Abstract
Sequestration of c-Fos at the nuclear envelope (NE) through interaction with A-type lamins suppresses AP-1–dependent transcription. We show here that c-Fos accumulation within the extraction-resistant nuclear fraction (ERNF) and its interaction with lamin A are reduced and enhanced by gain-of and loss-of ERK1/2 activity, respectively. Moreover, hindering ERK1/2-dependent phosphorylation of c-Fos attenuates its release from the ERNF induced by serum and promotes its interaction with lamin A. Accordingly, serum stimulation rapidly releases preexisting c-Fos from the NE via ERK1/2-dependent phosphorylation, leading to a fast activation of AP-1 before de novo c-Fos synthesis. Moreover, lamin A–null cells exhibit increased AP-1 activity and reduced levels of c-Fos phosphorylation. We also find that active ERK1/2 interacts with lamin A and colocalizes with c-Fos and A-type lamins at the NE. Thus, NE-bound ERK1/2 functions as a molecular switch for rapid mitogen-dependent AP-1 activation through phosphorylation-induced release of preexisting c-Fos from its inhibitory interaction with lamin A/C.
Introduction
c-Fos is a member of the dimeric activator protein 1 (AP-1) transcription factor family that regulates key cellular processes, including cell proliferation, death, survival differentiation, and oncogenic transformation (Shaulian and Karin, 2002; Eferl and Wagner, 2003). c-fos is an “early response” gene that undergoes rapid transcriptional activation in response to multiple pathophysiological stimuli (e.g., growth factors, chemical and physical stress, etc.) (Shaulian and Karin, 2002). Fine regulation of c-Fos activity is achieved via its interaction with regulatory proteins that can either enhance or inhibit AP-1 activity, and through posttranslational processing of preexisting or newly synthesized c-Fos protein (Piechaczyk and Blanchard, 1994; Shaulian and Karin, 2002). For example, c-Fos can be phosphorylated by protein kinases C and A, cdc2 (Abate et al., 1991), FRK (Deng and Karin, 1994), extracellular signal-regulated kinase 7 (ERK7) (Abe et al., 1999), and the mitogen-activated protein kinases (MAPK) ERK1/2 (Monje et al., 2003) and p38 (Tanos et al., 2005). Importantly, c-Fos transcriptional activity is modulated through reversible phosphorylation by some of these kinases, including ERK1/2 (Hunter and Karin, 1992; Hill and Treisman, 1995; Monje et al., 2003). Phosphorylation of c-Fos by ERK1 and RSK occurs soon after serum stimulation (Chen et al., 1993, 1996). Moreover, a docking site for ERK in c-Fos facilitates its sequential phosphorylation in multiple C-terminal sites upon prolonged ERK activation (Murphy et al., 2002). The binding of growth factors, differentiation stimuli, and cytokines to cell surface receptors promotes the activation of the small GTPase Ras and dual phosphorylation (activation) of ERKs. Active ERK1 and ERK2 translocate into the nucleus, where they phosphorylate several transcriptional regulators (e.g., c-Fos) that cause a rapid induction of immediate early genes (Sharrocks, 2001).
In addition to fulfilling structural functions at the nuclear envelope (NE), A-type lamins (lamin A and C) play important roles in the control of gene expression via their interaction with histones, transcription factors (e.g., SREBP1, MOK2, BAF, GCL, Mel18, and c-Fos), and cell cycle regulators (e.g., the retinoblastoma protein [pRb] and cyclin D3) (Taniura et al., 1995; Gruenbaum et al., 2005; Mariappan and Parnaik, 2005; Broers et al., 2006; Heessen and Fornerod, 2007; Vlcek and Foisner, 2007). We have recently shown that lamin A/C–dependent sequestration of c-Fos at the NE reduces AP-1 DNA-binding activity and cellular proliferation (Ivorra et al., 2006). However, the molecular mechanisms controlling this interaction remain unknown. Here we tested the hypothesis that ERK1/2 is a critical regulator of the interaction between lamin A/C and c-Fos. We show that active ERK1/2 directly interacts with lamin A and colocalizes with c-Fos at the NE. Therein, mitogen-induced ERK1/2-mediated phosphorylation of c-Fos releases it from the inhibitory interaction with lamin A/C before de novo synthesis of c-Fos, thus allowing a rapid induction of AP-1 activity.
Results
c-Fos accumulation at the extraction-resistant nuclear fraction is regulated by lamin A/C expression and the level of c-Fos phosphorylation
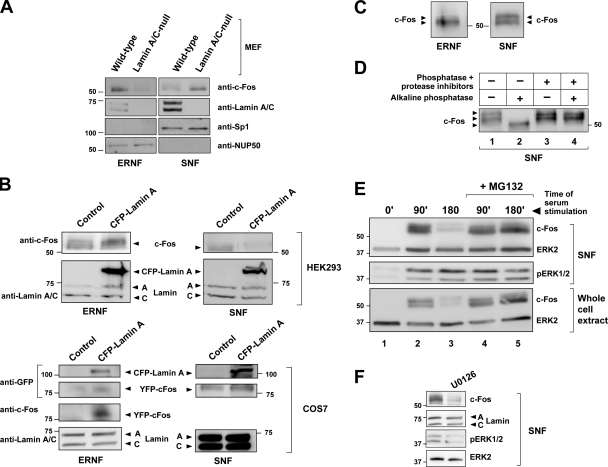
We first examined by Western blot the consequences of altering lamin A/C expression on the amount of c-Fos within the soluble nuclear fraction (SNF) and extraction-resistant nuclear fraction (ERNF) of cultured cells. Lamin A/C–null mouse embryonic fibroblasts (MEFs) exhibited decreased ERNF-associated and increased SNF-associated level of c-Fos compared with wild-type controls (Fig. 1 A), and ectopically expressed CFP-Lamin A increased the presence of both endogenous c-Fos and recombinant YFP–c-Fos within the ERNF of HEK293 and COS7 cells, respectively (Fig. 1 B). Consistent with our previous confocal microscopy studies in MEFs (Ivorra et al., 2006), CFP-lamin A overexpression in NIH-3T3 and COS7 cells recruited YFP–c-Fos to the perinuclear rim (unpublished data). These results demonstrated that c-Fos accumulation at the NE and ERNF is dependent on lamin A/C expression.
Figure 1.
Hypophosphorylated c-Fos preferentially accumulates in the ERNF. Western blot analysis of whole cell extracts and subcellular fractions. Numbers on the side of the blots indicate molecular weight (in kD). (A) Wild-type and lamin A/C–null MEFs were starved for 24 h and stimulated for 90 min with PDGF-BB. Anti-Sp1 and anti-NUP50 antibodies showed equal protein loading and the absence of cross-contamination for the subnuclear fractionation protocol used. (B) HEK293 cells were transfected with pcDNA3 (control) or CFP-Lamin A, starved for 24 h, and stimulated for 90 min with 10% FBS. Asynchronously growing COS7 cells were cotransfected with YFP–c-Fos and either pcDNA3 (control) or CFP-Lamin A. The used antibodies and the detected proteins are indicated on the left and right, respectively. (C) Serum-stimulated NIH-3T3 cells. Arrows point to c-Fos species with different degree of phosphorylation. (D) SNF of serum-stimulated NIH-3T3 cells. Lysates were prepared in buffer with or without protease and phosphatase inhibitors and treated with alkaline phosphatase when indicated. (E) SNF and whole cell extract of starvation-synchronized NIH-3T3 cells stimulated with serum for the indicated time and treated with the proteasome inhibitor MG132 when indicated. (F) SNF of serum-stimulated NIH-3T3 cells preincubated with the ERK1/2 pathway inhibitor U0126 when indicated.
We next sought to investigate the molecular mechanisms regulating the association of c-Fos with the NE. Although two main species of c-Fos with different electrophoretic mobilities were detected in the SNF of NIH-3T3 cells restimulated with serum after a period of starvation, the faster migrating band predominated in the ERNF (Fig. 1 C). Incubation of the SNF with alkaline phosphatase increased c-Fos mobility (Fig. 1 D, lane 1 vs. 2), but this change was not seen in the presence of phosphatase and protease inhibitors (Fig. 1 D, lanes 3 and 4). Thus, the SNF of serum-stimulated cells accumulates hyperphosphorylated and hypophosphorylated c-Fos, whereas the ERNF predominantly accumulates the hypophosphorylated form of c-Fos. These results suggested that mitogen-dependent phosphorylation of c-Fos facilitates its release from the NE.
