Abstract
Rad9 is conserved from yeast to humans and plays roles in DNA repair (homologous recombination repair, and base-pair excision repair) and cell cycle checkpoint controls. It has not previously been reported whether Rad9 is involved in DNA mismatch repair (MMR). In this study, we have demonstrated that both human and mouse Rad9 interacts physically with the MMR protein MLH1. Disruption of the interaction by a single-point mutation in Rad9 leads to significantly reduced MMR activity. This disruption does not affect S/M checkpoint control and the first round of G2/M checkpoint control, nor does it alter cell sensitivity to UV light, gamma rays or hydroxyurea. Our data indicate that Rad9 is an important factor in MMR and carries out its MMR function specifically through interaction with MLH1.
INTRODUCTION
Errors and damage of genomic DNA occur during normal cellular activities or when cells are exposed to genotoxins. Cells manage DNA errors and damage using an intricate network consisting of many proteins including cell cycle checkpoint proteins and DNA repair factors. The main DNA repair mechanisms include homologous recombination (HR), nonhomologous end-joining, base excision, nucleotide excision, direct reversal and mismatch repair (MMR) (1,2). When DNA is damaged, the progress of cells through the cell cycle is arrested or slowed down to allow time for DNA repair (2,3). Cell cycle checkpoint proteins Rad9, Rad1 and Hus1 play important roles in both cell cycle checkpoint control and DNA repair (2,4,5). These three proteins are evolutionarily conserved in eukaryotes, and can form a ring-shaped heterotrimer, dubbed the 9-1-1 complex (6–11). The deletion of each of the three genes for these proteins in the fission yeast Schizosaccharomyces pombe inactivates S/M, intra-S and G2/M checkpoint controls, and sensitizes fission yeast cells to killing by UV light, γ-rays and the replication inhibitor hydroxyurea (HU) (12–15). Disruption of the mouse ortholog of Rad9 or Hus1 also sensitizes mouse cells to UV light, γ-rays and HU, and leads to genome instability (16,17).
MMR is an important repair mechanism that maintains genomic stability. It plays key roles in the repair of base–base mismatches and insertion/deletion mispairs generated during DNA replication and recombination. MMR also functions in preventing HR and in DNA damage signaling in eukaryotic cells (18–21). In vitro reconstitution biochemical studies indicate that MutS, MutL, RPA, EXOI, HMGB1, PCNA, replication factor C (RFC), polymerase and DNA ligase I play important roles in MMR (22–24). EXOI is only partially responsible for excision enzyme activity in MMR (25,26). A recent study showed that regulatory factor X has a stimulatory role in mismatch-dependent 5′ to 3′ excision activity (27). hMRE11 was found to play a role in mismatch-dependent 3′ to 5′ excision (28). Furthermore, the excision of reconstituted MMR proteins is less specific than in cell extracts, suggesting that one or more additional regulatory factors are required for the accuracy of the excision step (22,29). Therefore, other protein components are required for MMR to work efficiently and accurately under various in vivo conditions.
In this study, Rad9 was found to physically interact with mammalian MLH1, a protein essential for DNA MMR. A single amino acid residue mutation (S160A) on Rad9 drastically weakened its interaction with MLH1 and cellular mismatch repair activity. The mutation did not affect cell sensitivity to UV light, gamma rays or HU, and neither S/M or G2/M checkpoint controls, the typical phenotypes of Rad9-deleted cells (17), suggesting that Rad9 functions in MMR specifically through its interaction with MLH1.
MATERIALS AND METHODS
Antibodies
Anti-hRad9 polyclonal antibody was obtained by immunizing mice with purified MBP-hRad9 protein. Anti-hMLH1 monoclonal antibody (554 073) was from BD Biosciences Pharmingen, monoclonal anti-His antibody, monoclonal anti-FLAG M2 antibody (F1804), polyclonal anti-FLAG antibody (F7425) and anti-γ-tubulin monoclonal antibody were obtained from Sigma–Aldrich, and anti-HA antibody was obtained from Santa Cruz Technology.
Cell culture
Human HeLa and HEK 293T cells were cultured in DMEM (Invitrogen, CA) supplemented with 10% fetal bovine serum (Hyclone) and 100 U/ml penicillin/streptomycin. The 293T cells stably expressing FLAG-hRad9 constructed in our laboratory were cultured in DMEM with 10% FBS and 50 μg/ml Hygromycin B to keep the plasmid pFLAG-CMV2-hRad9 in the cells.
Methyl methanesulfonate (MMS) and N-nitroso-N-methylurea (MNU), the alkylation agents for cell treatments were purchased from Sigma–Aldrich (St. Louis, MO). After HeLa cells reached 80% confluence and were rinsed twice with PBS, the cells were incubated with designated concentrations of MMS or MNU for 60 min in serum-free DMEM and cells were then harvested to prepare lysate for Western blot and co-immunoprecipitation analysis.
Culture of mouse ES cells was described previously (17). The antibiotic zeocin, at a final concentration of 100 μg/ml, was added to cell cultures to select stable transfectants. The selected stable transfectants were cultured in medium containing 25 μg/ml zeocin to maintain the transfected genes in the cells.
Mass spectrometry
FLAG-hRad9 was immmunopurified from 1 × 108 293 T cells stably expressing FLAG-hRad9 using 200 μl anti-FLAG M2 agarose (Sigma). After extensive washings with lysis buffer (150 mM NaCl, 50 mM Tris–HCl, pH 7.5, 10% glycerol, 0.5% NP-40) containing a protease inhibitor cocktail (Roche, Indianapolis, IN), the bound proteins were eluted with 200 μg/ml FLAG peptide (Sigma) in PBS. Eluted proteins were resolved by SDS–PAGE and revealed by Coomassie blue staining or silver staining. The apparent bands were excised, in-gel digested with trypsin, and analyzed by LC-MS/MS on a ProteomeX-LTQ mass spectrometer (Thermo Fisher Scientific, Waltham, USA) to identify the protein bands.
Co-immunoprecipitation
Two vectors, pCDNA3-6HA (30) and pFLAG-CMV2 (Sigma, St Louis, MO), were used to construct plasmids capable of expressing hMLH1 in human cells. To make pCDNA3-6HA-hMLH1, pFLAG-CMV2-hMLH1, the cDNA of the hMLH1 gene was amplified by PCR from the plasmid pEGFP-hMLH1 (a kind gift from Dr. Lene Juel Rasmussen, Roskilde University, Denmark). The sequences of forward and reverse primers for these PCR reactions are given in Table 1. The hMLH1 gene was cloned between the EcoRI and BamHI sites of pFLAG-CMV2, and in the EcoRI site of pCDNA3-6HA. The pCDNA3-6HA and pFLAG-CMV2 constructs bearing the Hus1, Rad1 or Rad9 open reading frame were constructed previously (30).
Table 1.
