Abstract
In Arabidopsis, canonical 21-nt miRNAs are generated by Dicer-like (DCL) 1 from hairpin precursors. We have identified a novel class of functional 23- to 25-nt long-miRNAs that is generated independently from the same miRNA precursors by DCL3. Long-miRNAs are developmentally regulated and in some cases have been conserved during evolution implying that they have biological functions. Plant microRNA genes (MIR) have been proposed to evolve by inverted duplication of the target gene. We found that recently evolved MIR genes consistently give rise to long-miRNAs, while ancient MIR genes give rise predominantly to canonical miRNAs. Transcripts from inverted repeats representing evolving proto-MIR genes were processed by DCL3 into long-miRNAs and also by DCL1, DCL2 or DCL4 depending on hairpin stem length to produce different sizes of miRNAs. Our results suggest that evolution of MIR genes is associated with gradual, overlapping changes in DCL usage resulting in specific size classes of miRNAs.
INTRODUCTION
Small RNAs (smRNA) approximately 19–40 nt in length are important regulators of gene expression in Eukaryotes. These smRNAs, termed small interfering RNAs (siRNA) when processed from double-stranded RNAs (dsRNA) or microRNAs (miRNA) when processed from single-stranded, hairpin-folded RNA precursors, are generated by the RNAse III activities of Dicer and Dicer-like (DCL) proteins (28). These smRNAs act either by guiding the cleavage of cognate RNAs, blocking productive translation of these RNAs, or inducing methylation of specific DNA targets (1,2).
Although the core mechanisms are evolutionarily conserved, some organisms, especially seed plants, have evolved multiple pathways with shared components and overlapping functions that generate smRNAs of specific sizes with dedicated functions (3–6). The major smRNA pathways in the seed plant Arabidopsis thaliana differ in the nature of their precursors and requirements for four DCLs. Thus, 20- to 21-nt miRNAs are generated by DCL1 from hairpin-folded primary miRNA (pri-miR) precursors; 24-nt natural antisense transcript-derived siRNAs (nat-siRNA) depend on DCL2; 23- to 25-nt repeat-associated siRNAs (ra-siRNA) are generated by DCL3 from RDR2-dependent dsRNAs associated with heterochromatin and DNA repeats; and, 21-nt trans-acting siRNAs (ta-siRNA) are generated by DCL4 from RDR6-dependent dsRNAs (3–6).
Studies with double, triple and quadruple dcl mutants have shown that DCL proteins act redundantly to produce certain smRNA classes; i.e. that one DCL can compensate, at least partially, for deficiencies in other DCLs (7–13). This led us and others to propose that DCLs act hierarchically on the various smRNA precursors, presumably as a result of differences in the affinity for these substrates. This raises the possibility that environmental and developmental factors, which might alter DCL expression, play a role in regulating the cross-talk and function of smRNA pathways. While the hierarchical action of DCLs has been clearly established for endogenous ta-siRNAs and ra-siRNAs as well as for viral smRNAs, studies of a small subset of miRNA families had led to the common belief that miRNA precursors are exclusively processed by DCL1 to generate a single size class of 20- to 21-nt miRNAs (7–9). However, although the molecular basis is unclear, 2 of 38 novel non-conserved miRNAs, miR822 and miR839, were recently reported to depend on DCL4 (14).
Here we report studies of large miRNA data sets showing that precursors representing most miRNA families are also independently processed by DCL3 to generate a new class of bona fide miRNAs 23–25 nt in length, which we call long-miRNAs. Plant MIR genes are proposed to arise from initial duplication events to create perfect inverted repeat (IR) loci, which evolve by random mutation into short, imperfectly paired, hairpin stems characteristic of MIR genes (15). We found that recently evolved MIR genes consistently gave rise to long-miRNAs, while ancient MIR genes gave rise predominantly to canonical 21-nt miRNAs. Transcripts from inverted repeats representing evolving proto-MIR genes were processed by DCL3 into long-miRNAs as well as by DCL1, DCL2 or DCL4. DCL usage, and hence the size of the miRNA produced, was correlated with hairpin stem-length. These results support the working hypothesis that evolutionary changes in the fold-back structure of precursors help determine DCL usage resulting in specific size classes of miRNA. Our finding that long-miRNAs are developmentally regulated and, in some cases, have been conserved during evolution suggests that long-miRNAs have dedicated biological functions with adaptive significance.
MATERIALS AND METHODS
Mutant plants and growth conditions
Arabidopsis thaliana and A. Suecica plants were grown in soil under long day conditions (16 h 10 000 lux light at 21°C/8 h dark at 16°C) in growth chambers at 65% relative humidity. Unless indicated otherwise, experiments were done with rosette leaves and inflorescences of 6-week-old plants.
The dcl1-9 (N3828), which we backcrossed 5-times to Col-0, dcl2-5 (SALK_123586), dcl3-1 (SALK_005512), dcl4-2 (GK-160G05) and hen1-5 (SALK_049197) mutants have been described previously (12,13,16–19). Physcomitrella patens patens (strain WT06) was cultured on plates for 7 days. All other plant species were grown in a greenhouse and collected 9 days post-germination.
RNA analysis
SmRNA was purified and RNA blot hybridization was performed as described in Ref. (12) except that the smRNA fraction was enriched by PEG precipitation, 15% polyacrylamide gels were used and blots were washed 5-times in a 2× SSC 0.5% SDS solution at 37°C. The probes used are listed in Supplementary Data S4. Blots were stripped by three consecutive washes with boiling 0.5% SDS and re-probed. Signals on smRNA blots were quantified using ImageQuant5.2 (Amersham Biosciences, Freiburg, Germany).