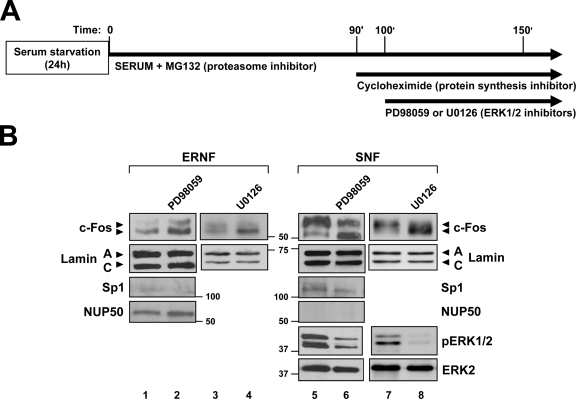
We then focused our attention on ERK1/2-dependent signaling, the main transduction pathway implicated in the rapid and transient phosphorylation and subsequent activation of c-Fos elicited by mitogens (Monje et al. 2003). As expected, Western blot analysis of the SNF of NIH-3T3 cells revealed the activation (phosphorylation) of ERK1/2 after serum stimulation (Fig. 1 E). Compared with serum-starved cells, the level of c-Fos protein was up-regulated in cultures stimulated for 90 min, which was markedly reduced after 180 min of serum stimulation (Fig. 1 E, lanes 1–3). Moreover, treatment with the proteasome inhibitor MG132 abrogated c-Fos down-regulation at 180 min after serum stimulation (Fig. 1 E, lane 5 vs. 3). These findings are in agreement with the concept that mitogen stimulation leads to de novo c-Fos expression, which then undergoes proteasome-dependent degradation (Bossis et al., 2003; Ito et al., 2005). In line with previous studies (Monje et al., 2003), we found that preincubation of serum-starved cells with the ERK1/2 inhibitor U0126 greatly reduces c-Fos level in cells stimulated with serum for 90 min (Fig. 1 F). Therefore, to circumvent the possible influence of ERK1/2 inhibitors on c-Fos synthesis and/or degradation, the effects of U0126 and PD98059 on c-Fos expression and subnuclear localization was investigated using the protocol schematized in Fig. 2 A. In brief, mitogen-depleted cells were stimulated with serum for 150 min in the presence of MG132 to avoid proteasome-dependent degradation of c-Fos. To prevent further protein synthesis after reaching a high level of c-Fos expression, cycloheximide was added at 90 min. Finally, U0126 or PD98059 was added at 100 min after serum stimulation. In agreement with the results of Fig. 1 C, the ERNF and SNF of NIH-3T3 cells manipulated as described above predominantly contained the fastest migrating hypophosphorylated and the two migrating forms of c-Fos, respectively (Fig. 2 B, ERNF: lanes 1 and 3; SNF: lanes 5 and 7). Treatment with either PD98059 or U0126 inhibited ERK1/2 activation, as revealed by the reduced amount of phosphorylated ERK1/2 (pERK1/2) in the SNF (Fig. 2 B, PD98059: lane 5 vs. 6; U0126: lane 7 vs. 8), and caused the accumulation of hypophosphorylated c-Fos both in the ERNF (Fig. 2 B, PD98059: lane 1 vs. 2; U0126: lane 3 vs. 4) and SNF (Fig. 2 B, PD98059: lane 5 vs. 6; U0126: lane 7 vs. 8). The pattern of expression of Sp1 and nucleoporin-50 + Npap60 (NUP50) suggested the absence of cross-contamination in these studies (Fig. 2 B). The results presented thus far indicated that the amount of c-Fos in the ERNF and perinuclear rim is regulated by the level of lamin A/C expression. They also suggested that inhibition of ERK1/2 activity reduces the extent of c-Fos phosphorylation and promotes its accumulation within the ERNF.
Figure 2.
ERK1/2 inhibition increases the amount of c-Fos associated to the ERNF. (A) Scheme of the protocol used to study the effects of post-transductional modifications of c-Fos on its accumulation within the ERNF and SNF, and on its interaction with lamin A. Cultures were serum starved for 24 h and then stimulated for 150 min with serum (U20S and COS7 cells: 10% FBS; NIH-3T3 cells: 10% NBS) to activate the MAPK pathway and to induce de novo c-Fos protein synthesis. The proteasome inhibitor MG132 (25 μM) was present throughout serum stimulation to prevent c-Fos degradation. After c-Fos induction (∼90 min), protein synthesis was inhibited with cycloheximide (10 μg/ml). When indicated, the ERK1/2 inhibitors PD98059 (20 μM) and U0126 (10 μM) were added for the last 50 min (B) Western blot analysis of the SNF and ERNF of NIH-3T3 cells treated as described above. Numbers on the side of the blots indicate molecular weight (in kD). Arrows point to c-Fos species with different degree of phosphorylation.
ERK1/2 inhibition increases the interaction of c-Fos with lamin A in vivo
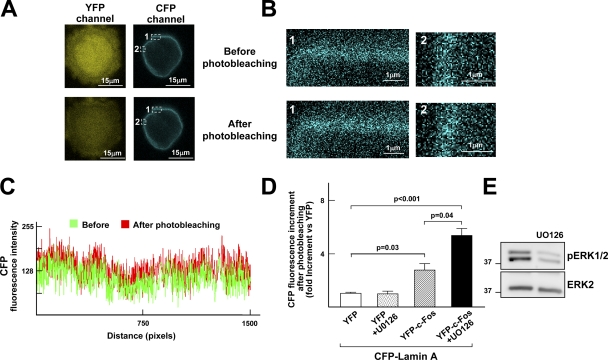
We next performed fluorescence resonance energy transfer (FRET) confocal microscopy studies to examine the in vivo interaction between c-Fos and lamin A. FRET efficiency determined as the increment in CFP fluorescence after YFP photobleaching (Kenworthy, 2001) was quantified in asynchronously growing COS7 cells cotransfected with CFP-lamin A and either YPF–c-Fos or YFP (as negative control). Fig. 3, A–C show representative images and intensity line profiles. In agreement with our previous FRET studies (Ivorra et al., 2006), cells cotransfected with CFP-lamin A/YFP–c-Fos exhibited a significant increase in FRET efficiency in the perinuclear rim compared with negative controls (Fig. 3 D). Addition of U0126 inhibited ERK1/2, as revealed by reduced ERK1/2 phosphorylation in the SNF (Fig. 3 E), and significantly increased FRET efficiency in cells cotransfected with CFP-lamin A/YFP but not with CFP-lamin A/YFP (Fig. 3 D). These results indicated that ERK1/2 inhibition increases the level of c-Fos/lamin A interaction in vivo.
Figure 3.
ERK1/2 inhibition increases the interaction between c-Fos and lamin A in vivo. FRET was measured in NIH-3T3 cells using the acceptor photobleaching method. Cells were transiently cotransfected with CFP-lamin A and either YFP (negative control) or YFP–c-Fos. Transfected cells were maintained in 10% NBS for 24 h and U0126 was added for the last 50 min when indicated. (A) Representative images of one YPF–c-Fos/CFP-lamin A–transfected U0126-treated cell before and after YFP photobleaching. The two perinuclear regions marked with white squares enlarged in B illustrate increased CFP fluorescence after photobleaching. (C) Fluorescence line profile analysis of CFP-lamin A intensity throughout the entire perinuclear rim shown in A before (green) and after (red) photobleaching of YFP. (D) Quantification of protein–protein interactions calculated as the percentage of CFP fluorescence increment after photobleaching in 20–30 cells from three independent experiments. (E) Western blot analysis of the SNF of cells maintained in 10% NBS treated or not with U0126. Numbers on the side of the blots indicate molecular weight (in kD).
Constitutive ERK1/2 activation reduces the presence of c-Fos in the ERNF
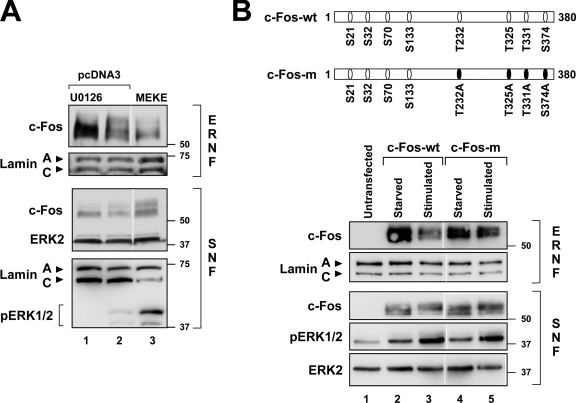
Consistent with the results of Fig. 2 B, treatment of U2OS cells with U0126 reduced the amount of hyperphosphorylated c-Fos in the SNF and ERNF fractions, and increased the amount of hypophosphorylated c-Fos within the ERNF (Fig. 4 A, lane 1 vs. 2). Overexpression of MEKE, a constitutively active form of MEK1, markedly augmented ERK1/2 phosphorylation in the SNF compared with controls transfected with empty vector (Fig. 4 A, pERK1/2, lane 2 vs. 3). This effect coincided with diminished retention of c-Fos in the ERNF, especially of the hyperphosphorylated species, which appeared to accumulate in the SNF (Fig. 4 A, lane 2 vs. 3). Thus, constitutive activation of ERK1/2 reduces the amount of ERNF-associated c-Fos.
Figure 4.
ERK1/2 constitutive activation reduces c-Fos accumulation in the ERNF and mutations that hinder ERK1/2-dependent phosphorylation of c-Fos impair its release from the ERNF. The SNF and ERNF of U2OS cells were analyzed by Western blot. Numbers on the side of the blots indicate molecular weight (in kD). (A) Cells were transfected with pcDNA3 or with a plasmid encoding MEKE, a constitutively active form of MEK1. 1 d after transfection, cells were subjected to the protocol shown in Fig. 2 A. (B) Scheme of the potentially phosphorylatable residues of wild-type c-Fos (c-Fos-wt) and c-Fos-m, which contains the following amino acid substitutions in residues phosphorylated by ERK1/2: T232A, T325A, T331A, S374A. Cells were transfected with a plasmid encoding c-Fos-wt or c-Fos-m. 1 d after transfection, cells were starved for 24 h and serum stimulated for 30 min when indicated. Of note, four putative sites for ERK1/2-dependent phosphorylation are intact in c-Fos-m (S21, S32, S70, S133).