Primers used to amplify cDNA for constructing plasmids
| Plasmids | Primer | Restriction sites |
|---|---|---|
| pFLAG-CMV2-hMLH1 | 5′-ATACGCGAATTCATCGTTCGTGGCAGGGGTTAT-3′ | EcoRI |
| 5′-ATACGCGGATCCTTAACACCTCTCAAAGACTT-3′ | BamHI | |
| pCDNA3-6HA-hMLH1 | 5′-ATACGCGAATTCTCGTTCGTGGCAGGGGT-3′ | EcoRI |
| 5′-ATACGCGAATTCTTAACACCTCTCAAAGAC-3′ | EcoRI | |
| pCDNA3-6HA-hRad9 | 5′-ATACCCCTCGAGAAGTGCCTGGTCACGGGCGG-3′ | XhoI |
| (1-270) | 5′GCGCGCGGATCCCTACGAGTCGGTGTCTGAGAGTG-3′ | XhoI |
| pCDNA3-6HA-hRad9 | 5′-ATACCCGAATTCCACTCCCAGGACCTGGGCTC-3′ | EcoRI |
| (271-391) | 5′-GCGCGCGGATCCTCAGCCTTCACCCTCACTGT-3′ | EcoRI |
| pGEX-6P-1-hRad9 | 5′-ATACCCCTCGAGAAGTGCCTGGTCACGGGCGG-3′ | XhoI |
| (1-130) | 5′-ATACCCCTCGAGCTCACAGTCCTGGAAGGACA-3′ | XhoI |
| pGEX-6P-1-hRad9 | 5′-ATACCCCTCGAGTCCCTGCAGGCCGTCTTCGA-3′ | XhoI |
| (131-270) | 5′-ATACCCCTCGAGCTAGTGCGAGTCGGTGTCTGAGA-3′ | XhoI |
| pGEX-6P-1-hRad9 | 5′-ATTCGGCTCGAGCCTGCACTGGCTG-3′ | XhoI |
| (161-270) | 5′-ATACCCCTCGAGCTAGTGCGAGTCGGTGTCTGAGA-3′ | XhoI |
| R9160A-F | 5′-GCTGTTCTGCCCTTCGCTCCTGCACTGGCTG-3′ | |
| R9160A-R | 5′-CAGCCAGTGCAGGAGCGAAGGGCAGAACAGC-3′ | |
| PET24a (+)-mMLH1 | 5′-ATTCGCGGATCCATGGCGTTTGTAGCAGGAGT-3′ | BamHI |
| 5′-ATACGC GAATTCTTAACACCGCTCAAAGACTT-3′ | EcoRI |
Human embryonic kidney 293T cells were transfected with the plasmids of interest using Lipofectamine plus (Invitrogen, USA). Cells were grown to 60–80% confluence in 60 mm tissue culture dishes and transfected with 1.5 μg pFLAG-CMV2 DNA and 1.5 μg pCDNA3-6HA DNA following the procedure described by Invitrogen. Immunoprecipitation and Western blotting were performed as described previously (31).
Construction of the hRad9 S160A mutant
pGEX-6P-1-hRad9 (S160A) was derived from pGEX-6P-1-hRad9 following the QuickChange mutagenesis procedure (Stratagene). The primers for the mutagenesis PCR of S160A were R9160A-F and R9160A-R in Table 1. The mutant was confirmed by DNA sequencing. The plasmid of pZeoSV2- hRad9 (S160A) was constructed by cutting from pGEX-6P-1-hRad9 (S160A) and inserting into the XhoI site of pZeoSV2 (Invitrogen, USA).
In vitro pull-down tests
The full-length hRad9 cDNA removed from pCDNA3-6HA-hRad9 using XhoI was inserted into the XhoI site of pGEX-6P-1 (GE Health) to make pGEX-6P-1-hRad9. The hMLH1 open reading frame was cut from pCDNA3-6HA-hMLH1 using EcoRI and inserted into the EcoRI site of pET24a (+) (Novagen) to make pET24a (+)-hMLH1, and inserted in pGEX-6P-1 to make pGEX-6P-1-hMLH1. The truncated hRad9 and hMLH1 constructs were amplified by PCR using template pGEX-6P-1-hRad9. The primers are listed in Table 1. The PCR products of hRad9 were digested with XhoI and ligated with XhoI-digested pGEX-6P-1 to yield pGEX-6P-1-hRad9 plasmids containing hRad9 (1–130), hRad9 (131–270) and hRad9 (161–270). The pGEX-6P-1-hRad9 (1–270) and pGEX-6P-1-hRad9 (271–391) plasmids were derived from pCDNA3-6HA-hRad9 (1–270) and pCDNA3-6HA-hRad9 (271–391) in the same way as described above. Cloned sequences were confirmed by DNA sequencing. Mouse MLH1 cDNA was amplified from mouse ES cell cDNA and inserted into BamHI and EcoRI sites of pET24a (+) to make pET24a (+)-mMLH1.
For the expression of GST tagged hRad9 fragments and His-hMLH1, Rosetta cells (Invitrogen) harboring expression plasmids were cultured in LB broth containing 100 μg/ml of ampicillin for GST tagged proteins or 50 μg/ml of kanamycin for His-hMLH1 at 37°C. Protein expression was induced at an A600 of 0.6 by the addition of isopropytlthiogalactoside (IPTG) to a final concentration of 0.4 mM and cells were grown at 16°C for 24 h and then harvested. The cell paste, from a 500-ml culture, was resuspended in 10 ml PBS containing 1 mM PMSF at 4°C. After sonication, the solution was centrifuged at 16 000 g for 30 min and the supernatant was saved.
For purification of the GST tagged protein, 1 ml of a 50% slurry of glutathione-Sepharose 4B (GE Healthcare) in PBS buffer with 1% Triton X-100 was added to 10 ml supernatant and incubated overnight at 4°C. The GST-tagged proteins bound to the beads were pelleted at 500 g for 5 min and washed three times with 1 ml PBS buffer containing 1% Triton X-100 and then eluted with 10 mM reduced L-glutathione in 50 mM Tris–HCl, pH8.0.
To purify His-hMLH1, the supernatant was added to a 50% slurry of Ni Sepharose High Performance (Amersham Biosciences) and eluted in a stepwise manner with 1.6 ml (0.4 ml × 4 times each) of 10, 25, 50, 100 and 250 mM imidazole in 50 mM Na-phosphate buffer, pH 7.5.
His-tagged hMLH1 expressed in Rosetta cells was added to the appropriate GST-tagged proteins (100 ng) immobilized on glutathione-Sepharose 4B and incubated for 1 h in 500 μl binding buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, 1% Triton X-100, 0.5 mM DTT, 1% BSA) at 4°C. After centrifugation at 500 g for 5 min, the pellets were washed five times with 500 μl of binding buffer at 4°C and fractionated on a 10% SDS–polyacrylamide gel. Western blots for hMLH1 were probed with anti-hMLH1 antibody (BD Biosciences Pharmingen, Franklin Lake, NJ). A control of immobilized GST alone was run concurrently.
Yeast two-hybrid analysis
The matchmaker LexA two-hybrid system from Clontech was used to determine the interaction of hMLH1 and hRad9. pLexA and pB42AD plasmids containing the hRad9 sequence were constructed previously (30). The full-length coding sequence of hMLH1 was amplified by PCR from pACJ112 (a gift from Dr Lene Juel Rasmussen, Roskilde University, Denmark). The PCR product was digested with EcoRI and inserted into the EcoRI sites of pLexA and pB42AD. Yeast two-hybrid analyses were performed using a previously published procedure (30).
