Abstract
Correct intranuclear organization of chromosomes is crucial for many genome functions, but the mechanisms that position chromatin are not well understood. We used a layered screen to identify Saccharomyces cerevisiae mutants defective in telomere localization to the nuclear periphery. We find that events in S phase are crucial for correct telomere localization. In particular, the histone chaperone Asf1 functions in telomere peripheral positioning. Asf1 stimulates acetylation of histone H3 lysine 56 (H3K56) by the histone acetyltransferase Rtt109. Analysis of rtt109Δ and H3K56 mutants suggests that the acetylation/deacetylation cycle of the H3K56 residue is required for proper telomere localization. The function of H3K56 acetylation in localizing chromosome domains is not confined to telomeres because deletion of RTT109 also prevents the correct peripheral localization of a newly identified S. cerevisiae “chromosome-organizing clamp” locus. Because chromosome positioning is subject to epigenetic inheritance, H3K56 acetylation may mediate correct chromosome localization by facilitating accurate transmission of chromatin status during DNA replication.
Introduction
The three-dimensional organization of DNA within the nucleus is related to chromatin function, and the genomic DNA is organized and actively positioned within the nuclear space (Gasser, 2002). Nucleosome packaging and histone modification have been implicated in chromatin domain positioning (Tanabe et al., 2002; Chambeyron and Bickmore, 2004; Zink et al., 2004; Williams et al., 2006), but the precise mechanisms determining physical organization of chromosomes are not well understood.
Telomeres are the nucleoprotein structures that protect chromosome ends from degradation and ensure the telomerase-mediated length maintenance of the terminal repeat sequences. The organization of the telomeres of budding yeast Saccharomyces cerevisiae offers a useful model system to study chromosome localization (Gasser, 2002). The 32 telomeres of haploid yeast cells are organized in three to six clusters at the nuclear periphery (Gotta et al., 1996). S. cerevisiae subtelomeric sequences are subject to transcriptional silencing (Gottschling et al., 1990) and replicate late during S phase (Raghuraman et al., 2001). Recombination between telomeres and other chromosomal loci is suppressed (Pryde et al., 1997). Therefore, budding yeast telomeres are “insulated” from other chromosomal regions both spatially and functionally.
Two partially redundant pathways have been identified that mediate tethering of telomeres to the nuclear periphery: the Sir4-dependent and Ku-dependent pathways (Hediger et al., 2002; Taddei et al., 2004). Sir4 is a telomere-binding protein that, along with Sir2 and Sir3, is essential for transcriptional silencing of telomere-proximal genes. Sir4 tethers telomeres to nuclear periphery through interactions with Esc1 and Mps3 on the nuclear membrane (Taddei et al., 2004; Bupp et al., 2007). The Ku protein complex is a heterodimer consisting of Yku70 and Yku80, which binds to telomeres and is essential for their late replication (Cosgrove et al., 2002), as well as playing a major role in telomere length regulation (Porter et al., 1996). The Ku heterodimer can also mediate peripheral tethering of telomeres, but the interaction partner of Ku on the nuclear membrane has not yet been identified. Mutants lacking both Ku and Sir4 function show random positioning of all telomeres tested, but single mutants compromise the peripheral localization of different telomeres to different extents, which suggests that the 32 telomeres vary in their dependence on the two pathways for peripheral tethering (Bystricky et al., 2005). Telomere peripheral tethering is cell cycle–regulated: telomeres generally show perinuclear localization in G1 and S phases but not in G2 or M phases.
Telomeres are not the only genomic sequences to show localization to the nuclear periphery. Other yeast genomic loci that display peripheral localization include transcriptionally activated GAL1 and INO1 genes (Brickner and Walter, 2004; Casolari et al., 2004; Schmid et al., 2006), and in Schizosaccharomyces pombe, the so-called “chromosome-organizing clamp” (COC) sites that behave as insulator elements (Noma et al., 2006). S. cerevisiae HMR-silenced chromatin is also peripherally localized, and Sir4 and Ku proteins contribute to HMR localization and confinement of its mobility within the nuclear space (Gartenberg et al., 2004). Therefore, mechanisms involved in telomere localization may act more generally in subnuclear organization, and identification of the factors involved in telomere localization may provide insight into mechanisms controlling the subnuclear organization of other chromosome sites.