Mutations that hinder ERK1/2-dependent phosphorylation of c-Fos impair its release from the ERNF and enhance its interaction with lamin A
We next sought to assess whether physiological levels of ERK1/2 activation can release c-Fos from the ERNF. To this end, we investigated in U2OS cells the subnuclear localization of ectopically expressed wild-type c-Fos (c-Fos-wt) and c-Fos-m, a mutant in which four residues that undergo ERK1/2-dependent phosphorylation are rendered unphosphorylatable (T232A, T325A, T331A, S374A) (Monje et al., 2003) (see scheme in Fig. 4 B). The level of serum-dependent ERK1/2 activation was similar in cells transfected with c-Fos-wt and c-Fos-m, as revealed by comparable increases in the amount of pERK1/2 in the SNF (Fig. 4 B, c-Fos-wt: lane 2 vs. 3; c-Fos-m: lane 4 vs. 5). Serum-inducible ERK1/2 activation coincided with reduced amount of ERNF-associated c-Fos-wt (Fig. 4 B, lane 2 vs. 3), but this effect was weaker for c-Fos-m (Fig. 4 B, lane 4 vs. 5). The mobility shift of mutated c-Fos-m upon stimulation is likely due to ERK1/2-dependent phosphorylation at site(s) that is(are) not important for lamin A/C association and possibly for c-Fos activation, per se (e.g., S21, S32, S70, S133; see scheme in Fig. 4 B). These findings suggested that serum-inducible phosphorylation of c-Fos through ERK1/2 activation is a physiological mechanism for releasing c-Fos from the ERNF.
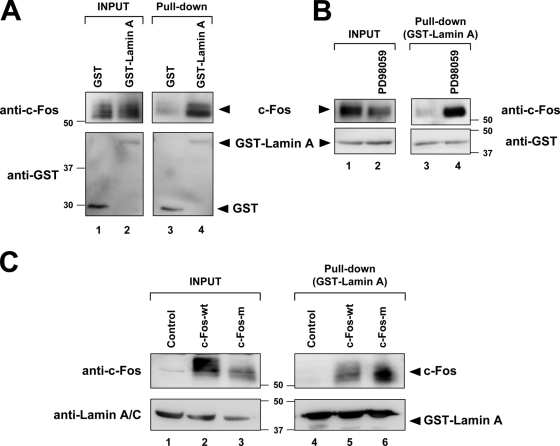
To directly examine whether ERK1/2-dependent phosphorylation of c-Fos affects its affinity for lamin A, we performed in vitro pull-down assays using a GST-lamin A fusion protein containing amino acids 37–244 of rat lamin A, which interacts with recombinant c-Fos (Ivorra et al., 2006). GST-lamin A specifically interacted with the endogenous c-Fos protein present in extracts from serum-stimulated NIH-3T3 cells (Fig. 5 A, lane 3 vs. 4). Moreover, the amount of GST-lamin A–bound c-Fos was greatly increased upon treatment of cells with PD98059 (Fig. 5 B, lane 3 vs. 4). We also compared the binding of GST-lamin A to c-Fos-wt and c-Fos-m, which were ectopically expressed in serum-stimulated U2OS cells. Examination of input samples suggested the presence of phosphorylation activity in these cultures because c-Fos-wt exhibited slower electrophoretic mobility than c-Fos-m (Fig. 5 C, lane 2 vs. 3). Moreover, GST-lamin A interacted more efficiently with c-Fos-m compared with c-Fos-wt (Fig. 5 C, lane 5 vs. 6). These results suggested that the interaction of c-Fos with lamin A is facilitated by inhibiting ERK1/2-mediated phosphorylation of c-Fos, either by mutational blockade of ERK1/2 phosphorylation sites in c-Fos or by pharmacological inhibition of the ERK1/2 pathway.
Figure 5.
ERK1/2-dependent phosphorylation of c-Fos affects its interaction with lamin A. GST-lamin A fusion protein containing amino acid residues 37–244 of rat lamin A was tested for its interaction with either endogenous c-Fos present in the SNF of NIH-3T3 cells (A and B) or with c-Fos-wt or c-Fos-m ectopically expressed in U2OS cells (C). Nuclear extracts were incubated with GST proteins and 1/20 of the reaction mixture was examined by Western blot (INPUT). The rest was precipitated with gluthatione-Sepharose 4B, washed and analyzed by Western blot. The used antibodies and the detected proteins are indicated. Numbers on the side of the blots indicate molecular weight (in kD). In B, cells were subjected to the protocol schematized in Fig. 2 A. In C, cells were transfected with c-Fos-wt or c-Fos-m. 24 h after transfection, cells were starved for 24 h and stimulated with serum for 30 min.
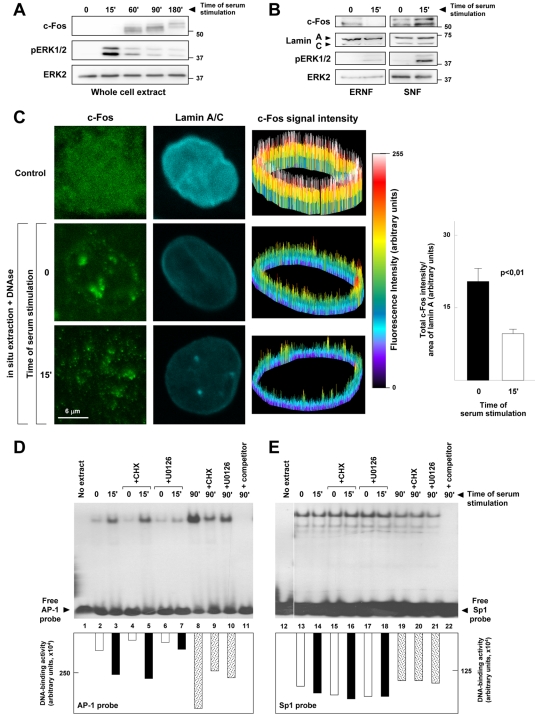
Serum-dependent activation of ERK1/2 rapidly releases preexisting c-Fos from the ERNF and induces AP-1 DNA-binding activity
The results presented thus far suggested that serum-inducible ERK1/2-dependent phosphorylation of c-Fos releases is from the inhibitory interaction with A-type lamins. To assess whether this process might be physiologically relevant, we investigated the effects of serum stimulation on the subnuclear localization of endogenous c-Fos and on AP-1 DNA-binding activity. In agreement with numerous studies, Western blot analysis of NIH-3T3 whole cell extracts demonstrated the activation of pERK1/2 and up-regulation of c-Fos after 15 and 60 min of serum stimulation, respectively (Fig. 6 A). Coinciding with the peak of ERK1/2, c-Fos expression diminished in the ERNF and accumulated within the SNF (Fig. 6 B). The presence of c-Fos in the NE and its release after 15 min of serum stimulation was also observed in double immunofluorescence confocal microscopy experiments with U2OS cells submitted to in situ extraction of soluble cytoplasmic and nucleoplasmic proteins (Fig. 6 C). Moreover, electrophoretic mobility shift assays (EMSA) revealed that 15 min of serum induction causes in NIH-3T3 cells a rapid increase in AP-1 DNA-binding activity in the SNF (Fig. 6 D, lanes 2 vs. 3), which was not affected by inhibiting de novo protein synthesis (lanes 4 vs. 5) but was blunted upon pharmacological inhibition of ERK1/2 (lanes 6 vs. 7). AP-1 DNA-binding activity was further increased after 90 min of serum stimulation (Fig. 6 D, lane 8) through a mechanism requiring both de novo protein synthesis (lane 9) and ERK1/2 activation (lane 10). Sp1 DNA-binding activity remained essentially unaffected by all the above manipulations (Fig. 6 E).
Figure 6.
Serum-inducible ERK1/2 activation correlates with the release of preexisting c-Fos from the ERNF and causes a rapid induction of AP-1 DNA-binding activity. (A and B) NIH-3T3 cells were starved for 24 h and stimulated with 10% NBS for the indicated times. Whole cell extracts (A), or SNF and ERNF (B) were analyzed by Western blot. Of note, the amount of protein analyzed in the blots of B was ∼14 times higher than in A. Numbers on the side of the blots indicate molecular weight (in kD). (C) Double immunofluorescence of c-Fos and lamin A/C in U2OS cells. Top images: Asynchronously growing control cells. Middle and bottom images: Starved and serum-stimulated cells in situ extracted and treated with DNase. Images in the right column show 3D representation of c-Fos signal intensity in lamin A/C–containing perinuclear rim. Graph shows the total intensity of c-Fos signal per total lamin A/C–containing perinuclear area of in situ extracted cells (n = 30 cells of each condition). (D and E) NIH 3T3 were starved for 24 h and stimulated with 10% NBS for the indicated times. When indicated, cycloheximide (CHX, 10 μg/ml) or U0126 (10 μM) was added 1 h before serum stimulation. EMSA was performed using 10 fmol of a consensus AP-1 (D) or Sp1 (E) radiolabeled probe and SNF (5 μg total protein). The graphs show the quantification of DNA-binding activity as assessed by densitometric analysis. Reactions in lanes 11 and 22 contained a 50-fold molar excess of competitor (unlabeled AP-1 or Sp1 consensus oligonucleotide, respectively).