Tests for MMR activity
A method developed by Lei et al. (32) to detect MMR activity in living cells was used with minor modifications. The DNA substrates, homoduplex and heteroduplex EGFP plasmids, were prepared in the same way as Lei et al. (32). Lipofectamine reagents (Invitrogen) were used to transfect DNA into cultured cells. Cells were plated on 60-mm dishes at a density of 5 × 105 cells/dish. Transfection was performed the following day according to the manufacturer's protocols. Homoduplex or heteroduplex EGFP plasmid measuring 0.2 μg was co-transfected with 0.2 μg red fluorescent protein (RFP) expression plasmid pmRFP-C1 (a kind gift from Dr. Liusheng He, St Jude Children's Research Hospital, Memphis, TN, USA). pmRFP-C1 was used to ascertain that the transfection efficiencies of the EGFP plasmids were comparable. The cells were trypsinized 20 h after transfection, and resuspended in PBS at a concentration of 5 × 105 cells/ml. 30 000 cells were measured for their fluorescence intensities at wavelengths of 530/30 nm (green) and above 670 nm (red) using a FACSCalibur flow cytometer (Becton Dickinson). MMR activity was calculated using a formula developed by Lei et al. (32). HCT116 cells do not express functional hMLH1 and therefore have an impaired MMR system. HCT116 cells transfected with pFLAG-CMV2 and pFLAG-CMV2 -hMLH1 were used as negative and positive MMR controls. Mouse ES cells were transfected in the same way as described above for HCT116 cells except for the use of a specialized medium for culture of ES cells (17).
ES cell survival and cell cycle checkpoint assays
Mouse ES cells were seeded at various concentrations in duplicate on gelatinized tissue culture dishes. Sensitivities to 250 μM HU, 254-nm UV light at 10 J/m2 and 60Co gamma rays at 6 Gy were tested as described previously (17). Survival against MMS and MNU were evaluated by counting colonies formed with and without mutagen treatment. Cells were incubated for 24 h after seeding in duplicated 60-mm dishes and then exposed to various concentrations of MMS or MNU for 1 h at 37°C in an incubator. The medium was removed, the cells were rinsed once with PBS, and fresh medium was added. Colonies formed after 1 week of growth were stained with Giemsa and counted. Relative colony formation (%) was expressed as colonies per treatment level/colonies that appeared in the control.
The assay for G2/M checkpoint control of mouse ES cells induced by gamma rays was conducted as described in a previous study (17). A previously developed method was utilized to evaluate S/M checkpoint function (33). Briefly, 1 mM HU was added to ES cells when they were grown to 70% confluence. Cells were incubated at 37°C under 5% CO2 for designated times, then processed and suspended in PBS. The cells were probed with rabbit anti-phospho-histone H3 (Upstate)/FITC-conjugated anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, INC), and stained with PI followed by flow cytometric analysis.
RESULTS
hRad9 interacts physically with hMLH1
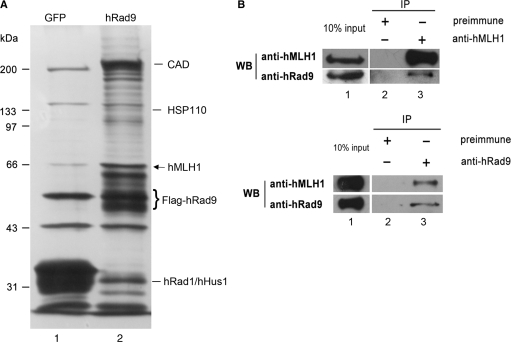
Lindsey-Boltz et al. (34) reported that when FLAG tagged hRad9 expressed in HEK293T cells was purified with anti-FLAG agarose, carbamoyl phosphate synthetase/aspartate transcarbamoylase/dihydroorotase (CAD) and HSP110 were copurified with hRad9. Since mammalian Rad9 plays critical roles in multiple formats of DNA repair and cell cycle checkpoint control, Rad9 is very likely to interact with other proteins that are important for DNA repair and cell cycle checkpoint control. Therefore, we performed a similar but more careful immunoprecipitation assay to search for human proteins capable of interacting with hRad9. To enhance the sensitivity and reduce background noise, we established HEK293T cell clones stably expressing FLAG-tagged hRad9 and picked a clone that expressed FLAG-hRad9 at a level lower than the endogenous hRad9 level (data not shown). The affinity-purified proteins associated with FLAG-hRad9 were fractioned by SDS–PAGE and revealed by silver staining (Figure 1A). The bands of interest which were not seen in the affinity-purified extracts from control HEK293T cells stably expressing FLAG-GFP, were cut out, digested and analyzed with LC-MS/MS. LC-MS/MS analysis of hRad9-copurified bands revealed that besides CAD and HSP110, there were other proteins not previously reported to interact with hRad9. As expected, hHus1 and hRad1 were also purified together with hRad9. One of these proteins was hMLH1, a protein essential for DNA MMR. hRad9 has been reported to interact with several DNA repair proteins involved in HR repair and base-excision repair (35–45), but none of the proteins involved in MMR have previously been found to associate with Rad9. Therefore, we chose to characterize this interaction and its functional significance further.
Figure 1.
hRad9 associates with hMLH1. (A) Silver-staining of hRad9-associated proteins. 293T cells stably expressing FLAG-hRad9 were immunoprecipitated with antibody against FLAG M2. Samples were resolved by 10% SDS–PAGE and visualized by silver staining (lane 2). Lane 1 is a negative control in which the immunoprecipitation was performed in 293T cells stably expressing FLAG-GFP. The bands of interest were cut and analyzed by a Mass-Spectrometer, hMLH1 (arrow) was identified to be one of the partners of hRad9. The protein bands of CAD, HSP110, hRad9, hHus1 and hRad1 are also indicated. (B) Co-immunoprecipitation of hRad9 and hMLH1. In the upper panel, endogenous hRad9 was immunoprecipitated with an anti-hMLH1 antibody from cell extracts of 106 HeLa cells, but not with pre-immune antiserum (lane 2). The immunoprecipitated proteins were detected with antibodies against hMLH1 (upper) and hRad9 (lower). Ten percent of the lysate was used for the loading control and the remaining 90% for co-immunoprecipitation. In the lower panel, endogenous hMLH1 was immunoprecipitated from cell extracts of 106 HeLa cells with an anti-hRad9 antibody (lane 3), but not with pre-immune serum (lane 2). The immunoprecipitated proteins were visualized by western blot analysis with antibodies against hMLH1 (upper) and hRad9 (lower).
To confirm the association of hMLH1 and hRad9 in vivo, co-immunoprecipitation was performed using an anti-hMLH1 monoclonal antibody or anti-hRad9 polyclonal antibodies. hRad9 and hMLH1 were immunoprecipitated together from the extracts of HeLa cells by either the anti-hMLH1 antibody or the anti-hRad9 antibodies, but not by pre-immune serum (Figure 1B). This result indicates that hRad9 and hMLH1 are associated together in cells.
