase Dicer enzymes provide some insight into these previous results and show the Dicer helicase domain restricts the processivity of the enzyme, thereby limiting the effectiveness of long hairpins for intracellular applications in which multiple siRNAs are desired.
Steric hindrance of helicase domain function by the inserted residues could provide the mutant helicase Dicer its’ unique sh/lhRNA processing properties. It is possible that the inserted amino acid residues disrupt required conformational changes that take place after cleavage to bring together the hDicer C-terminal domain, which mediates dsRNA binding and cleavage, with the helicase-containing N-terminus of hDicer. Such an interaction could promote the release of the newly cleaved siRNA duplex. This interaction between the C- and N-termini of wild-type hDicer is expected to prevent the Dicer C-terminal domain from immediately initiating another round of cleavage resulting in poor processing of lhRNA substrates which require more than one Dicer cleavage event. In the absence of a functional Dicer helicase domain, however, the C-terminus of the mutant Dicer enzyme is capable of multiple rounds of hairpin cleavage on long hairpin substrates. In support of this possible mechanism, the recent studies of Ma et al. (30) demonstrate that the major consequence of eliminating the helicase domain from human dicer is a substantial increase in the kcat for cleavage of a perfectly matched 37-nt linear duplex RNA. They suggest that the helicase domain is involved in a rearrangement of the enzyme following catalysis, which restricts the enzyme's ability to reinitiate cleavage on the long substrates. In the absence of a functional helicase domain this rearrangement does not occur, thereby allowing the enzyme to rapidly reinitiate cleavage on a long substrate.
In contrast to the enhanced cleavage processivity of the mHelicase on long hairpins, short hairpin and pre-miRNA substrates that harbor mismatches in the stem region are cleaved poorly in our system. It is possible that that the complete processing of pre-miRNAs with poor thermodynamic stability properties by Dicer may require interaction with an accessory protein, such as TRBP or PACT. This seems unlikely though since human Dicer can effectively recognize and cleave pre-miRNAs and other hairpin RNA substrates in vitro in the absence of accessory proteins (Supplementary Figure 5). Perhaps a more reasonable explanation for our observations, as well as those of Ma et al. (30), is that once cleavage of an imperfect stem region takes place, the helicase domain helps mediate a conformational rearrangement of the hDicer enzyme to release the guide strand for hand-off to RISC. In contrast, perfectly matched helices in shRNAs are directly handed off to Ago 2 wherein the passenger strand is cleaved to create an active RISC (39). Thus, a plausible model for the role of the helicase domain in the human Dicer enzyme is to facilitate product release from substrates with imperfect helices. This may take place via interactions with accessory proteins such as TRBP or PACT or through interactions between N- and C-terminal domains of the same Dicer molecule. Further structural analyses of the Dicer helicase domain should facilitate a better understanding of why the helicase domain is required for unstable substrate processing and hand-off but is not required for thermodynamically stable duplex processing and handoff.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (HL07470, AI29329 to J.J.R.); a Beckman Fellowship from the Arnold and Mabel Beckman Foundation (to H.S.S.). Funding for open access charges: NIH grants HL07470 and AI29329 to JJR
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Bert Vogelstein and Jordan Cummins for the generous gifts of the HCT116 wild-type and Dicer helicase mutant cell lines and the NIH AIDS Research & Reference Reagent Program for providing a pNL4-3.Luc plasmid. Dr. Patrick Provost generously provided the plasmid expressing the wild-type human Dicer cDNA.
REFERENCES
- 1.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 2.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 3.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson RH, Nicholson AW. Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm. Genome. 2002;13:67–73. doi: 10.1007/s00335-001-2119-6. [DOI] [PubMed] [Google Scholar]
- 7.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 10.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lund E, Dahlberg E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 12.Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J. Mol. Biol. 2007;374:106–120. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 14.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc. Natl Acad. Sci. USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 22.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz L.A., Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sano M, Li H, Nakanishi M, Rossi JJ. Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: advantages and limitations. Mol. Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- 25.Aagaard L, Amarzguioui M, Sun G, Santos LC, Ehsani A, Prydz H, Rossi JJ. A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol. Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- 26.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Sali A, Blundell TL. Comparative protein modeling by satisfication of spatial constraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Ma E, Macrae IJ, Kirsch JF, Doudna JA. Autoinhibition of human Dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 33.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 34.Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb. Symp. Quant. Biol. 2006;71:73–80. doi: 10.1101/sqb.2006.71.042. [DOI] [PubMed] [Google Scholar]
- 35.Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol. Microbiol. 2008;67:143–154. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- 36.Ji X. Structural basis for non-catalytic and catalytic activities of ribonuclease III. Acta Crystallogr. D Biol. Crystallogr. 2006;62:933–940. doi: 10.1107/S090744490601153X. [DOI] [PubMed] [Google Scholar]
- 37.Konstantinova P, de Vries W, Haasnoot J, ter Brake O, de Haan P, Berkhout B. Inhibition of human immunodeficiency virus type 1 by RNA interference using long-hairpin RNA. Gene Ther. 2006;13:1403–1413. doi: 10.1038/sj.gt.3302786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15(3):534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 39.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.