Lamin A/C inhibits AP-1 activity and facilitates c-Fos phosphorylation
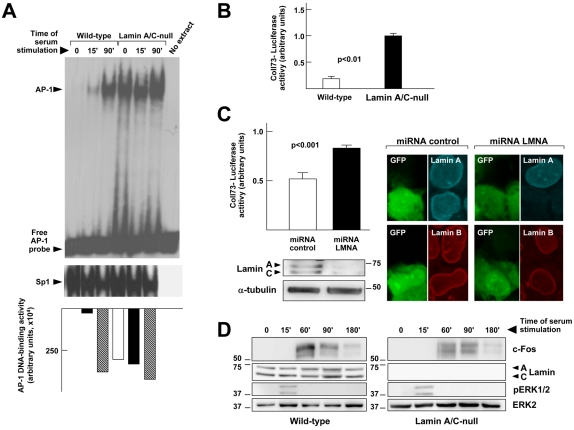
We have previously shown that lamin A/C overexpression suppresses AP-1 DNA binding and transcriptional activity (Ivorra et al., 2006). To gain further insight into the role of A-type lamins on AP-1 function, we compared the kinetics of AP-1 DNA binding and transcriptional activity in wild-type versus lamin A/C–null MEFs. Consistent with the results in NIH-3T3 cells (Fig. 6 D), EMSAs demonstrated a progressive increase in AP-1 DNA-binding activity in wild-type MEFs stimulated with serum for 15 and 90 min (Fig. 7 A). Compared with wild-type controls, two dramatic alterations in AP-1 DNA-binding activity were observed in lamin A/C–null MEFs: a marked induction under serum starvation, and a lack of activation at 15 min of serum restimulation; in contrast, Sp1 DNA-binding activity remained largely unaffected upon lamin A/C ablation (Fig. 7 A). As shown in Fig. 7 B, lamin A/C–null MEFs exhibited significantly higher AP-1-dependent transcriptional activity, as revealed in transient transfection assays with the AP-1–responsive coll73-luciferase reporter gene. Silencing lamin A/C expression with miRNA also increased activity of coll73-luciferase in U2OS cells (Fig. 7 C). Moreover, we found that both the maximum level of c-Fos protein and the relative abundance of its hyperphosphorylated form were reduced in serum-stimulated lamin A/C compared with wild-type MEFs (Fig. 7 D).
Figure 7.
Lamin A/C inhibits AP-1 activity and facilitates c-Fos phosphorylation. (A) EMSA using 10 fmol of a consensus AP-1 or Sp1 radiolabeled probe and SNF (15 μg total protein) from wild-type and lamin A/C–null MEFs starved for 48 h and stimulated with 10% FBS for different times. Graph shows the quantification of AP-1 DNA-binding activity after normalization with Sp1 DNA-binding activity as assessed by densitometric analysis. (B) Luciferase activity in extracts from wild-type and lamin A/C–null MEFs cotransfected with the AP-1–responsive coll73-luciferase and pGL4-Renilla luciferase reporters (n = 12). (C) Luciferase activity in extracts from U2OS cells cotransfected with coll73-luciferase and either miRNA-control/GFP or miRNA-LMNA/GFP (n = 12). The Western blot and the confocal microscopy images demonstrate efficient lamin A/C silencing by miRNA-LMNA/GFP. (D) Western blot analysis of whole cell extracts from starved and restimulated wild-type and lamin A/C–null MEFs. Numbers on the side of the blots indicate molecular weight (in kD).
ERK1/2 interacts with lamin A in vitro and in vivo
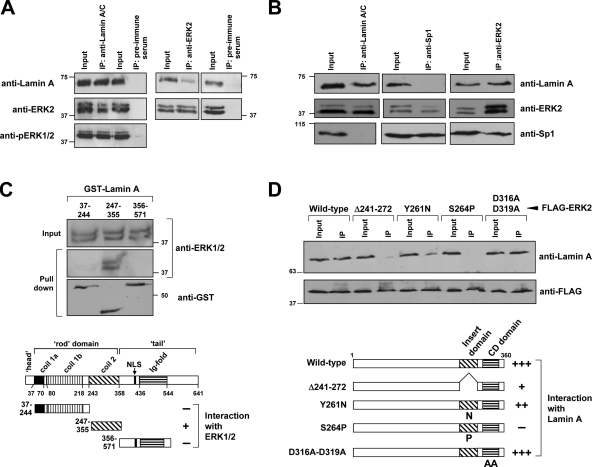
We next performed immunoprecipitation-coupled Western blot analysis to test whether ERK1/2 could directly interact with lamin A/C. Anti-lamin A serum, but not preimmune serum, coprecipitated lamin A and ERK1/2 from asynchronously growing HEK293 cells (Fig. 8 A). Moreover, anti-ERK2, but not preimmune serum, coprecipitated lamin A and ERK1/2 (Fig. 8 A). Of note, the anti-lamin A serum precipitated phosphorylated ERK1/2 (Fig. 8 A), suggesting that active ERK1/2 can interact with lamin A/C in vivo. In contrast, lamin A and the transcription factor Sp1 did not coimmunoprecipitate, whereas Sp1-ERK2 complexes could be immunoprecipitated with either anti-Sp1 or anti-ERK2 antibodies, in agreement with previous studies (Milanini-Mongiat et al., 2002).
Figure 8.
ERK1/2 interacts with lamin A in vitro and in vivo. (A, B) Lysates from HEK293 cells (stimulated 5 min with EGF after serum deprivation) were immunoprecipitated with either anti-lamin A/C (sc-20680), anti-ERK2 or anti-Sp1 antibodies. Preimmune serum was used as control. Both the input and the whole immunoprecipitated material were subjected to Western blot analysis using anti-ERK2, anti-pERK1/2, anti-Sp1 and anti-lamin A/C (sc-20680) antibodies as indicated. (C) GST-lamin A fusion proteins containing amino acid residues 37–244, 243–388 or 453–571 of rat lamin A were tested for their interaction with endogenous ERK1/2 from whole HEK293 cell extracts. Approximately 90% of the reaction mixture was precipitated with gluthatione-Sepharose 4B, washed and analyzed by Western blot. (D) Lysates from HEK293 cells overexpressing FLAG-tagged ERK2 (wild-type and several mutants) were immunoprecipitated with an anti-FLAG antibody. Both the input and the whole immunoprecipitated material were subjected to Western blot analysis using anti-FLAG and anti-lamin A/C (sc-20680) antibodies. In the scheme in C and D, (+) and (−) indicate positive and negative interactions, respectively. NLS: Nuclear localization signal. Numbers on the side of the blots indicate molecular weights (in kD).
GST pull-down assays revealed that amino acids 247–355 of lamin A/C, but not the fragments spanning amino acids 37–244 and 356–571, associated to ERK1/2 from HEK293 cells (Fig. 8 C). We extended these studies by determining the region in ERK2 responsible for binding to lamin A/C. In particular, we tested two interaction “hot spots”: the CD domain and the Insert region (Robinson et al., 1998). We found that mutations D316,319A that abrogate interactions mediated through the CD domain (Tanoue et al., 2000) did not affect the binding of ERK2 to lamin A/C. Conversely, an ERK2 Insert region deletion mutant (Δ241-272) (Whitehurst et al., 2004) and point mutants within this region, Y261N and S264Y, were impaired in their association with lamin A/C (Fig. 8 D).
Lamin A/C, ERK1/2, and c-Fos colocalize at the mammalian NE
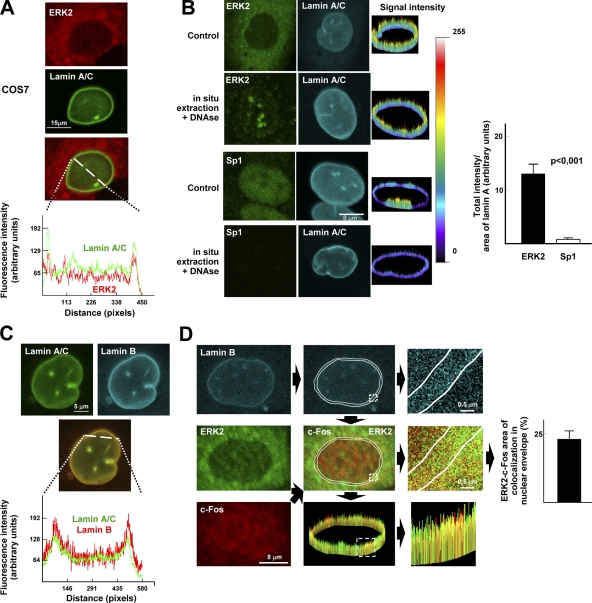
Double confocal immunofluorescence microscopy revealed areas of colocalization between endogenous ERK2 and lamin A/C at the perinuclear rim in COS7, U2OS, and HEK293 cells (Fig. 9 A, and unpublished data). Perinuclear colocalization of lamin A/C–ERK2 was also manifest after in situ extraction plus DNase treatment (Fig. 9 B). In contrast, perinuclear localization of Sp1 was negligible under the same experimental conditions (Fig. 9 B). We next examined the possible colocalization of c-Fos and ERK1/2 at the NE. Lamin A/C entirely colocalized with lamin B at the perinuclear rim (Fig. 9 C); therefore, NE-associated lamin A/C could be unequivocally recognized by localizing lamin B expression. Analysis of asynchronously growing U2OS cells co-immunostained for c-Fos, ERK2, and lamin B revealed partial colocalization of these proteins in the perinuclear rim (Fig. 9 D).