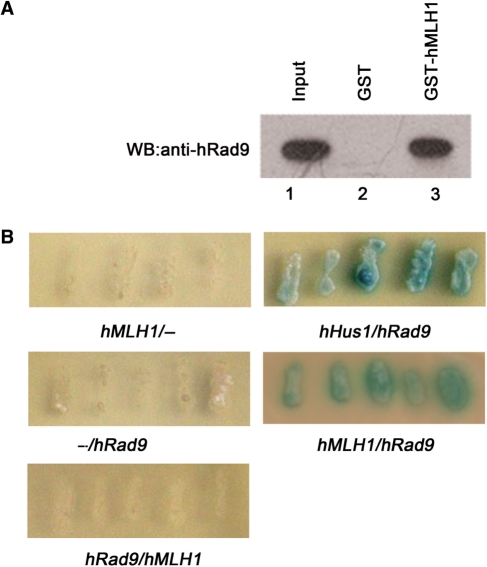
We next determined whether the interaction between hRad9 and hMLH1 was direct, rather than being mediated via accessory proteins. GST pull-down was performed to test whether purified recombinant hRad9 and hMLH1 could interact directly in vitro. GST-hMLH1 fusion protein bound to glutathione-Sepharose was incubated with purified His-hRad9 protein. As shown in Figure 2A, hRad9 could be pulled down by GST-hMLH1 (lane 3), while hRad9 did not bind to GST alone (lane 2). Furthermore, a yeast two-hybridization assay was also performed to assess the interaction of hRad9 and hMLH1. Co-expression of pLexA-hMLH1 and pB42AD-hRad9 supported yeast growth and turned colorless X-gal into blue product (Figure 2B), while the yeast cells containing pLexA-hMLH1 and pB42AD or pLexA and pB42AD-hRad9 did not grow and remained colorless. As a positive control, yeast cells bearing pLexA-hHus1 and pB42AD-hRad9 grew and turned blue as expected (30). These results again support the idea that hRad9 interacts with hMLH1 directly. It is noteworthy that the yeast bearing pLexA-hRad9 and pB42AD-hMLH1 did not grow and remained colorless, suggesting that hRad9 and hMLH1 interact physically only when the protein contact is in the right direction.
Figure 2.
hRad9 interacts with hMLH1 directly. (A) GST pull-down assay. One microgram GST-hMLH1 (lane 3) or an equal amount of GST alone (lane 2) was immobilized on glutathione-Sepharose and incubated with purified His-hRad9 (100 ng) in binding buffer for 2 h at 4°C. The Sepharose was washed five times with binding buffer. The purified proteins were subjected to western blot analysis and probed with the hRad9 antibody. Lane 1 contains 10 ng (10% of the total input) of His-hRad9. Unlike GST (lane 2), GST-hMLH1 (lane 3) was able to pull down His-hRad9. (B) Interactions of hRad9 and hMLH1 in a yeast two-hybrid assay. The pairs of two-hybrid recombinant proteins tested in each population of Saccharomyces cerevisiae EGY48 host cells were arranged as follows: (left) pLexA-gene 1/(right) pB42AD-gene 2. BD-hMLH1 was able to interact specifically with AD-hRad9.
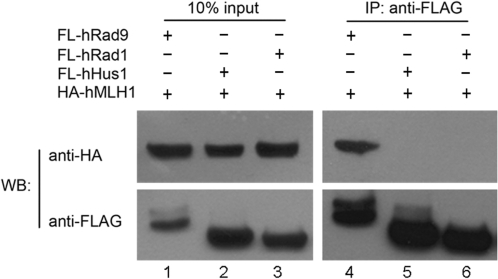
hMLH1 does not interact with hRad1 and hHus1
Given that hMLH1 interacts with hRad9, we asked whether hMLH1 interacted with hRad1 and hHus1, the two other proteins that form a heterotrimer (the 9-1-1 complex) with hRad9 (7–11,30,46–48). A co-immunoprecipitation assay showed that overexpressed HA-tagged hRad1 and hHus1 did not interact with FLAG-tagged hMLH1, while as a positive control, HA-hRad9 was found to interact with FLAG-hMLH1 (Figure 3). These findings suggest that hMLH1 binds to free hRad9 but not to hRad9 within the 9-1-1 complex.
Figure 3.
hMLH1 is not associated with hRad1 or hHus1. pCDNA3.0-6HA-hMLH1 (1.5 μg) was transfected into 293T cells along with 1.5 μg pFLAG-CMV2-hRad1 (lanes 1 and 4), 1.5 μg pFLAG-CMV2-hRad9 (lanes 2 and 5) and 1.5 μg pFLAG-CMV2-hHus1 (lanes 3 and 6), respectively. Ten percent of the lysate was used for the loading control and the remaining 90% for co-immunoprecipitation. The lysate was immunoprecipitated with anti-FLAG antibody, and the western blot membrane was probed with an antibody against HA (upper) and against FLAG (lower). The anti-FLAG antibody was able to precipitate HA-hMLH1 mediated only by FLAG-hRad9.
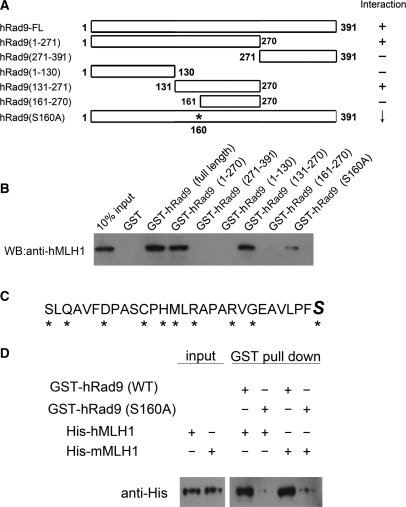
Mapping hRad9 domains required for its interaction with hMLH1
To investigate which region of the hRad9 protein was responsible for mediating the interaction with hMLH1, a series of hRad9 deletion mutants were generated and tested for their abilities to interact with full-length hMLH1 in a GST pull-down assay, in which GST-tagged fragments of hRad9 were immobilized on glutathione-Sepharose and incubated with His-hMLH1, and as a negative control, GST alone with His-hMLH1 was included. Experimental results show that the hMLH1-interacting region of hRad9 is localized to residues 131–160 (Figure 4A and B). To narrow down which of these amino acid residues was important for the interaction of hRad9 with hMLH1, each of the polar amino acids on hRad9 in this region was mutated into alanine residues separately, and the mutants were subjected to a GST pull-down assay with His-hMLH1 (Figure 4C). We chose to only mutate polar residues to avoid disrupting the integrity of protein domains in hRad9. The results from the GST pull-down assay indicate that S160 of hRad9 (marked with a star in Figure 4A) is important for the interaction, and all the other polar amino acid residues in the 131–160 region have no or only very minor impact on the hRad9–hMLH1 interaction (data not shown). In one of the following sections, we used mRad9–/– mouse ES cells to test the functional importance of hRad9 in MMR, thus we also examined the interaction between human hRad9 and mouse mMLH1. As with hMLH1, mouse mMLH1 interacted directly with wild type hRad9, but not with mutant S160A hRad9 (Figure 4D), suggesting conservation of Rad9–MLH1 interaction from mouse to human.
Figure 4.