Computational analysis of publicly available smRNA data sets
SmRNA data sets of Ref. (14) were filtered to retrieve sequences 23 nt or longer that only match miRNA loci. The miRNA sequences arising from MIR163 were excluded. Long-miRNA sequences were positioned relative to their respective miRNA duplex. The first 5′ nucleotide of either miRNA or miRNA* were used as coordinates. For long-miRNAs located on the 3′ arm, the position of the last 3′ nucleotide was deduced from their length. For MIR genes for which a unique precursor was known to yield several phased miRNAs, coordinates were defined for each miRNA duplex.
For analyses of tissue-specific smRNAs, data sets, sequences were retrieved from GEO (http://www.ncbi.nlm.nih.gov/geo) via accession numbers GSM118372, GSM118373, GSM154336 and GSM154370. The inflorescence and leaf data sets were combined and filtered for sequences in the range of 20–25 nt. FASTA-formatted data sets were queried to a BLAST database of the assembled Arabidopsis chromosome sequences (ftp://ftp.arabidopsis.org/Sequences/whole_chromosomes) using NCBI-BLAST v2.2.16. SmRNAs perfectly matching the genome >5 times were excluded since they could be of repetitive-element origin. The 184 MIR genes obtained from http://microrna.sanger.ac.uk/cgi-bin/sequences/ were queried to the BLAST database. MIR gene coordinates and genomic coordinates of non-repetitive smRNA matches were compared to tally smRNAs of MIR origin.
For inverted repeat (IR) sequence analyses, separate BLAST databases were generated for each IR region queried to smRNA data sets. Unique and duplicated matches to forward (+) and reverse (−) chromosomal strands were quantified.
RESULTS
Identification of 23- to 25-nt long-miRNA species in Arabidopsis
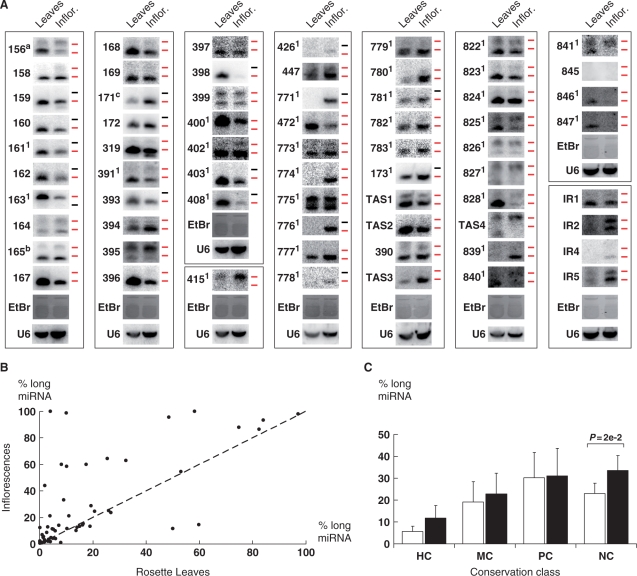
We used smRNA-blot hybridization to measure the size of miRNAs that accumulate in rosette leaves and inflorescences of 6-week-old Arabidopsis plants. The blots were hybridized with probes for 58 of the 94 recognized miRNA families (Figure 1A). For 16 miRNA families, e.g. miR160, miR162 or miR393, only the 20- to 21-nt size class was detected. For miR163, only the expected 24-nt miRNA, known to depend on DCL1 (20), was detected. Strikingly, probes for 41 miRNA families detected a miRNA species that migrated in the 23- to 25-nt region of the gels in addition to the expected 20- to 21-nt miRNAs (Figure 1A). Visual inspection of the smRNA blots showed that the relative abundance of the two size classes varied considerably. In 14 of the 41 miRNA families that accumulated both size classes, e.g. miR156, miR395 or miR399, the 23- to 25-nt size class was more abundant than was the 20- to 21-nt size class in at least the inflorescence sample. Thus, our results show that accumulation of a 23- to 25-nt smRNA species is a major, but family-dependent feature of Arabidopsis miRNA biogenesis.
Figure 1.
The abundance of Arabidopsis 23- to 25-nt long-miRNAs is developmentally regulated and is correlated with the evolutionary age of MIR genes. (A) RNA gel blot analysis of 58 miRNA families, 4 TAS families, and 4 IR-miRNAs in rosette leaves (Leaves) and inflorescences (Inflor.) of Col-0 plants. Each square block shows the hybridizations obtained for an individual blot probed and stripped several times. Probed smRNAs are indicated on the left side of the blots. The loading controls are U6 snRNA and EtBr-stained smRNAs. Accumulation of U6 snRNA was higher in inflorescences than in rosette leaves as previously observed (51). The upper and lower ticks to the right of each lane mark the positions of the 24- and 21-nt size markers, respectively, and are colored red when smRNA species of that size class are detected. amiR156 stands for the miR156/157 family. bmiR165 stands for the miR165/miR166. cmiR171 stands for the miR170/171 family. 1indicates that miRNAs are encoded by a single MIR gene. miR173, miR390, miR828 and their TAS target family were probed on the same blot. (B) The correlation of long-miRNA abundance in inflorescences and leaves. The data are expressed as percentage of long-miRNA of total miRNA detected by RNA blot hybridization using probes for 58 miRNA families. The dashed line indicates equal abundance of long-miRNA in both tissues. The raw data for the individual miRNA families are shown in Supplementary Data S3. (C) The mean percent long-miRNA representing highly- (HC), moderately- (MC), poorly- (PC) and nonconserved (NC) miRNA families in rosette leaves (white bars) and inflorescences (black bars). The error bars represent the SEM. The relative abundance of long-miRNAs in both leaves and inflorescences is inversely correlated (two-way analysis of variance, P = 5e − 2) with miRNA conservation as classified by (14,27). The percent long-miRNA is significantly higher in inflorescences than in leaves in nonconserved miRNA families (P = 2e − 2).