Figure 9.
ERK1/2 and c-Fos partially colocalize at the NE. Representative images of cells examined by confocal microscopy. (A) ERK2-lamin A/C double immunofluorescence in COS7 cells. Graph shows a line profile illustrating areas of colocalization. (B) Control and in situ–extracted plus DNase-treated U2OS cells were subjected to ERK2–lamin A/C or Sp1–lamin A/C double immunofluorescence. Images in the right column show 3D representation of ERK2 or Sp1 signal intensity in the lamin A/C–containing perinuclear rim of control and in situ–extracted/DNase-treated cells, and the graph shows the total intensity of ERK2 and Sp1 signal per total lamin A/C–containing perinuclear area in the in situ–extracted/DNase-treated cells (n = 30). (C) Double lamin A/C–lamin B immunofluorescence in U2OS cells. The graph shows a line profile illustrating nearly total colocalization. (D) ERK2–c-Fos–lamin B triple immunofluorescence in U2OS cells (left). Lamin B signal was used to delineate the nuclear envelope (white lines in middle column) to allow the quantification of perinuclear ERK2-c-Fos colocalization (yellow signal) shown in the graph, as determined by analyzing 50 cells.
Discussion
We recently reported that sequestration of c-Fos at the NE through its interaction with A-type lamins is a mechanism of suppressing AP-1–dependent transcription in mammalian cells (Ivorra et al., 2006). In the present study we provide evidence that disruption of lamin A/C constitutively activates AP-1–dependent DNA-binding and transcriptional activity. We also demonstrate that the interaction of c-Fos with lamin A/C is a reversible process controlled by mitogenic stimuli through ERK1/2-dependent signaling. First, we show that mitogen-dependent ERK1/2-mediated phosphorylation of c-Fos releases it from the inhibitory interaction with lamin A/C, allowing the rapid activation of AP-1 before de novo c-Fos protein synthesis. Second, we demonstrate that phosphorylated (active) ERK1/2 can interact with lamin A/C in vitro and in vivo, and colocalize with c-Fos at the NE. This scaffold role of lamin A/C appears to facilitate the phosphorylation of c-Fos elicited by ERK1/2 in cells stimulated with serum.
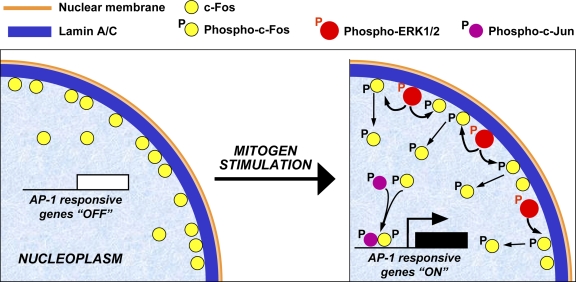
It is generally accepted that mitogen-dependent activation of ERK1/2 in fibroblasts is a biphasic process consisting of a rapid and strong burst of kinase activity peaking at 5–15 min, followed by a second wave of lower activity that persists throughout the G1 phase of the cell cycle (Kahan et al., 1992; Meloche et al., 1992; Meloche, 1995). c-Fos is expressed minimally in most nongrowing cells as well as during the initial phase of mitogen-induced ERK activation, with a peak of de novo c-Fos protein synthesis and AP-1 activation occurring between 1–2 h after stimulation (Kruijer et al., 1984; Kerr et al., 1988; Monje et al., 2003; Ivorra et al., 2006; Murphy and Blenis, 2006). In line with this notion, we observed maximum c-Fos protein expression at 60–90 min after serum stimulation (Fig. 1 E, Fig. 6 A), coinciding with a peak of AP-1 DNA-binding activity that could be attenuated by inhibitors of either protein synthesis or ERK1/2 activity (Fig. 6 D). Our novel observation is the induction of AP-1 DNA-binding activity as early as 15 min after serum addition concomitantly with maximum ERK1/2 activity and the release of c-Fos from the ERNF to the nucleoplasm in a manner dependent on ERK1/2 activation but independent of de novo protein synthesis (Fig. 6). Based on these findings and the observations that gain-of and loss-of ERK1/2 activity reduces and enhances, respectively, c-Fos accumulation within the ERNF and its interaction with lamin A, and that regulation of these processes is impaired by hindering ERK1/2-dependent phosphorylation of c-Fos, we propose the model depicted in Fig. 10. In serum-deprived cells, sequestration of c-Fos at the NE via its interaction with lamin A/C limits transcription of AP-1 target genes. Subsequent to mitogen stimulation, lamin A–bound active ERK1/2 phosphorylates c-Fos and releases it from the NE, thus allowing the rapid transcriptional activation of AP-1–responsive genes before de novo c-Fos protein synthesis. This mechanism permits the continuous existence in the cell of a certain level of inactive c-Fos (e.g., in quiescent cells) that can be rapidly activated by mitogen stimulation via ERK1/2-dependent phosphorylation without requiring de novo protein synthesis.
Figure 10.
Model for the rapid activation of AP1 upon mitogen stimulation. In serum-deprived cells, the c-Fos protein is expressed at low level predominantly in a hypophosphorylated state and associated to the NE through its interaction with A-type lamins. Under these conditions, transcription of AP-1 target genes is off. Upon mitogen stimulation, phosphorylated (active) ERK1/2 bound to lamin A/C phosphorylates c-Fos, causing its release from the NE. Hyperphosphorylated nucleoplasmic c-Fos can then heterodimerize with AP-1 family members (e.g., c-Jun), thus allowing the transcriptional activation of AP-1–responsive genes before de novo c-Fos synthesis.
Intracellular compartmentalization through interactions among MAPKs and scaffold proteins plays an important role in the regulation of signal transduction pathways (Morrison and Davis, 2003; Kolch, 2005). For example, active MEK–ERK complexes are retained in the cytoplasm and in sites of focal adhesion through interaction with the transmembrane protein Sef (Torii et al., 2004) and paxilin (Ishibe et al., 2004), respectively. Moreover, kinase suppressor of Ras 1 (Muller et al., 2001) and the complex p14-MEK-partner 1 (Teis et al., 2002; Pullikuth et al., 2005) are recruited to the plasma membrane and the endosome, respectively, where they enhance MEK and ERK activity. On the other hand, the association between ERKs and structural nuclear proteins, such as kinetochores, may serve anchoring purposes (Shapiro et al., 1998). To the best of our knowledge, we show here for the first time that A-type lamins function as a nuclear docking platform for EKR1/2, and that NE-bound ERK1/2 contributes to the rapid activation of AP-1 via releasing c-Fos from the interaction with A-type lamins (see above). Indeed, AP-1–dependent DNA-binding and transcriptional activity were significantly higher in lamin A/C–null versus wild-type MEFs and in U2OS cells in which lamin A/C expression was silenced with miRNA (Fig. 7, A–C). Our results also suggest that the efficiency of serum-inducible activation (phosphorylation) of the small preinduction levels of c-Fos elicited by ERK1/2 is aided by their colocalization at the NE (Fig. 7 D). We are currently investigating whether ERK1/2–lamin A/C complex formation may help prolong the activation state of ERK1/2 by maintaining these signal transducers in a reservoir out of the reach of soluble nuclear phosphatases. It will be also of interest to examine whether A-type lamins serve a scaffolding function only for ERK1/2-dependent activation of c-Fos, or whether induction of other transcription factors also requires the activity of NE-associated signal transducers.
The Insert and the CD domains of ERK1/2 are involved in protein–protein interactions. ERK1/2 substrate docking can be selectively dissociated in vitro by single point mutations without perturbing ERK activation or its intrinsic catalytic activity (Dimitri et al., 2005). Indeed, mutations affecting the Insert region (Δ241-272, Y261N, and S264P) or the CD domain (D316,319A) alter the interaction of ERK2 with some substrates (Tanoue et al., 2000; Whitehurst et al., 2004; Casar et al., 2007). We have shown here that the ERK2–lamin A/C interaction requires the Insert domain of ERK2 and the coil 2 of lamin A/C. Other proteins such as PEA15 (Whitehurst et al., 2004) and Mxi2 (Casar et al., 2007) also interact with ERK through the Insert region. The determinant mediating in this interaction is a “reverse D domain”, with a consensus sequence R/K-φa-X3/4-φb (where φ are Leu, Ile, or Val) (Callaway et al., 2005). Notably, the R/K-φa-X3/4-φb motif is present in the region of A-type lamins, which we found to intervene in their interaction with ERK1/2 (RIRISL at residues 296–301).