Mapping of the hRad9 domain and amino acid residues that interact with hMLH1. (A) The map schematically illustrates the regions and amino acid residues of hRad9 that interact with hMLH1. The results are derived from the experimental data in Figure 4B–D. The intact hRad9 contains 391 amino acid residues. The point mutation is marked with a star. ‘+’ stands for positive, ‘—’for negative and ‘↓’ for reduced interactions with hMLH1 or mMLH1. (B) Mapping the hRad9 region that interacted with His-hMLH1. Immobilized GST-hRad9, GST-hRad9 (1–270), GST-hRad9 (271–390), GST-hRad9 (1–130), GST-hRad9 (131–270), GST-hRad9 (161–270) and GST-hRad9 (S160A) (1 μg in each case) were incubated with 100 ng purified His-hMLH1 and the western blot was probed with anti-hMLH1 antibody. A region between 131 and 160 was shown to be essential for hRad9–hMLH1 interaction, and Ser160 was shown to be critical for the interaction. (C) Graphic depiction of the polar amino acid residues in the 131–160 region of hRad9 that were mutated into alanine residues. Mutated residues are marked with stars. The italic and large sized ‘S’ is used to label the Ser160 that is critical for the hRad9-hMLH1 interaction. (D) The hRad9 S160A point mutation also significantly reduced the interaction of human hRad9 with mouse mMLH1. The experimental procedure was the same as in Figure 4B.
DNA mismatch-damage stimulates the hRad9–hMLH1 interaction
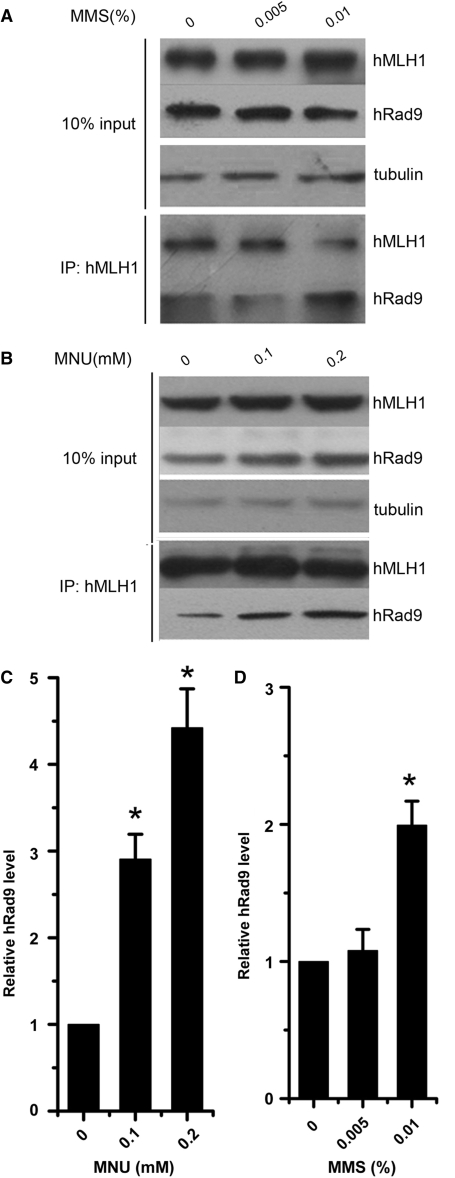
N-nitroso-N-methylurea (MNU) is an SN1 DNA-alkylating agent which produces high levels of O6-methylguanine (O6-meG) on DNA in cells. The resulting O6-meG lesions are recognized and/or repaired by MMR proteins (49). Methyl methanesulfonate (MMS), a SN2 DNA-alkylating-agent, produces high levels of N7-methylguanine (N7-meG) and N3-methyladenine (N3-meA), which can lead to the formation of promutagenic abasic sites. MMR pathways also play a role in the repair of MMS-induced DNA lesions (50,51). To find out whether the activation of DNA MMR influences the interaction between hRad9 and hMLH1, we tested whether increases in cellular DNA lesions incurred by incubating HeLa cells with MNU and MMS would enhance hRad9–hMLH1 interactions. Indeed, the interaction between hRad9 and hMLH1 was enhanced by either MNU or MMS treatment (Figure 5). The protein loading control using 10% lysate indicates that hRad9 and hMLH1 did not change within the experimental period, and thus the higher amount of hRad9 copurified with hMLH1 was due to higher affinity in the presence of MMS or MNU. This enhancement of hMLH1-hRad9 interaction in response to MMS or MNU treatment implies that the hMLH1–hRad9 interaction is an important step in MMR.
Figure 5.
Interaction between hRad9 and hMLH1 is enhanced in response to MMS or MNU treatment. (A) hRad9 displays increased affinity to hMLH1 after MMS treatment. HeLa cells were mock-treated or treated with 0.005 and 0.01% MMS (g/solution volume) for 1 h, and the lysate was used for protein concentration controls (upper panel) and immunoprecipitation by anti-hMLH1 antibody (lower panel). (B) MNU treatment induced the interaction between hRad9 and hMLH1 in HeLa cells. HeLa cells were exposed to 0, 0.1 and 0.2 mM MNU in serum-free medium for 1 h. Immunoprecipitation and western blot analysis protocols were the same as in (A). Ten percent of the lysate was used for the loading control and the remaining 90% for co-immunoprecipitation. (C) The interaction between hRad9 and hMLH1 was significantly induced by MMS. Data were derived from three independent experiments as in (A). (D) The interaction between hRad9 and hMLH1 was significantly induced by MNU. The relative hRad9 levels were presented as mean ± SD from three independent experiments as described in (B). Asterisk indicates the statistical significance (P < 0.01) between precipitated hRad9 levels from cells treated or untreated with MMS or MNU.
Rad9-deleted ES cells are defective in MMR
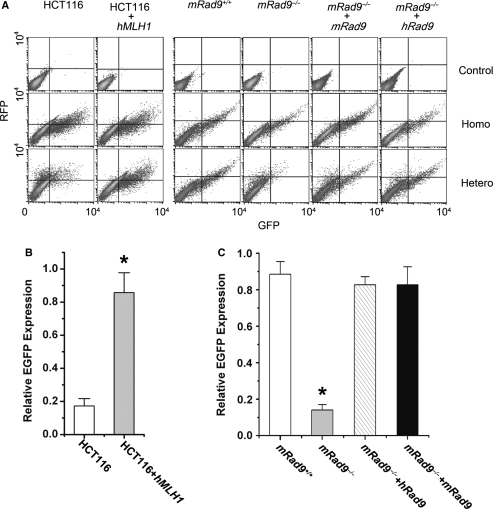
To address the question of whether hRad9 acts together with MLH1 in MMR, an assay using a T:G mismatch-corrupted start codon in the EGFP gene transfected into tested cells was performed to quantitatively measure MMR activity in living cells (32). The T:G mismatch can be corrected to T:A by MMR, thus MMR activity can be determined by flow cytometry measuring the number of EGFP-positive cells and the intensity of EGFP expression (refer to ‘Materials and methods’ section). Lei et al. (32) reported that HCT116 cells did not express hMLH1 and did not repair the T:G mismatch efficiently, but transfection with hMLH1 into the cells compensated for the defect, leading to an increased number of EGFP-positive cells and enhanced intensity of EGFP expression. We were able to repeat this experiment as a positive control (Figure 6A and B). The MMR activity reflected by EGFP expression efficiency in FLAG-hMLH1-expressing HCT116 cells was 4.7 times that of parent HCT116 cells containing empty plasmid pFLAG-CMV2 (Figure 6B). Using this assay, we found that mRad9-deleted mouse ES cells (mRad9–/– cells) had a significantly lower MMR activity than wild-type mouse ES cells (mRad9+/+ cells), and ectopic expression in mRad9–/– ES cells of mRad9 at a level comparable to that in mRad9+/+ cells (data not shown) restored the MMR activity to the level of mRad9+/+ cells (Figure 6A and C). Ectopic expression of human Rad9 in mRad9–/– cells also complemented the loss of MMR activity resulting from mRad9 deletion (Figure 6A and C). Taken together, these data demonstrate that Rad9 plays an important role in MMR in mammalian cells.