Long-miRNAs represent a new class of miRNAs
Twenty-six of the 41 miRNA families that accumulated both smRNA size classes are encoded by a single MIR gene. This suggests that both size classes arise from processing of the same pri-miR. As the first step in determining the nature and origin of the 23- to 25-nt miRNA species, we analyzed the informative, large smRNA databases of Rajagopalan and colleagues (14). By filtering these data sets, we found that, of 1296 unique sequences matching exclusively miRNA loci, 149 were 23 nt or longer (Table 1 and Supplementary Data S1A). To avoid biasing the analyses we had excluded the highly abundant, 24-nt miRNA sequences arising from MIR163 that are produced by DCL1 (20). These 149 smRNAs represented 51% of the 140 MIR genes and 60% of the 68 miRNA families in the data sets. This indicates that the long miRNA species detected on our RNA blots represent a class of 23- to 25-nt long miRNAs arising specifically from MIR genes rather than the result of modifications of canonical miRNAs. Moreover, these findings confirm our results of RNA blot analyses that production of long miRNA species is a major feature of Arabidopsis miRNA biogenesis.
Table 1.
Distribution of miRNA and long-miRNA sequencesa in public smRNA data sets
| 16- to 27-nt miRNAs | 23- to 27-nt miRNAs | 23- to 27-nt miRNAs matching (+) strand | 23- to 27-nt miRNAs matching (−) strand | |
|---|---|---|---|---|
| Unique sequences | 1296 | 149 | 148 | 1 |
| Number of reads | 145 388 | 809 | 808 | 1 |
| Percent of reads (%) | 16.4 of total smRNAs | 0.6 of total miRNAs | 99.9 of long-miRNAs | 0.1 of long-miRNAs |
| miRNA families | 68 | 41 | 41 | 1 |
aAnalysis of data sets of Ref. (14).
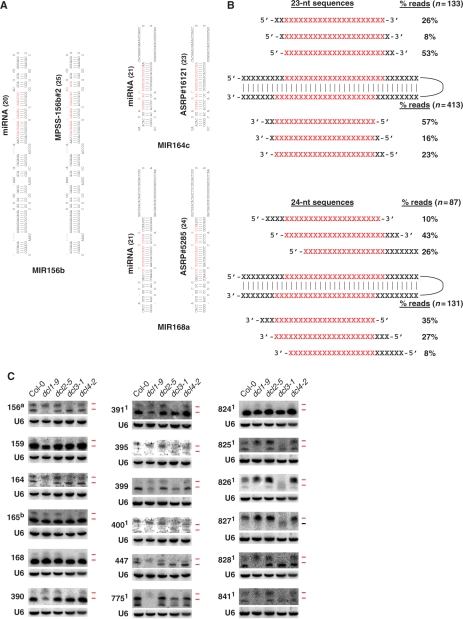
Of these long miRNA sequences, 99.9% (148 unique sequences representing 808 sequencing reads) matched exclusively the sense strand of miRNA precursors; a bias towards the precursor strand which shows that these smRNAs are directly generated from the hairpin-folded transcripts. Therefore, 23- to 25-nt miRNAs do not depend on the synthesis of a complementary strand. Similar analysis of the public databases MPSS PLUS and ASRP confirmed the conclusion that long-miRNAs arise from the miRNA precursor strand (Figure 2A, Supplementary Figure S1A, B and Supplementary Data S1B, C) (21–23). In summary, the 23- to 25-nt miRNA species detected on RNA blots are apparently processed directly from the hairpin folds of miRNA precursors and define a new class of miRNAs, which we call long-miRNAs.
Figure 2.
Long-miRNAs represent a class of bona fide miRNAs dependent on DCL3 for their biogenesis. (A) Representative canonical miRNAs (left) and long-miRNAs (right) identified in ASRP or MPSS PLUS data sets as indicated. The miRNAs and long-miRNAs shown in red are positioned on their respective hairpin precursors. The length in nucleotides of the miRNA and long-miRNA are indicated in parentheses. The position of all remaining long-miRNAs represented in the ASRP and MPSS PLUS data sets are shown in Supplementary Figure S1A, B. (B) Cartoons showing the position of 23-nt (upper panel) and 24-nt (lower panel) long-miRNAs relative to the canonical 21-nt miRNA processed from the same hairpin. Twenty-three or twenty-four nucleotide long-miRNA sequences present in the data sets of Rajagopalan and colleagues (14) (listed in Supplementary Data S1A) were positioned with reference to the first 5′ nucleotide of either miRNA or miRNA* depending on which hairpin arm these were located. Positions (X) with identical nucleotides in the canonical and long-miRNAs are in red. The percentage of the reads with the displacement shown are indicated on the right along with the number of reads in parentheses.(C) RNA gel blot analysis of miRNAs and long-miRNAs of 18 miRNA families. SmRNAs (20 µg) from inflorescences of dcl mutants and of Col-0 wild-type were loaded on the gels. Five individual blots were probed and stripped several times. Probed miRNAs are indicated on the left side of the blots. The loading control is U6 snRNA. The upper and lower ticks to the right of each lane mark the positions of the 24- and 21-nt size markers, respectively, and are colored red when miRNA species of that size class are detected. amiR156 stands for the miR156/157 family. bmiR165 stands for the miR165/miR166. 1Indicates the miRNAs encoded by a single MIR gene.