In this study, we have focused our attention on how mitogenic signals regulate the interaction between c-Fos and lamin A/C. Further research is required to assess whether additional pathophysiological conditions regulate this interaction and the underlying molecular mechanisms. Of notable interest in this regard are the inherited diseases termed laminopathies, which are caused by either mutations in the LMNA gene (which encodes for A-type lamins) or defective posttranslational processing of prelamin A (Worman and Bonne, 2007). Notably, lamin A/C and lamin-associated polypeptides can physically interact with histones, chromatin, and transcription factors (e.g., c-Fos, SREBP1, MOK2, BAF, GCL, Mel18), suggesting that altered gene expression contributes to the pathogenesis of laminopathies (Taniura et al., 1995; Gruenbaum et al., 2005; Broers et al., 2006; Heessen and Fornerod, 2007; Vlcek and Foisner, 2007). Indeed, microarray studies using fibroblast from Hutchinson-Gilford progeria syndrome patients revealed differential transcription factor expression (Ly et al., 2000; Csoka et al., 2004). Moreover, certain pathogenic lamin A mutations cause alterations in the transcription factors MOK2 (Dreuillet et al., 2008) and SREBP1 (Lloyd et al., 2002; Hubner et al., 2006) and in the cell cycle regulator pRb (Hubner et al., 2006). Interestingly, transgenic mice overexpressing either wild-type MEK5 or constitutively active MEK1 exhibit excessive ERK1/2-dependent signaling and dilated cardiomyopathy (Bueno et al., 2000; Nicol et al., 2001), a clinical manifestation of several laminopathies. Moreover, expression of the Emery-Dreifuss muscular dystrophy–causing H222P–lamin A mutant protein in homozygous lmnaH222P/H222P knock-in mice causes in the heart aberrant ERK1/2 activity and myopathy, and leads to activation of several MAPKs—including ERK1/2—and downstream target genes in cultured cells (Muchir et al., 2007). It is also noteworthy that MEK/ERK pathway inhibition improves defective myogenic factor expression and differentiation of C2C12 myoblasts expressing the Emery-Dreifuss muscular dystrophy–causing R453W–lamin A mutant (Favreau et al., 2004, 2008). In the light of the aforementioned studies and the findings reported herein, it will be of interest to assess whether pathogenic lamin A/C mutants alter the interaction of c-Fos and/or ERK1/2 at the NE, and if so, whether they affect the regulation and function of AP-1 target genes.
Materials and Methods
Antibodies
Primary antibodies directed against c-Fos (sc-52), lamin A/C (sc-7292 in Figs. 1 B, 4, 6 C, 7 C and 9; sc-7293 in Figs. 1 A, C–F, 2, 5, 6 A and B, and 7 D; and sc-20680 in Fig. 8), lamin B (sc-6217), GST (sc-138), ERK2 (sc-154 and sc-1647 only in Fig. 9 D), pERK1/2 (sc-7383), Sp1 (sc-59-G), α-tubulin (sc-8035), anti-FLAG, and HRP-coupled secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-NUP50 and anti-GFP were from Abcam (ab4005) and Invitrogen (A6455), respectively.
Plasmids
pECFP-YFP was constructed by inserting YFP into pECFP-C1 (gift from J. López-Giménez, University of Glasgow, Scotland). The following plasmids are described elsewhere: pGEX-lamin A (37–244), pGEX-lamin A (247–355), and pGEX-lamin A (356–571) (gift from T. Ozaki, Chiba Cancer Center Research Institute, Japan) (Ozaki et al., 1994); pEYFP-c-Fos and pECFP-lamin A (Ivorra et al., 2006); pmycCMV-ERK2-MEK1 (Robinson et al., 1998); pCDNAI-HA-ERK2 (Crespo et al., 1994); pCEFL-MEKE (Sanz-Moreno et al., 2003); pCMV-FLAG-ERK2; pCMV-FLAG-ERK2-Y261N; pCMV-FLAG-ERK2-Δ241-272; pCMV-FLAG-ERK2-S264P and pCMV-FLAG-ERK2-D316A-D319A (Robinson et al., 2002); pCEFL-AU5-c-Fos (c-Fos-wt); and pCEFL-AU5-c-Fos-mut (c-Fos-m, which contains the following mutations: T232A, T325A, T331A, and S374A; gift from S. Gutkind, National Institutes of Health, MD; see Fig. 4 B) (Monje et al., 2003).
Cell culture
NIH-3T3, COS-7, HEK293, U20S, and HeLa cells were obtained from the American Type Culture Collection. Lmna-null and littermate wild-type MEFs are described elsewhere (Sullivan et al., 1999). Cells maintained in DME supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mmol/L l-glutamine (Invitrogen) and 10% FBS (or 10% NBS for NIH-3T3 cells) were incubated at 37°C in a 5% CO2/95% O2 atmosphere. Cultures were serum starved for 24 h and then stimulated with either 10% FBS, 10% NBS, PDGF-BB (20 ng/ml; Sigma-Aldrich), or EGF (100 ng/ml; Sigma-Aldrich). For ERK1/2 inhibition, PD98059 (20 μM; Tocris) or U0126 (10 μM; Promega) were added before serum stimulation. MG132 and cycloheximide were from Sigma-Aldrich. When indicated, cells were treated as schematized in Fig. 2 A.
In vitro pull-down assays
GST proteins were purified using glutathione-Sepharose 4B (GE Healthcare) and eluted with 50 mM Tris-Cl (pH 8.0). For the experiments of Fig. 5, GST proteins and cell lysates from NIH-3T3 cells were incubated in 20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF supplemented with complete protease inhibitor cocktail (Roche). After 16 h of incubation at 4°C, glutathione-Sepharose 4B was added to a final concentration of 10% and agitated at 4°C for 45 min. The beads were collected by centrifugation and washed three times with 20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF. For the experiments of Fig. 8 C, whole cell lysates from HEK293 cells were incubated with GST-lamin A bound to glutathione-Sepharose 4B beads. After 4 h of incubation at 4°C, the beads were collected by centrifugation and washed twice with 1% NP-40/PBS. In all cases, pellets were air-dried, resuspended in 2x Laemmli buffer, boiled for 5 min, and separated onto 12% SDS–polyacrylamide gels (SDS-PAGE).
Subcellular fractionation, immunoprecipitation, and immunoblot experiments
Immunoprecipitation and Western blot analysis were performed as previously described (Ivorra et al., 2006; Casar et al., 2007). Subcellular fractionation was performed as described by Schreiber et al. (1989) with minor modifications (Ivorra et al., 2006). In brief, cells were washed with PBS and scraped into TEN buffer (150 mM NaCl, 1 mM EDTA, and 40 mM TrisCl at pH 7.4). Cells were collected by brief centrifugation in microfuge tubes and resuspended in 10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and 0.5 mM PMSF. After 15 min on ice, Nonidet NP-40 (Fluka) was added to a final concentration of 10%, and tubes were vortexed. Lysates were centrifuged at 4°C in a microfuge set at maximum speed to obtain the soluble cytoplasmic fraction (supernatant) and the nuclear pellet, which was resuspended in ice-cold 20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF, and agitated at 4°C for 15 min. The nuclear lysate was centrifuged for 45 min at 4°C to obtain the SNF and the pellet containing the ERNF.
In situ nuclear matrix isolation and indirect immunofluorescence analysis
In situ nuclear matrix isolation was performed as described previously (Fey et al., 1984). In brief, cells grown on coverslips were washed in PBS and extracted twice in cytoskeleton buffer (CSK: 100 mM NaCl, 300 mM sucrose, 10 mM PIPES (pH 6.8), 3 mM MgCl2, 0.5% Triton X-100, and 1.2 mM PMSF) for 10 min at 0°C. The resulting soluble fraction was removed. Extraction buffer (250 mM ammonium sulfate, 300 mM sucrose, 10 mM PIPES (pH 6.8), 3 mM MgCl2, 1.2 mM PMSF, and 0.5% Triton X-100) was added to the Triton X-100 insoluble structures for 10 min at 0°C and the cytoskeleton fraction was removed. DNase digestion was performed twice in digestion buffer (CSK buffer containing 100 μg/ml DNase I and 50 mM NaCl) followed by extraction in digestion buffer containing 0.25 M (NH4)2SO4. In situ– extracted and control cells were fixed in 4% formaldehyde/PBS and permeabilized with 0.5% Triton X-100. All samples were blocked for 5 min with 10 mM glycine (pH 8.5) and 1 h with 5% dry milk in 10% FBS, 0.5% BSA, 0.1% Triton X-100, and PBS, followed by an overnight incubation at 4°C with anti-c-Fos (1:100), anti-ERK2 (1:100), or anti-Sp1 (1:100) antibodies. Samples were then incubated with species-appropriate FITC-conjugated secondary antibody. After washes and incubation with anti-lamin A/C (1:100; sc-7292) for 1 h at room temperature, specimens were washed and incubated with an anti–mouse secondary antibody conjugated to Alexa 633 (1:300).