Asf1 is an evolutionarily conserved histone H3–H4 complex (H3–H4) chaperone that is implicated in several chromatin-remodeling pathways (Mousson et al., 2007). Asf1 is involved along with other histone chaperones in both replication-independent and replication-coupled histone deposition pathways (Mousson et al., 2007). Asf1 stimulates acetylation of the histone H3 lysine 56 (H3K56) and loading of H3 acetylated at K56 into newly assembled chromatin (Recht et al., 2006; Schneider et al., 2006), acting along with the histone chaperones CAF-1 and Rtt106, which preferentially load H3 acetylated at K56 during replication-coupled chromatin assembly (Li et al., 2008). Asf1 is also required under certain circumstances for integrity of the replication machinery (Mousson et al., 2007), and the replicative clamp-loading complex replication factor C binds Asf1 and can recruit it to DNA in vitro (Franco et al., 2005). These observations strongly suggest that Asf1 acts in replication-coupled histone deposition through direct interaction with the replication machinery. Asf1 has also been implicated in telomere-specific chromatin organization because either disruption or overexpression of ASF1 causes defective transcriptional silencing at telomeres (Singer et al., 1998).
Rap1 is a telomere-binding protein involved in telomere length regulation and recruitment of the Sir protein complex to telomeres (Kanoh and Ishikawa, 2003). Here, we have used GFP-Rap1 as a marker protein to screen for mutants with defective telomere organization. After secondary testing of these mutants, we identified eight new genes that are required for telomere localization to the nuclear periphery. The genes identified suggest that an event in S phase is important for telomere localization. In particular, we identify the chromatin-packaging histone chaperone Asf1 as one of the components required for telomere localization, and show that its stimulation of H3K56 acetylation is crucial for correct telomere positioning at the nuclear periphery. We demonstrate the existence of a peripherally localized S. cerevisiae COC site, and show that its positioning also depends on H3K56 acetylation. Collectively, our results suggest that H3K56 acetylation is crucially important for correct three-dimensional organization of chromosomes within the nucleus.
Results
Rap1 organization mutants
To identify mutants defective in telomere localization, we used a candidate-based layered screening approach. Telomere repeat sequences contain multiple Rap1-binding sites, and in cells expressing a GFP-Rap1 fusion protein, telomere clusters are detected as bright fluorescent foci at the nuclear periphery (Hayashi et al., 1998; Hiraga et al., 2006). We began by screening for mutants defective in spatial organization of Rap1 in the nucleus. 262 knockout mutants in nonessential genes were selected for screening based on the proposed function of their gene products in chromosome metabolism processes such as telomere maintenance, transcriptional silencing, or DNA replication, recombination, or repair, as well as localization to nuclear membrane or nuclear periphery (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1).
We transformed this panel of mutants with an episomal plasmid expressing GFP-Rap1 (Hiraga et al., 2006), and screened for those mutants that have aberrant Rap1 organization. The strains were observed microscopically, and each strain tested was categorized according to the type of GFP-Rap1 organization it displayed. Four different basic phenotypes were observed, as illustrated in Fig. 1.
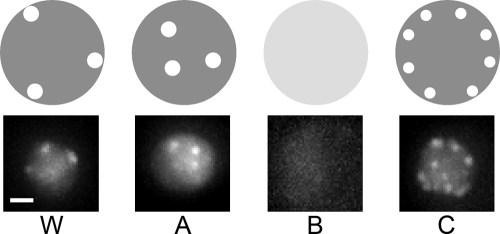
Figure 1.
Categories of mutants with defective Rap1 organization. The four observed categories of Rap1 organization phenotype are illustrated in the top panels. W, WT-like (i.e., no distinct Rap1 organization defect); A, internal foci; B, diffused fluorescence in entire nucleus; C, larger number of foci mainly still localized at nuclear periphery. Microscopic images on the bottom show strains typical for each category: W, WT; A, ace2Δ; B, ctf18Δ; C, chl1Δ. Bar, 1 μm.
Type W mutants displayed no significant difference from the wild type (WT), as would be expected if the mutation carried does not affect telomere localization or spatial Rap1 organization. 169 of the mutants tested fell in this category, and these mutants were not further examined.
For type A mutants, the internal Rap1 dots observed were similar to WT in number and intensity, but the dots were located in the interior of the nucleus rather than correctly positioned at the periphery. ace2Δ was a typical example of this mutant category. Mutants that are defective in perinuclear telomere localization but not in telomere clustering or length control may potentially fall in this category.