Canonical miRNAs and long-miRNAs are generated by independent processing events
Maturation of canonical miRNAs in Arabidopsis involves two sequential processing steps that depend on DCL1 (24). In the first step, DCL1 usually cuts at the base of the hairpin stem of the pri-miR to release the shorter, pre-miRNA with a 2-nt overhang at the 3′ end of the hairpin. In the second step, the pre-miRNA is cleaved to release the double-stranded miRNA intermediate (24). To establish how long-miRNAs are generated, we determined where in the hairpin the first 5′ nucleotide of the long-miRNAs on the 5′ arm and the last 3′ nucleotide of the long-miRNAs on the 3′ arm are located. For sequences matching several genes of a family, we analyzed only those at the same position on different precursors. We also restricted our analyses to the 133 long-miRNAs of the 23- and 24-nt classes since the 25- to 27-nt-long sequences were only represented by 37 sequencing reads of 12 unique sequences. The positioning of long-miRNAs relative to that of the corresponding canonical miRNAs is summarized in Figure 2B. The complete set of raw data is in Supplementary Data S1A. In every case, the 23- or 24-nt long-miRNA duplexes reconstituted from sequences in opposing arms had the canonical 1- to 2-nt 3′ overhangs expected for bona fide miRNA duplexes (Figure 2B).
Canonical miRNAs show a high degree of fidelity for the first processing step: namely, 95% of the 16- to 22-nt-long reads matching the 5′ arm, and 94.5% of the 16- to 22-nt-long reads matching the 3′ arm (Supplementary Data S2). In contrast, a large proportion of long-miRNAs are at position shifted by one or more nucleotides (Figure 2B) indicating that their biogenesis depends on a DCL activity with less fidelity than that of DCL1. The remaining long-miRNAs have their first processing step at the same position as that of the canonical miRNAs: 53% of the 23-nt long-miRNAs and 43% of the 24-nt long-miRNAs arising from the 5′ arm, and, 23% of the 23-nt long-miRNAs and 27% of the 24-nt long-miRNAs arising from the 3′ arm. This indicates that the size of these long-miRNAs, unlike that of miR163, does not depend on small bulges in the processed region. Therefore, both types of long-miRNAs are bona fide 23- to 24-nt miRNAs produced independently from canonical miRNAs encoded by the same MIR gene.
Different DCLs independently produce miRNAs and long-miRNAs
To identify the DCL required for biogenesis of long-miRNAs, we compared the accumulation of canonical miRNAs and long-miRNAs in rosette leaves and inflorescences of the dcl deficiency mutants dcl1-9, dcl2-5, dcl3-1 and dcl4-2 (Figure 2C and Supplementary Figure S2). As expected from earlier studies (7–9,14,25), accumulation of most 20- to 21-nt miRNAs was affected in the dcl1-9 mutant, but not in the dcl2-5, dcl3-1 or dcl4-2 mutants. In contrast, accumulation of the 23- to 25-nt long-miRNAs was impaired in the dcl3-1 mutant but was not affected in the dcl1-9, dcl2-5 or dcl4-2 mutants (Figure 2C and Supplementary Figure S2). These results establish that pri-miRNAs are processed independently by DCL1 and DCL3 to generate the 20- to 21-nt canonical miRNAs and the 23- to 25-nt long-miRNAs, respectively. Moreover, more than half of the miRNA families tested are encoded by a single gene showing that this dual processing is a major feature of Arabidopsis miRNA biogenesis.
DCL compensation in the processing of certain pri-miRNAs
Our smRNA-blot analyses in dcl mutants also indicate that there is partial redundancy among DCLs in the processing of certain pri-miRNAs (Figure 2C). In the case of miR825, miR826 and miR827, the long-miRNA size class was replaced by a 22-nt long-miRNA species in the dcl3-1 mutant. These 22-nt long-miRNA species were detected neither in wild-type Col-0 plants nor in other dcl mutants. This argues that in the absence of DCL3, a surrogate DCL, presumably DCL2, can process certain pri-miRNAs and generate 22-nt long-miRNAs. Thus, as shown for ta-siRNAs, ra-siRNAs and viral smRNAs (7–10,12), different DCLs can compete for the same precursor of miRNA.