Phosphatase treatment
NIH-3T3 cells were rapidly washed with cold PBS and collected for subcellular fractionation as described above. When indicated, fractionation was performed in the absence of phosphatase inhibitors, DTT and PMSF. Aliquots of the SNF were incubated in the absence or presence of 1 U of alkaline phosphatase (Roche) for 1 h at 37°C. Reactions were stopped by adding SDS sample buffer and processed for immunoblotting.
EMSA
Double-stranded oligonucleotides containing the AP-1 (5′-CGCTTGATGAGTCAG-3′; AP-1 site underlined) and the Sp1 (5′-ATTCGATCGGGGCGGGGCGAGC-3′; Sp1 site underlined) consensus sites were labeled with γ[32P]dATP using polynucleotide kinase (New England Biolabs, Inc.) and purified on a Sephadex G-50 column. EMSA was performed using the SNF of NIH-3T3 cells (5 μg total protein) and wild-type and lamin A/C–null mice MEFs (15 μg total protein) as previously described (Ivorra et al., 2006).
Confocal microscopy
Images were acquired on a laser confocal microscope (TCS/SP2; Leica) with a 63x oil immersion objective (NA 1.4). For single lamin A/C (Fig. 7 C) and lamin B (Fig. 7 C), double ERK2–lamin A/C (Fig. 9 A) and lamin B–lamin A/C (Fig. 9 C), and triple ERK2–c-Fos–lamin B immunofluorescence (Fig. 9 D): cells were fixed with 4% PFA/PBS at room temperature (RT) on glass coverslips and permeabilized using 0.5% Triton X-100. Samples were blocked for 5 min with 10 mM glycine (pH 8.5) and 1 h with 5% dry milk in 10% FBS, 0.5% BSA, 0.1% Triton X-100, and PBS before an overnight incubation at 4°C with anti-lamin A/C (1:100; sc-7292) or anti-lamin B antibody (1:100). In double immunofluorescence experiments, samples were incubated with anti-ERK2 (1:100) or anti-lamin B (1:100) antibodies for 1 h at room temperature after anti-lamin A/C antibody incubation. For triple immunofluorescence, samples were incubated simultaneously with anti-c-Fos (1:100) and anti-ERK2 (1:100; sc-1647) antibodies after anti-lamin B antibody incubation. Finally, secondary antibodies conjugated to Alexa 488, Texas red, and Alexa 633 (Molecular Probes, Inc.) were used (1:300). Image quantification was done using MetaMorph software (MDS Analytical Technologies).
FRET
NIH-3T3 cells were cotransfected with pECFP-lamin A + pEYFP-c-Fos or pECFP-Lamin A + EYFP as a negative control (1 μg each plasmid) using Lipofectamine (Invitrogen). Cotransfection of pECFP-YFP + pcDNA3 (0.5 μg each) was used as positive control to calibrate the system. Images were acquired on a confocal microscope (TCS/SP2; Leica) with a 63x oil immersion objective (NA 1.4). An argon laser line of 458 nm was used to excite CFP (PMT window 465–505 nm) and a 514-nm line (20% laser intensity for acquisition and 65% for photobleaching) to excite YFP (PMT window: 525–600 nm). Studies were performed in 4% paraformaldehyde-fixed cells using the acceptor-photobleaching method (Kenworthy, 2001) as previously described (Ivorra et al., 2006), in which FRET is calculated as the relative increase in donor fluorescence as a result of the reduction or elimination of energy transfer when the acceptor is photobleached. Specifically, we used the following equation: FRET = (Cafter − Cbefore)/Cafter x 100, where Cbefore and Cafter are the total fluorescence intensity of the CFP channel before and after photobleaching, respectively. For negative values, this parameter was considered 0.
Luciferase gene reporter assays
Wild-type and lamin A/C–null MEFs were transiently transfected with 5 μg of AP-1–dependent coll 73-luciferase reporter plasmid and pGL4-Renilla luciferase using the calcium phosphate method. U2OS were transfected with 5 μg of coll73-luciferase plus either a plasmid encoding for a miRNA-control/GFP or miRNA-LMNA/GFP (BLOCK-IT; Invitrogen). After 48 h in 10% FBS, cells were harvested and luciferase activity was measured following the manufacturer's instructions (dual luciferase reporter assay system; Promega). Luciferase activity and GFP expression were measured in a luminometer (Victor). miRNA-transfected cells were also fixed in 4% PFA and studied by immunofluorescence confocal microscopy, or lysed for Western blot analysis.
Statistical analysis
Results are reported as mean ± SE. In experiments with two groups, differences were evaluated using a two-tail, unpaired Student's t test. One-way ANOVA and Bonferroni's post hoc test was used for experiments involving more than two groups.
Acknowledgments
We thank M.J. Andrés-Manzano for help with the preparation of figures, and D. Barettino for critical reading of the manuscript. We are also grateful to colleagues who generously provided reagents (S. Gutkind: c-Fos-wt, c-Fos-m; T. Ozaki: pGEX-lamin A; J. López-Giménez: pECFP-YFP; R. Foisner, and C.L. Stewart: wt and lamin A–null MEFs). V. Andrés' laboratory is supported by grants SAF2004-03057 and SAF2007-6211 (Spanish Ministry of Science and Innovation–MICINN, and European Regional Development Fund -FEDER), and RD06/0014/0021 (Red Temática de Investigación Cooperativa en Enfermedades Cardiovasculares, Instituto de Salud Carlos III –ISCIII).
P. Crespo's laboratory is supported by grants BFU2005-00777 and GEN2003-20239-C06-03 (MICINN), the EU Sixth Framework Program under the GROWTHSTOP (LSHC CT-2006-037731) and SIMAP (IST-2004-027265) projects, and RD06/0020/0105 (Red Temática de Investigación Cooperativa en Cáncer, ISCIII). J.M. González received salary support from ISCIII and a research grant from Generalitat Valenciana (GVPRE/2008/163).
Abbreviations used in this paper: AP-1, activator protein 1; c-Fos, cellular homologue of the Finkel-Biskis-Jinkins osteosarcoma virus gene; EMSA, electrophoretic mobility shift assay; ERK, extracellular signal-regulated kinase; ERNF, extraction-resistant nuclear fraction; FRET, fluorescence resonance energy transfer; MEF, mouse embryonic fibroblast; NE, nuclear envelope; pERK1/2, phosphorylated ERK1/2; pRb, retinoblastoma protein; SNF, soluble nuclear fraction.
References
- Abate, C., D.R. Marshak, and T. Curran. 1991. Fos is phosphorylated by p34cdc2, cAMP-dependent protein kinase and protein kinase C at multiple sites clustered within regulatory regions. Oncogene. 6:2179–2185. [PubMed] [Google Scholar]
- Abe, M.K., W.L. Kuo, M.B. Hershenson, and M.R. Rosner. 1999. Extracellular signal-regulated kinase 7 (ERK7), a novel ERK with a C-terminal domain that regulates its activity, its cellular localization, and cell growth. Mol. Cell. Biol. 19:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis, G., P. Ferrara, C. Acquaviva, I. Jariel-Encontre, and M. Piechaczyk. 2003. c-Fos proto-oncoprotein is degraded by the proteasome independently of its own ubiquitinylation in vivo. Mol. Cell. Biol. 23:7425–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers, J.L., F.C. Ramaekers, G. Bonne, R.B. Yaou, and C.J. Hutchison. 2006. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 86:967–1008. [DOI] [PubMed] [Google Scholar]
- Bueno, O.F., L.J. De Windt, K.M. Tymitz, S.A. Witt, T.R. Kimball, R. Klevitsky, T.E. Hewett, S.P. Jones, D.J. Lefer, C.F. Peng, et al. 2000. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 19:6341–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, K., M.A. Rainey, and K.N. Dalby. 2005. Quantifying ERK2-protein interactions by fluorescence anisotropy: PEA-15 inhibits ERK2 by blocking the binding of DEJL domains. Biochim. Biophys. Acta. 1754:316–323. [DOI] [PubMed] [Google Scholar]
- Casar, B., V. Sanz-Moreno, M.N. Yazicioglu, J. Rodriguez, M.T. Berciano, M. Lafarga, M.H. Cobb, and P. Crespo. 2007. Mxi2 promotes stimulus-independent ERK nuclear translocation. EMBO J. 26:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., C. Abate, and J. Blenis. 1993. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA. 90:10952–10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., P.C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene. 12:1493–1502. [PubMed] [Google Scholar]
- Crespo, P., N. Xu, W.F. Simonds, and J.S. Gutkind. 1994. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature. 369:418–420. [DOI] [PubMed] [Google Scholar]
- Csoka, A.B., S.B. English, C.P. Simkevich, D.G. Ginzinger, A.J. Butte, G.P. Schatten, F.G. Rothman, and J.M. Sedivy. 2004. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 3:235–243. [DOI] [PubMed] [Google Scholar]
- Deng, T., and M. Karin. 1994. c-Fos transcriptional activity stimulated by H-Ras-activated protein kinase distinct from JNK and ERK. Nature. 371:171–175. [DOI] [PubMed] [Google Scholar]
- Dimitri, C.A., W. Dowdle, J.P. MacKeigan, J. Blenis, and L.O. Murphy. 2005. Spatially separate docking sites on ERK2 regulate distinct signaling events in vivo. Curr. Biol. 15:1319–1324. [DOI] [PubMed] [Google Scholar]
- Dreuillet, C., M. Harper, J. Tillit, M. Kress, and M. Ernoult-Lange. 2008. Mislocalization of human transcription factor MOK2 in the presence of pathogenic mutations of lamin A/C. Biol. Cell. 100:51–61. [DOI] [PubMed] [Google Scholar]