Figure 6.
mRad9–/– ES cells are deficient in MMR. (A) Experiments for detecting MMR activity. The reporter plasmid containing a T:G heteroduplex mismatch and a control plasmid containing homoduplex T:A in the start codon of EGFP were transfected into cells to measure MMR activity. MMR-mediated repair of T:G to T:A restored the starting codon and thus EGFP expression. Control plasmid containing intact mRFP-C1 sequence was co-transfected into the cells to monitor transfection efficiency. HCT116 cells were used as the MMR-negative control and HCT116 cells transfected with pZeoSV2-hMLH1 as the positive control; only a small population of HCT116 cells transfected with the reporter plasmid containing the T:G heteroduplex appeared in the lower-right and upper-right regions (EGFP-positive) and a significant portion of the same cells co-transfected with pZeoSV2-hMLH1 were located in the lower-right and upper-right regions. This MMR activity detection system was applied to monitor MMR activities of ES cells with and without Rad9. The cells containing Rad9 were significantly more efficient in repairing T:G into T:A than cells without Rad9. (B) Relative EGFP expressions of HCT116, HCT116+hMLH1 after co-transfection of nicked homo- or hetero-duplex EGFP plasmid and RFP plasmid (pmRFP-C1), derived from three independent experiments described in (A). The relative EGFP expression levels were calculated using a method developed by Lei et al. (32) and reflected MMR activities of these cells. (C) Relative EGFP expressions of mRad9+/+, mRad9–/–, mRad9–/– + hRad9 and mRad9–/– + mRad9 cells, derived from three independent experiments as described in (A).
The Ser160-mediated hRad9–hMLH1 interaction is important in MMR activity
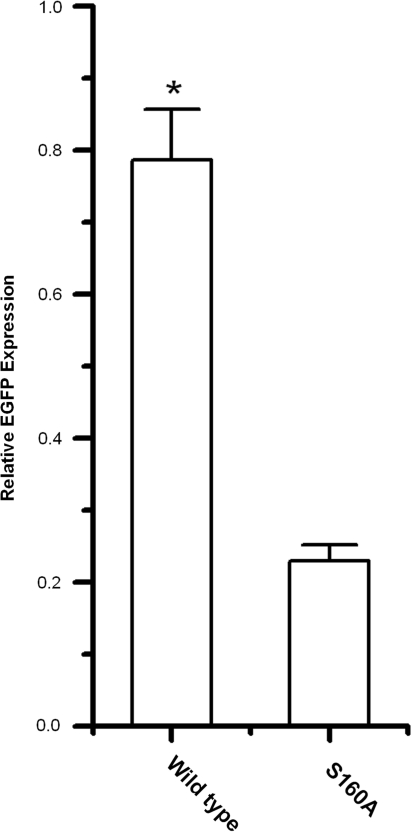
The functional significance of the interaction between hRad9 and hMLH1 was tested with the assay described above. We picked mRad9–/– cell clones that expressed wild type hRad9 (3 clones) and Ser160Ala hRad9 (3 clones) at levels close to the endogenous mRad9 level in mRad9+/+ cells (data not shown), and compared their MMR activities with those of mRad9–/– and mRad9+/+ cells. Ectopic expression of wild type hRad9 in mRad9–/– cells resulted in dramatically increased MMR activity as shown previously (Figures 6C and Figure 7), but expression of Ser160Ala hRad9 only increased MMR activity slightly (Figure 7). As shown previously, the Ser160Ala mutation severely demolished the hRad9–mMLH1 interaction (Figure 4). Taken together, these results suggest that hRad9 functions in the MMR pathway through its interaction with MLH1.
Figure 7.
The Ser160 of hRad9 plays a significant role in MMR. The relative EGFP expression of mRad9–/– + hRad9-WT (0.786 ± 0.07) is significantly higher than that of mRad9–/– + hRad9-S160A (0.23 ± 0.022). Three hRad9-WT-expressing and 3 hRad9-S160A-expressing mRad9–/– ES cell clones were tested for MMR activity, and similar results were obtained. In this figure, the result from one representative set of clones is given.
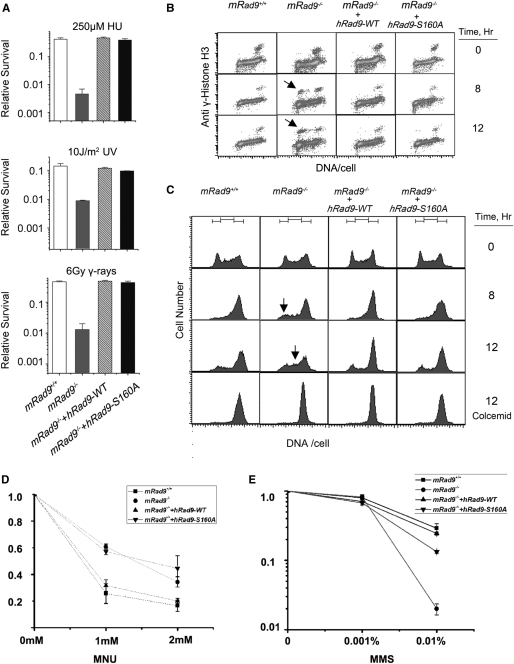
It has been reported that cells deficient in MMR are more tolerant to SN1 type DNA methylation agents than their parental cells (18,19,52–55). To investigate whether mRad9-deleted mouse ES cells are resistant to such DNA methylators or not, the ES cells mentioned above were examined for sensitivity to MNU. As expected, mRad9–/– cells were more resistant to MNU than mRad9+/+ cells (Figure 8D), consistent with previous studies (18,19,52–55). On the other hand, mRad9–/– cells were more sensitive to MMS than mRad9+/+ cells (Figure 8E), this may be due to the fact that MMS is a SN2 type methylating agent producing far fewer O6mG adducts (0.3%) and a greater proportion of N3mA (10%) and N7mG adducts (87%) than SN1 families (56,57). N3mA (10%) and N7mG adducts are repaired through base-excision repair pathway (55), so the damage induced by MMS is repaired mainly by BER rather than by MMR.
Figure 8.