The accumulation of long-miRNAs is developmentally regulated and might depend on organ-specific expression of DCL3
Visual inspection of our RNA blots suggested that long-miRNAs are generally more abundant in inflorescences than in leaves (e.g. miR395 and miR156) (Figure 1A). Quantification of the percentage of long-miRNAs relative to the total amount of miRNA in the two size classes showed that a significant proportion (binomial proportions, P = 1.2e−2), 74%, of the miRNA families tested accumulated higher amounts of long-miRNAs in inflorescences than in leaves (Figure 1B and Supplementary Data S3). Moreover, the average percentage of long-miRNAs was significantly higher in inflorescences, 28%, than in leaves, 20% (t-test, P = 3.3e−3). These results were consistent with the frequency of the two miRNA size classes represented in smRNA databases obtained with leaves and inflorescences (14,21). Supplementary Table S1 shows that while the number of reads obtained for canonical miRNAs was equal in leaves and inflorescences, the reads obtained for long-miRNAs were 3-fold higher in inflorescences than in leaves. Therefore, the proportion of long-miRNAs to the total miRNAs is developmentally regulated in the organs tested, and may depend on relative processing by DCL1 and DCL3.
To examine the possibility that the patterns of miRNA accumulation we observed depend on DCL expression, we compared the normalized expression patterns of DCL1, DCL2, DCL3 and DCL4 mRNAs in rosette leaves and inflorescences available in the AtGenExpress Visualization Tool (DCL1, DCL2, DCL3, DCL4) (26). The fold expression in inflorescences relative to the average for six rosette leaf stages showed that whereas the expression of DCL1 mRNA did not differ significantly (t-test) in the two organs; there was a highly significant (P < 0.0005) ∼10-fold higher expression of DCL3 mRNA in inflorescences relative to leaves (Table 2). This correlation of DCL expression and miRNA accumulation is consistent with the hypothesis that long-miRNA biogenesis is regulated, at least in part, by organ-specific expression of DCL3.
Table 2.
Expression of Arabidopsis DCL genesa indicates an organ-specific expression of DCL3
| Expression (AU) |
|||||
|---|---|---|---|---|---|
| DCL | Gene | Inflorescences | Rosette leavesb Mean ± SEM | *P< | Fold changec |
| DCL1 | At1g01040 | 1.39 | 0.81 ± 0.11 | NS | 1.72 |
| DCL2 | At3g03300 | 1.59 | 0.66 ± 0.09 | 0.02 | 2.41 |
| DCL3 | At3g43920 | 2.65 | 0.26 ± 0.04 | 0.0005 | 10.39 |
| DCL4 | At5g20320 | 1.87 | 0.90 ± 0.03 | 0.0005 | 2.09 |
aAnalysis of data of AtGenExpress Visualization Tool (26).
bMean expression for rosette leaves numbered 2, 4, 6, 8, 10 and 12 ± SEM.
cFold change in expression in inflorescences relative to leaves.
*Probability t-test of a single sample (inflorescences) and mean for rosette leaves, df = 5.
DCL usage is correlated with the evolutionary age of MIR genes
We examined the relationship between long-miRNA accumulation and evolutionary conservation of miRNA families. The data set comprised the subset of 30 recognized miRNAs grouped by Zhang and colleagues (27) into highly-, moderately-, poorly-, and nonconserved classes plus 28 non-conserved miRNAs subsequently identified by Rajagopalan and colleagues (14). The relative abundance of long-miRNAs in both leaves and inflorescences is inversely correlated (two-way analysis of variance, P = 5e−2) with the level of miRNA conservation in the classes defined (Figure 1C and Supplementary Data S3). Specifically, the average percent long-miRNA ranged from 5.67 ± 2.43% (average, SEM) for highly conserved miRNA families to 30.1 ± 11.7% for poorly conserved miRNA families in leaves and from 11.8 ± 5.69% for highly conserved miRNA families to 33.58 ± 6.69% for non-conserved miRNA families in inflorescences. The average percentage of long-miRNA was significantly greater (t-test, P = 2e−2) in inflorescences than in leaves for the non-conserved miRNA families but not for the other families. Thus, accumulation of long-miRNAs is developmentally regulated and affects preferentially long-miRNAs representing the less conserved miRNA families. Assuming that conservation in the data set we surveyed reflects antiquity, these results suggest that the relative abundance of canonical miRNAs and long-miRNAs, and hence, the DCL used for processing depends on the evolutionary age of the MIR gene.
DCL usage is correlated with the length of the hairpin precursor
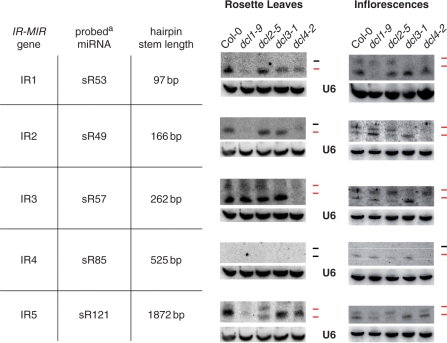
We examined the biogenesis of smRNAs derived from inverted-repeat (IR) transcripts. The rationale for this approach is that IR genes differing in hairpin stem length are thought to be intermediates in the evolution of a perfect IR ancestor to a modern MIR gene (15). Our analysis focused on a set of five putative IR loci (22). The analysis of the distribution of smRNA sequences retrieved from public data sets showed that the smRNAs matching these loci have an origin biased towards the same DNA strand (Supplementary Table S2). Thus, together with folding predictions of their precursor (Supplementary Figure S3), this shows that these five IR loci, termed IR1-IR5, produce hairpin-folded transcripts that are processed without synthesis of a complementary strand. These IR loci are designated IR-MIR genes since they produce long hairpin precursors that are processed to generate bona fide miRNAs (28). These loci, which encode hairpins differing in stem length from 97 bp to 1872 bp, are thus believed to represent intermediates at different stages in the evolution of MIR genes (15).