- Eferl, R., and E.F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 3:859–868. [DOI] [PubMed] [Google Scholar]
- Favreau, C., D. Higuet, J.C. Courvalin, and B. Buendia. 2004. Expression of a mutant lamin A that causes Emery-Dreifuss muscular dystrophy inhibits in vitro differentiation of C2C12 myoblasts. Mol. Cell. Biol. 24:1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau, C., E. Delbarre, J.C. Courvalin, and B. Buendia. 2008. Differentiation of C2C12 myoblasts expressing lamin A mutated at a site responsible for Emery-Dreifuss muscular dystrophy is improved by inhibition of the MEK-ERK pathway and stimulation of the PI3-kinase pathway. Exp. Cell Res. 314:1392–1405. [DOI] [PubMed] [Google Scholar]
- Fey, E.G., K.M. Wan, and S. Penman. 1984. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J. Cell Biol. 98:1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., A. Margalit, R.D. Goldman, D.K. Shumaker, and K.L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31. [DOI] [PubMed] [Google Scholar]
- Heessen, S., and M. Fornerod. 2007. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 8:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C.S., and R. Treisman. 1995. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 80:199–211. [DOI] [PubMed] [Google Scholar]
- Hubner, S., J.E. Eam, A. Hubner, and D.A. Jans. 2006. Laminopathy-inducing lamin A mutants can induce redistribution of lamin binding proteins into nuclear aggregates. Exp. Cell Res. 312:171–183. [DOI] [PubMed] [Google Scholar]
- Hunter, T., and M. Karin. 1992. The regulation of transcription by phosphorylation. Cell. 70:375–387. [DOI] [PubMed] [Google Scholar]
- Ishibe, S., D. Joly, Z.X. Liu, and L.G. Cantley. 2004. Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol. Cell. 16:257–267. [DOI] [PubMed] [Google Scholar]
- Ito, Y., D. Inoue, S. Kido, and T. Matsumoto. 2005. c-Fos degradation by the ubiquitin-proteasome proteolytic pathway in osteoclast progenitors. Bone. 37:842–849. [DOI] [PubMed] [Google Scholar]
- Ivorra, C., M. Kubicek, J.M. Gonzalez, S.M. Sanz-Gonzalez, A. Alvarez-Barrientos, J.E. O'Connor, B. Burke, and V. Andres. 2006. A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev. 20:307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan, C., K. Seuwen, S. Meloche, and J. Pouyssegur. 1992. Coordinate, biphasic activation of p44 mitogen-activated protein kinase and S6 kinase by growth factors in hamster fibroblasts. Evidence for thrombin-induced signals different from phosphoinositide turnover and adenylylcyclase inhibition. J. Biol. Chem. 267:13369–13375. [PubMed] [Google Scholar]
- Kenworthy, A.K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 24:289–296. [DOI] [PubMed] [Google Scholar]
- Kerr, L.D., J.T. Holt, and L.M. Matrisian. 1988. Growth factors regulate transin gene expression by c-fos-dependent and c-fos-independent pathways. Science. 242:1424–1427. [DOI] [PubMed] [Google Scholar]
- Kolch, W. 2005. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6:827–837. [DOI] [PubMed] [Google Scholar]
- Kruijer, W., J.A. Cooper, T. Hunter, and I.M. Verma. 1984. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 312:711–716. [DOI] [PubMed] [Google Scholar]
- Lloyd, D.J., R.C. Trembath, and S. Shackleton. 2002. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 11:769–777. [DOI] [PubMed] [Google Scholar]
- Ly, D.H., D.J. Lockhart, R.A. Lerner, and P.G. Schultz. 2000. Mitotic misregulation and human aging. Science. 287:2486–2492. [DOI] [PubMed] [Google Scholar]
- Mariappan, I., and V.K. Parnaik. 2005. Sequestration of pRb by cyclin D3 causes intranuclear reorganization of lamin A/C during muscle cell differentiation. Mol. Biol. Cell. 16:1948–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche, S. 1995. Cell cycle reentry of mammalian fibroblasts is accompanied by the sustained activation of p44mapk and p42mapk isoforms in the G1 phase and their inactivation at the G1/S transition. J. Cell. Physiol. 163:577–588. [DOI] [PubMed] [Google Scholar]
- Meloche, S., K. Seuwen, G. Pages, and J. Pouyssegur. 1992. Biphasic and synergistic activation of p44mapk (ERK1) by growth factors: correlation between late phase activation and mitogenicity. Mol. Endocrinol. 6:845–854. [DOI] [PubMed] [Google Scholar]
- Milanini-Mongiat, J., J. Pouyssegur, and G. Pages. 2002. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 277:20631–20639. [DOI] [PubMed] [Google Scholar]
- Monje, P., M.J. Marinissen, and J.S. Gutkind. 2003. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol. Cell. Biol. 23:7030–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, D.K., and R.J. Davis. 2003. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19:91–118. [DOI] [PubMed] [Google Scholar]
- Muchir, A., P. Pavlidis, V. Decostre, A.J. Herron, T. Arimura, G. Bonne, and H.J. Worman. 2007. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 117:1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D.K. Morrison. 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell. 8:983–993. [DOI] [PubMed] [Google Scholar]
- Murphy, L.O., and J. Blenis. 2006. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 31:268–275. [DOI] [PubMed] [Google Scholar]
- Murphy, L.O., S. Smith, R.H. Chen, D.C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4:556–564. [DOI] [PubMed] [Google Scholar]
- Nicol, R.L., N. Frey, G. Pearson, M. Cobb, J. Richardson, and E.N. Olson. 2001. Activated MEK5 induces serial assembly of sarcomeres and eccentric cardiac hypertrophy. EMBO J. 20:2757–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, T., M. Saijo, K. Murakami, H. Enomoto, Y. Taya, and S. Sakiyama. 1994. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene. 9:2649–2653. [PubMed] [Google Scholar]
- Piechaczyk, M., and J.M. Blanchard. 1994. c-fos proto-oncogene regulation and function. Crit. Rev. Oncol. Hematol. 17:93–131. [DOI] [PubMed] [Google Scholar]
- Pullikuth, A., E. McKinnon, H.J. Schaeffer, and A.D. Catling. 2005. The MEK1 scaffolding protein MP1 regulates cell spreading by integrating PAK1 and Rho signals. Mol. Cell. Biol. 25:5119–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, F.L., A.W. Whitehurst, M. Raman, and M.H. Cobb. 2002. Identification of novel point mutations in ERK2 that selectively disrupt binding to MEK1. J. Biol. Chem. 277:14844–14852. [DOI] [PubMed] [Google Scholar]
- Robinson, M.J., S.A. Stippec, E. Goldsmith, M.A. White, and M.H. Cobb. 1998. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 8:1141–1150. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno, V., B. Casar, and P. Crespo. 2003. p38alpha isoform Mxi2 binds to extracellular signal-regulated kinase 1 and 2 mitogen-activated protein kinase and regulates its nuclear activity by sustaining its phosphorylation levels. Mol. Cell. Biol. 23:3079–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, E., P. Matthias, M.M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, P.S., E. Vaisberg, A.J. Hunt, N.S. Tolwinski, A.M. Whalen, J.R. McIntosh, and N.G. Ahn. 1998. Activation of the MKK/ERK pathway during somatic cell mitosis: direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J. Cell Biol. 142:1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks, A.D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2:827–837. [DOI] [PubMed] [Google Scholar]
- Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131–E136. [DOI] [PubMed] [Google Scholar]
- Sullivan, T., D. Escalante-Alcalde, H. Bhatt, M. Anver, N. Bhat, K. Nagashima, C.L. Stewart, and B. Burke. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura, H., C. Glass, and L. Gerace. 1995. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 131:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos, T., M.J. Marinissen, F.C. Leskow, D. Hochbaum, H. Martinetto, J.S. Gutkind, and O.A. Coso. 2005. Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J. Biol. Chem. 280:18842–18852. [DOI] [PubMed] [Google Scholar]
- Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110–116. [DOI] [PubMed] [Google Scholar]
- Teis, D., W. Wunderlich, and L.A. Huber. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell. 3:803–814. [DOI] [PubMed] [Google Scholar]
- Torii, S., M. Kusakabe, T. Yamamoto, M. Maekawa, and E. Nishida. 2004. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell. 7:33–44. [DOI] [PubMed] [Google Scholar]
- Vlcek, S., and R. Foisner. 2007. A-type lamin networks in light of laminopathic diseases. Biochim. Biophys. Acta. 1773:661–674. [DOI] [PubMed] [Google Scholar]
- Whitehurst, A.W., F.L. Robinson, M.S. Moore, and M.H. Cobb. 2004. The death effector domain protein PEA-15 prevents nuclear entry of ERK2 by inhibiting required interactions. J. Biol. Chem. 279:12840–12847. [DOI] [PubMed] [Google Scholar]
- Worman, H.J., and G. Bonne. 2007. “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 313:2121–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]