The Ser160Ala mutation of hRad9 does not affect the known roles of hRad9. (A) Comparison of the sensitivities of mRad9+/+ ES cells, mRad9–/– ES cells and the two mRad9–/– ES cells that stably express hRad9-WT and hRad9-S160A, respectively. The four types of cells were treated with mock treatment, 250 μM HU for 24 h, 10 J/m2 and 6 Gy gamma rays, respectively, and then allowed to grow for 10 days to form cell colonies. The survival level stands for the ratio of the number of colonies formed from treated cells compared with the number of colonies formed from untreated cells. Results presented are the averages of three independent experiments, and each experimental data point is the average of duplicate cell samples. (B) The S/M checkpoint. The four types of cells described above were incubated in medium containing 1 mM HU for 0, 8 and 12 h, and then prepared for S/M checkpoint analysis by flow cytometry. The mRad9–/– ES cells stably expressing either hRad9-WT or hRad9-S160A showed intact S/M checkpoint while mRad9–/– ES cells containing the empty vector demonstrated prematured chromosome condensation (arrows) after incubation with HU for 8 or 12 h. (C) G2/M checkpoint. The four types of cells described above were irradiated with 10 Gy gamma rays and then grown for 0, 8 and 12 h. mRad9+/+ ES cells were arrested in the G2/M border 8 h after irradiation, and only a small population of the cells from the last cell cycle moved into the G1 phase 12 h after irradiation. mRad9–/– ES cells were unable to arrest at the G2/M border and moved into the G1 phase (arrow) 8 h after irradiation, and cells from the last cell cycle had already moved into the S phase (arrow) 12 h after irradiation. To confirm that the mRad9–/– ES cells in G1 and S phases 8 and 12 h after irradiation were from the last cycle, 50 ng/ml colcemid was added to one set of cells to block cells in the M phase. Indeed, colcemid was able to block the cells in the M phase 8 and 12 h after irradiation, suggesting that they came from the last cell cycle. The mRad9–/– ES cells stably expressing either hRad9-WT or hRad9-S160A showed intact G2/M checkpoint control. (D) The different cell lines were incubated in serum-free medium containing various concentrations of MNU at 3°C for 1 h. After rinsing, fresh medium was added to form colonies, and survival rates were determined. Results presented are the means of three separate experiments performed in duplicate. (E) The MMS-resistant experiment was carried out in the same way as that in (D).
In order to confirm whether the S160A mutation of hRad9 specifically influences MMR function, we tested the effect of the S160A mutation on the known functions of hRad9, such as sensitivity to HU, 254-nm UV light and gamma rays as well as S/M and G2/M checkpoint controls. This mutation did not sensitize ES cells to DNA-damaging agents (Figure 8A), nor lead to S/M or G2/M checkpoint deficiency (Figure 8B and C), suggesting that the overall structural integrity of hRad9 is not affected by the S160A mutation, and that hRad9 plays a role in MMR specifically via the hRad9–hMLH1 interaction.
DISCUSSION
hRad9 is critical for cell cycle checkpoint control (17,58) and for HR repair (40) and base-pair excision repair (35,38). In this study we have shown that hRad9 directly interacts with hMLH1 (Figures 1 and 2), and this interaction is important for DNA MMR (Figures 4, 6 and 7). Recent studies have shown that mismatch proteins function as sensors for cell cycle checkpoint controls. Mismatch protein MSH2 (MutS homolog 2) can interact with ATR, and MutSalpha and MutLalpha are required to activate ATR to phosphorylate Chk1 and promote cell cycle checkpoint control in response to MNNG-caused DNA methylation damage (59,60). This study shows that Rad9 plays an important role in MMR via a direct interaction with MMR protein MLH1. Whether there is a signaling flow from cell cycle checkpoint via checkpoint protein Rad9 toward mismatch repair is currently unclear.
The current prevailing view is that hRad9, hRad1 and hHus1 form a PCNA-like ring structure, the 9-1-1 complex (6–11), and in this heterotrimer form function in cell cycle checkpoint control and DNA repair (61,62). PCNA itself can also interact with hMLH1 and play a role in 3′ to 5′ excision in MMR (22). However, unlike either hRad 1 or hHus1, hRad9 is able to immunoprecipitate hMLH1 (Figure 3). This implies that hRad9's function in MMR is not in the 9-1-1 complex form. A hRad9 mutant that contains point mutations that affect interactions with hHus1 and/or hRad1 would be useful to further clarify if hRad9 plays a role in MMS independent of the 9-1-1 complex.
hRad9 has been reported to possess 3′ to 5′ nuclease activity (63) and this feature of hRad9 is compatible with its role in MMR. Four redundant exonucleases (I, VII, X and RecJ) have been identified in Escherichia coli MMR (19), whereas only ExoI has been experimentally implicated as an MMR-associated 5′-directed exonuclease in eukaryotic cells (26,64). The effects of ExoI deficiency on MMR in both yeast and mice are moderate compared to those of Msh2 or Mlh1, and this suggests the existence of more functionally redundant exonucleases in these organisms (26,64). Recent studies have shown that regulatory factor X (RFX) and MRE11 play roles in MMR. RFX stimulates mismatch-dependent 5′ to 3′ excision activity (27), and MRE11 possesses mismatch-dependent 3′ to 5′ excision activity (65). Since hRad9 has 3′ to 5′ nuclease activity (63), hRad9 is likely to also function in mismatch-dependent 3′ to 5′ excision. However, the 3′ to 5′ nuclease activity of hRad9 is weaker than would be expected if hRad9's 3′ to 5′ nuclease activity is important in MMR. We cannot exclude the possibility that hRad9 may function in MMR through the other unknown ways. Nevertheless, this hypothesis that hRad9 plays a role in MMR-dependent 3′ to 5′ nuclease activity merits future experimental studies.
The Rad9–MLH1 interaction might be a hot spot for mutation in tumor cells. The hMLH1 mutation leads to hereditary nonpolyposis colorectal cancer (HNPCC) and various other types of tumor (66–69). Mice with an mRad9 deletion in skin keratinocytes are susceptible to developing skin tumors (70). The deletion of mRad9 in all cell types causes mouse embryonic lethality, possibly due to the loss of its many important functions including cell cycle checkpoint control, base-excision repair and DNA double-strand break repair. In this study, we have shown that the Ser160Ala mutation of hRad9 leads to drastic reduction of the hRad9–hMLH1 interaction and MMR activity (Figures 4 and 6), but has no impact on cell cycle checkpoint control and cellular sensitivity to gamma rays, UV-light and DNA replication inhibitorHU. Cells with this type of mutation may easily survive and possess a mutator phenotype, thus developing into tumors.
FUNDING
National Natural Science Foundation of China (30530180); National Protein Project of Ministry of Science and Technology (2006CB910 902); and Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-R63). National Key Basic Research Program of China (973) of Ministry of Science and Technology (2004CB720004). Funding for open access charges: National Natural Science Foundation of China (30530180).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Liusheng He for providing pmRFP-C1 plasmid and Dr Lene Juel Rasmussen for the kind gift of the plasmid of pEGFP-hMLH1.