Figure 3 shows by RNA-blot hybridization that the size of the miRNA that accumulates in wild-type Col-0 depends on the IR-MIR gene. IR1, IR2 and IR3 give rise to 21-nt miRNAs and 24-nt long-miRNAs; the weakly expressed IR4 gives rise to 21-nt miRNAs; and IR5 gives rise to 22-nt miRNAs and 24-nt long-miRNAs. Accumulation of 24-nt long-miRNAs, but not the shorter miRNAs was impaired specifically in the dcl3-1 mutant (Figure 3). Moreover, confirming our earlier observations, inflorescences accumulated more long-miRNAs than did rosette leaves (Figures 1A, 3 and Supplementary Data S3).
Figure 3.
DCL usage for processing hairpins of IR-MIR genes correlates with stem length.RNA gel blot analysis of smRNAs (20 µg) from rosette leaves or inflorescences of dcl mutants and Col-0 wild-type. The IR-MIR gene, the miRNA probed and the length of the hairpin stem are indicated. The loading control is U6 snRNA. The upper and lower ticks to the right of each lane mark the positions of the 24- and 21-nt size markers, respectively, and are colored red when miRNA species of that size class are detected. aThe names of probed miRNAs are according to the MPSS PLUS database online.
Our RNA blots also show that for IR-MIR genes, DCL usage is correlated with the length of the hairpin stem. In both tissues the accumulation of 21-nt IR-miRNAs arising from IR1, the shortest hairpin precursor, was impaired in the dcl1-9 mutant; that of 21-nt IR-miRNAs arising from IR2, IR3 and IR4, hairpin precursors of intermediate lengths, was impaired in the dcl4-2 mutant; and that of 22-nt IR-miRNAs arising from IR5, the longest hairpin available, was impaired in dcl2 (Figure 3). In rosette leaves, there was some weak redundancy in DCL usage. Thus, accumulation of 21-nt IR-miRNAs arising from IR1, which depends on DCL1, was also partially impaired in the dcl4-2 mutant and that of 21-nt IR-miRNAs arising from IR2, the shortest of the DCL4-dependent IR-miRNA precursors, was also impaired in the dcl1-9 mutant (Figure 3). We conclude that all four known DCLs are able to process hairpin precursors. Both DCL usage and the size of the miRNAs generated are correlated with the length of the hairpin stem of the precursor. These findings support our working hypothesis that evolutionary changes in the structure of the hairpin can change their affinity for DCLs, and hence the size of the miRNAs.
Evolutionary conservation of long-miRNAs
Our data indicate that evolution of Arabidopsis MIR genes is associated with a shift in DCL usage, which eventually leads to the exclusive use of DCL1 for production of 21-nt miRNAs. Unexpectedly, the miR156/157, miR164 and miR390 families, although highly conserved, also use DCL3 to accumulate relatively high levels of long-miRNAs (Figures 1A and 2C). Probing for these families in widely divergent plant species showed that, in addition to the canonical 21-nt miRNA forms, the 23- to 25-nt long-miRNA species of miR156/157, miR164 and miR390 accumulated in members of the Brassicaceae, Solanaceae and Poaceae families (Figure 4). In contrast, no long-miRNAs were detected for the highly conserved miR160 family in the same plant species. Our data show that the biogenesis of long-miRNAs from certain highly conserved miRNA families is conserved in widely divergent plant families.
Figure 4.
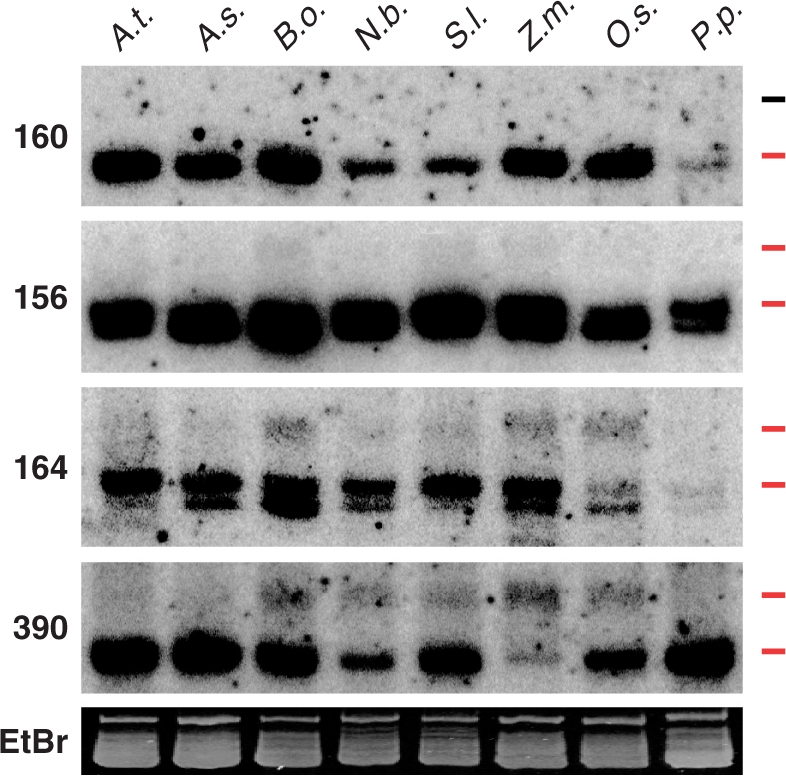
Conserved accumulation of long-miRNA of highly conserved miRNA families in widely divergent plant species. SmRNAs (8 µg) from rosette leaves of the Bassicaceae A. thaliana (A.t.), A. suecica (A.s.), Brassica oleracea (B.o.), the Solanaceae Nicotiana benthamiana (N.b.), Solanum lycopersicum (S.l.), the Poaceae Zea mays (Z.m.), Oryza sativa (O.s.) and the Bryophyte P. patens (P.p.) were loaded on the gel. Probed miRNAs are indicated on the left side of the blots. EtBr-stained smRNAs serves as the loading control. The upper and lower ticks to the right of each lane mark the positions of the 24- and 21-nt size markers, respectively, and are colored red when miRNA species of that size class are detected.
DISCUSSION
Independent processing of pri-miRNAs by different DICER activities
The common belief was that miRNAs, unlike other well-characterized plant smRNAs, comprise a single class generated exclusively by the dedicated activity of DCL1 (7–12). We show here that the same miRNA precursor can be processed independently by DCL1 and DCL3 to generate two size classes of miRNA. Specifically, we describe a novel class of bona fide miRNAs. These 23- to 25-nt long-miRNAs are generated by DCL3 directly from hairpins without synthesis of a complementary strand. Our findings can explain both the weak 23- to 25-nt miRNA signals obtained with wild-type plants (20,29–35) as well as the strong 23- to 25-nt signals for miRNA and miRNA duplexes obtained with transgenic lines overexpressing individual MIR genes in earlier studies (25,36–38).
Most miRNA families, 71%, comprised both long-miRNAs and canonical miRNAs. Of these 41 families, more than 60% are encoded by a single MIR gene. Therefore, independent processing of miRNA precursors by DCL1 and DCL3 is a general feature of Arabidopsis miRNA biogenesis.
Analysis of smRNAs encoded by IR genes showed that production of additional size classes of miRNAs depends on other DCLs. IR1 gives rise to 21-nt miRNAs dependent on DCL1; IR2 and IR3, which we found correspond to the recently recognized MIR839 and MIR822 (14), as well as IR4 give rise to 21-nt miRNAs dependent on DCL4; and IR5, which we found corresponds to the IR71 loci (9), gives rise to 22-nt miRNAs dependent on DCL2. Because DCL3 also processes these precursors, we conclude that all known DCL activities can process hairpin miRNA precursors, and that DCL redundancy affects all classes of smRNAs, including miRNAs. Our detection of long-miRNAs in diverse plant families (Figure 4) and the fact that numerous plant species have multiple DCLs (39) suggests that dual processing of pre-miRNAs is a general feature of plants.
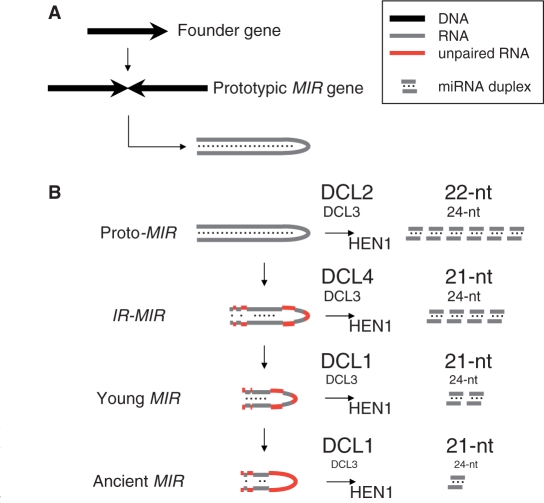
A model relating DCL usage, hairpin structure and MIR-gene evolution
Our findings bear directly on the relationship between DCL usage and the evolution of MIR genes. Allen and colleagues (15) proposed that MIR genes arise from an initial duplication of the target gene to create a perfect, long IR locus, which we designate here as a proto-MIR gene. Subsequent mutations shorten the hairpin stem, increase the number of unpaired bases in the stem, increase the size of the distal loop and eventually give rise to modern MIR genes with IR-MIR genes as intermediates (Figure 5). This implies that the hairpin of more conserved, ancient MIR genes should be shorter and show more mismatching than the hairpin of non-conserved, young MIR genes. We found a highly significant correlation between the relative abundance of long-miRNAs and the evolutionary conservation of MIR gene families. This suggests that DCL3 is less able to process conserved hairpins than less conserved ones and, hence, that evolution of hairpin structure affects DCL processing. We could not test this hypothesis by comparing hairpin-stem length, MIR-gene conservation and abundance of long-miRNAs because none of the known moderately- or highly conserved miRNAs are encoded by single genes.
Figure 5.
A model relating DCL usage, hairpin structure and MIR-gene evolution. (A) A current model for the evolution of MIR genes by inverted duplication of the founder target gene (black arrow) to give a long IR (head-to-head black arrow) (15), which we designate a proto-MIR gene. (B) Our model for changes in hairpin precursor structure, DCL usage and miRNA size as MIR genes evolve. Hairpin sequences that have diverged in evolution are shown in red; conserved sequences are shown in gray. The approximate contribution of different DCLs to processing is reflected by their font size. The miRNA size generated is indicated on the right.