REFERENCES
- 1.Errol C, Friedberg GCW, Wolfram S. DNA Repair and Mutagenesis. Washington, D.C: ASM press; 1995. [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Gary S, Stein RB, Giodano Antonio, Denhardt DT. The Molecular Basis of Cell Cycle and Growth Control. New York: John Wiley, & Sons; 1999. [Google Scholar]
- 4.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 5.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 6.Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc. Natl Acad. Sci. USA. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roos-Mattjus P, Vroman BT, Burtelow MA, Rauen M, Eapen AK, Karnitz LM. Genotoxin-induced Rad9-Hus1-Rad1 (9-1-1) chromatin association is an early checkpoint signaling event. J. Biol. Chem. 2002;277:43809–43812. doi: 10.1074/jbc.M207272200. [DOI] [PubMed] [Google Scholar]
- 9.Griffith JD, Lindsey-Boltz LA, Sancar A. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J. Biol. Chem. 2002;277:15233–15236. doi: 10.1074/jbc.C200129200. [DOI] [PubMed] [Google Scholar]
- 10.Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-1-1 and Rad17-RFC. Genes Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 11.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunnerhagen P, Seaton BL, Nasim A, Subramani S. Cloning and analysis of a gene involved in DNA repair and recombination, the rad1 gene of Schizosaccharomyces pombe. Mol. Cell. Biol. 1990;10:3750–3760. doi: 10.1128/mcb.10.7.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray JM, Carr AM, Lehmann AR, Watts FZ. Cloning and characterisation of the rad9 DNA repair gene from Schizosaccharomyces pombe. Nucleic Acids Res. 1991;19:3525–3531. doi: 10.1093/nar/19.13.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman HB, Hopkins KM, Laverty M, Chu HM. Molecular cloning and analysis of Schizosaccharomyces pombe rad9, a gene involved in DNA repair and mutagenesis. Mol. Gen. Genet. 1992;232:367–376. doi: 10.1007/BF00266239. [DOI] [PubMed] [Google Scholar]
- 15.Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 16.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 17.Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 19.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 20.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 22.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 25.Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H., Jr, Kneitz B, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Yuan F, Wang D, Gu L, Li GM. Identification of regulatory factor X as a novel mismatch repair stimulatory factor. J. Biol. Chem. 2008;283:12730–12735. doi: 10.1074/jbc.M800460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vo AT, Zhu F, Wu X, Yuan F, Gao Y, Gu L, Li GM, Lee TH, Her C. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 2005;6:438–444. doi: 10.1038/sj.embor.7400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 30.Hang H, Lieberman HB. Physical interactions among human checkpoint control proteins HUS1p, RAD1p, and RAD9p, and implications for the regulation of cell cycle progression. Genomics. 2000;65:24–33. doi: 10.1006/geno.2000.6142. [DOI] [PubMed] [Google Scholar]
- 31.Hang H, Zhang Y, Dunbrack R.L., Jr, Wang C, Lieberman HB. Identification and characterization of a paralog of human cell cycle checkpoint gene HUS1. Genomics. 2002;79:487–492. doi: 10.1006/geno.2002.6737. [DOI] [PubMed] [Google Scholar]
- 32.Lei X, Zhu Y, Tomkinson A, Sun L. Measurement of DNA mismatch repair activity in live cells. Nucleic Acids Res. 2004;32:e100. doi: 10.1093/nar/gnh098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hang H, Fox MH. Analysis of the mammalian cell cycle by flow cytometry. Methods Mol. Biol. 2004;241:23–35. doi: 10.1385/1-59259-646-0:23. [DOI] [PubMed] [Google Scholar]
- 34.Lindsey-Boltz LA, Wauson EM, Graves LM, Sancar A. The human Rad9 checkpoint protein stimulates the carbamoyl phosphate synthetase activity of the multifunctional protein CAD. Nucleic Acids Res. 2004;32:4524–4530. doi: 10.1093/nar/gkh789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan X, Madabushi A, Chang DY, Fitzgerald ME, Shi G, Drohat AC, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 2007;35:6207–6218. doi: 10.1093/nar/gkm678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gembka A, Toueille M, Smirnova E, Poltz R, Ferrari E, Villani G, Hubscher U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi G, Chang DY, Cheng CC, Guan X, Venclovas C, Lu AL. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 40.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, Gupta A, Wellinger RJ, Zhang J, Powell SN, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Shell SM, Yang Z, Zou Y. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006;66:2997–3005. doi: 10.1158/0008-5472.CAN-05-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Lindsey-Boltz LA, Sancar A, Bambara RA. Mechanism of stimulation of human DNA ligase I by the Rad9-rad1-Hus1 checkpoint complex. J. Biol. Chem. 2006;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- 44.Smirnova E, Toueille M, Markkanen E, Hubscher U. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St Onge RP, Udell CM, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol. Biol. Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J. Biol. Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 48.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karran P, Bignami M. Self-destruction and tolerance in resistance of mammalian cells to alkylation damage. Nucleic Acids Res. 1992;20:2933–2940. doi: 10.1093/nar/20.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 51.Biade S, Sobol RW, Wilson SH, Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- 52.Aquilina G, Ceccotti S, Martinelli S, Hampson R, Bignami M. N-(2-chloroethyl)-N′-cyclohexyl-N-nitrosourea sensitivity in mismatch repair-defective human cells. Cancer Res. 1998;58:135–141. [PubMed] [Google Scholar]
- 53.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 54.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 55.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Liu L, Taverna P, Whitacre CM, Chatterjee S, Gerson SL. Pharmacologic disruption of base excision repair sensitizes mismatch repair-deficient and -proficient colon cancer cells to methylating agents. Clin. Cancer Res. 1999;5:2908–2917. [PubMed] [Google Scholar]
- 57.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 58.Chunbo Zhang CXZ, He Y, Hang H. Phosphorylation sites on Tyr28 and the C-terminus of Rad9 are required for inhibition of premature chromosomal condensation across the entire s phase. Cell. Physiol. Biochem. 2008;22:295–306. doi: 10.1159/000149808. [DOI] [PubMed] [Google Scholar]
- 59.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl Acad. Sci. USA. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 63.Bessho T, Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J. Biol. Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- 64.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl Acad. Sci. USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao N, Zhu F, Yuan F, Haick AK, Fukushige S, Gu L, Her C. The interplay between hMLH1 and hMRE11: role in MMR and the effect of hMLH1 mutations. Biochem. Biophys. Res. Commun. 2008;370:338–343. doi: 10.1016/j.bbrc.2008.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, McKinnon W, Duraisamy S, et al. Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129:537–549. doi: 10.1016/j.gastro.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J. Mol. Diagn. 2008;10:301–307. doi: 10.2353/jmoldx.2008.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Avdievich E, Reiss C, Scherer SJ, Zhang Y, Maier SM, Jin B, Hou H., Jr, Rosenwald A, Riedmiller H, Kucherlapati R, et al. Distinct effects of the recurrent Mlh1G67R mutation on MMR functions, cancer, and meiosis. Proc. Natl Acad. Sci. USA. 2008;105:4247–4252. doi: 10.1073/pnas.0800276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition – Update of ICG-HNPCC/INSiGHT mutation database. Dis. Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhishang Hu YL, Zhang C, Zhao Y, He W, Han L, Yang L, Hopkins KM, Yang X, Lieberman HB, Hang H. Targeted deletion of Rad9 in mouse skin keratinocytes enhances genotoxin-induced tumor development. Cancer Res. 2008;68:1–10. doi: 10.1158/0008-5472.CAN-07-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]