We were, however, able to correlate hairpin length with DCL usage in a series of IR-MIR genes. Assuming hairpin length decreases with evolutionary age, our results show that long hairpins, potentially encoded by proto-MIR genes, are processed primarily by DCL2; hairpins encoded by somewhat older intermediate IR-MIR genes are processed primarily by DCL4; and short hairpins encoded by MIR genes are processed primarily by DCL1 (Figure 5). Based on this assumption, hairpins at every stage of MIR-gene evolution are also processed to some degree by DCL3. The graded differences in long-miRNA abundances associated with young, non-conserved and older, conserved MIR genes suggests that their length, and presumably additional structural features of hairpins, contribute to the relative processing by DCL1 and DCL3.
Functional and biological significance
Several lines of evidence suggest that long-miRNAs have biological functions. Long-miRNAs appear to be genuine, functional miRNAs. RNA blot analysis of hen1-5 mutants confirmed that long-miRNAs, like other functional smRNAs, require HEN1 for their biogenesis (data not shown). Functional smRNAs are incorporated into AGO–RISC effector complexes (40). Recent studies have shown by co-immunoprecipitation that 21-nt forms of miR391 and miR393 associate with specific AGO proteins (38). We noticed that the corresponding long-miRNAs in this study were also associated with AGO proteins suggesting that both forms of miRNA are incorporated into effector complexes. Determining to what extent long-miRNAs contribute to cleavage of the mRNA targets is difficult to ascertain because of the extensive overlaps between canonical and long-miRNA sequences. Nevertheless, certain long-miRNAs are highly conserved in several divergent Angiosperm families (Figure 4). This conservation together with their developmental regulation supports the view that long-miRNAs have biological functions.
The exact biological function of long-miRNAs and other non-DCL1 miRNAs is unknown. Strong alleles of the dcl1 mutant are lethal (41) indicating that long-miRNAs cannot compensate for deficiencies in canonical miRNAs with essential functions. This probably reflects the fact that these functions depend on ancient, highly conserved MIR genes that produce low levels of long-miRNAs. A possible function for long-miRNAs is suggested by the observation that a double dcl1-9 dcl3-1 mutant exhibits a specific, late-flowering phenotype involving the epigenetically regulated FLC gene while the dcl1-9 single mutant does not (42). This late-flowering phenotype was not observed in dcl3-1 and rdr2-1 deficiency mutants, and therefore does not depend on 24-nt ra-siRNAs. This raises the possibility that long-miRNAs are involved and provides possible links between miRNA-mediated regulation based on mRNA cleavage or translational repression and more stable forms of epigenetic regulation.
Hypothetically, DCL4-dependent IR-miRNAs might act like ta-siRNAs, which depend on DCL4, and mediate the cleavage of target mRNAs in trans (16,43). While canonical miRNAs are thought to act primarily in a cell-autonomous fashion (44), DCL4-dependent smRNAs derived from hairpins encoded by IR transgenes can spread from cell to cell (45). Thus, we speculate that the DCL4-dependent miRNA class might be capable of cell-to-cell movement as well.
In conclusion, the picture that emerges is that all known types of Arabidopsis smRNAs show redundancy in DCL usage and generate multiple classes of smRNA products. We show that in the case of miRNAs, these parameters are correlated with the stage of MIR gene evolution and hairpin structure. MiRNAs are an ancient form of regulation established in plants before multicellularity (46,47) and some individual miRNAs appear to have conserved functions reaching back to the mosses and the ferns (27,48–50). While their precise functions remain to be explored, the novel miRNA classes we describe open a new dimension in studying the diversity of miRNA functions and how they evolved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novartis Research Foundation and Swiss National Science Foundation [grant number 3100A0-105852 to Th.B.]. Funding for open access charge: Friedrich Miescher Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Thomas Hohn, Witold Filipowicz, Helge Grosshans and Mikhail Pooggin for critical comments, James Carrington for the dcl4-2 mutant, Claudia Kutter for the dcl2-5 mutant and Pierre-François Perroud for Physcomitrella patens patens strain WT06.
REFERENCES
- 1.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 2.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 3.Meins F, Jr, Si-Ammour A, Blevins T. RNA silencing systems and their relevance to plant development. Annu. Rev. Cell Dev. Biol. 2005;21:297–318. doi: 10.1146/annurev.cellbio.21.122303.114706. [DOI] [PubMed] [Google Scholar]
- 4.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 5.Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 10.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 11.Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl Acad. Sci. USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F., Jr, Hohn T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F., Jr, Hohn T, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 16.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 19.Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 20.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 28.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 36.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 37.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 38.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PM. The evolution and diversification of Dicers in plants. FEBS Lett. 2006;580:2442–2450. doi: 10.1016/j.febslet.2006.03.072. [DOI] [PubMed] [Google Scholar]
- 40.Vaucheret H. Plant ARGONAUTE. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz RJ, Hong L, Fitzpatrick KE, Amasino RM. DICER-LIKE 1 and DICER-LIKE 3 redundantly act to promote flowering via repression of FLOWERING LOCUS C in Arabidopsis thaliana. Genetics. 2007;176:1359–1362. doi: 10.1534/genetics.107.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tretter EM, Alvarez JP, Eshed Y, Bowman JL. Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol. 2008;147:58–62. doi: 10.1104/pp.108.117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 46.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 47.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 49.Barakat A, Wall K, Leebens-Mack J, Wang YJ, Carlson JE, Depamphilis CW. Large-scale identification of microRNAs from a basal eudicot (Eschscholzia californica) and conservation in flowering plants. Plant J. 2007;51:991–1003. doi: 10.1111/j.1365-313X.2007.03197.x. [DOI] [PubMed] [Google Scholar]
- 50.Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 51.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.