Type B mutants displayed loss of any obvious GFP-Rap1 dots. Typical examples of mutants falling in this category are asf1Δ and ctf18Δ. Various factors could cause this phenotype, such as telomere localization and/or clustering defects, severe telomere shortening causing loss of Rap1 binding sites, or a defect in telomere binding by Rap1. Partly because the Rap1 fluorescence of a single unclustered telomere is so weak as to be hardly detectable, the telomere localization phenotype cannot be unequivocally assessed for type B mutants on the basis of the Rap1 localization pattern alone, and requires further investigation (see the following sections).
Type C mutants displayed an increased number of Rap1 dots at nuclear periphery, with individual dots less intense than normal. Typical examples in this category are chl1Δ and tdp1Δ mutants. Mutants in this category are likely to be proficient in telomere localization to the nuclear periphery, with the mutant phenotype instead probably reflecting a defect in telomere clustering. However, mutants with a very severe telomere clustering defect may not fall in this category, because if a mutant exhibits a clustering defect so severe that GFP-Rap1 foci are not detectable, the mutant will be classified as type B.
Mutants of types A, B, and C were also numerically scored (1–3) for severity and uniformity of the phenotype. Mutants with a uniformly severe phenotype (i.e., the majority of the cells in the population displaying the defect) were assigned a score of 3. Mutants with less severe phenotypes and/or mutants in which only a small proportion of the population showed defects were given a lower score. The complete list of phenotype scores and assignments is shown in Table S1.
The exact nature of the defect in each mutant cannot be determined simply from the observed Rap1 organization patterns, and further tests were required to understand the nature of the abnormality and to ascertain whether telomere peripheral localization is compromised. We decided to examine type A and B mutants further, as well as mutants with mixed phenotypes that were partly A or B. Type B mutants were of particular interest because previously identified telomere localization-defective mutants (including yku70Δ and sir4Δ) fell into this category. A type B GFP-Rap1 pattern does not, however, necessarily imply a telomere localization defect, as this phenotype could be caused by other abnormalities (such as telomere shortening or a severe clustering defect). Type A mutants also display a phenotype that is suggestive of telomere mislocalization and were good candidates for further study, although this category is prone to false positives arising from technical difficulty in judging whether GFP-Rap1 dots located close to the poles in a z series are perinuclear. The type C pattern suggests proficient telomere localization to the nuclear periphery, and these mutants have not as yet been further examined. 23 phenotypically A, B, or “mixed” mutants with numerical scores of 2 or 3 were chosen for further investigation (listed in Table I).
Table I. Summary of Rap1 organization mutants further examined.
| Gene | ORF name | Rap1 phenotype | Score |
|---|---|---|---|
| ACE2 | YLR131C | A | 2 |
| ASF1 | YJL115W | B | 3 |
| BRE2 | YLR015W | B | 3 |
| CSM4 | YPL200W | B | 2 |
| CTF18 | YMR078C | B | 3 |
| CTF4 | YPR135W | B | 3 |
| ELP3 | YPL086C | B | 2 |
| ELP4 | YPL101W | B | 2 |
| GBP2 | YCL011C | A | 2 |
| IMD4 | YML056C | A | 2 |
| JJJ1 | YNL227C | A/C | 2 |
| MRE11 | YMR224C | B | 2 |
| PGD1 | YGL025C | B | 3 |
| PHO23 | YNL097C | B | 2 |
| RAD27 | YKL113C | B | 2 |
| RRM3 | YHR031C | B | 3 |
| RSC2 | YLR357W | B | 2 |
| SIC1 | YLR079W | A/B | 2 |
| SIR3 | YLR442C | B | 3 |
| SPE1 | YKL184W | A | 2 |
| SWI3 | YJL176C | B | 2 |
| TEL1 | YBL088C | B | 2 |
| YKU80 | YMR106C | B | 3 |
Mutants were categorized as described in the text. Only mutants with a phenotype score of 2 or 3 are listed for further examination.
Telomere localization mutants
We wished to assess directly whether telomere peripheral localization was affected in the mutants chosen for further analysis. A telomere can be microscopically visualized using the “chromosome dot” system, in which a chromosome site is “tagged” using GFP-LacI bound to an array of lacO operator repeats inserted at the required locus (Straight et al., 1996). Deletion alleles for the various genes were transferred into a strain background where the chromosomal locus 11 kb away from telomere XIV-left (XIV-L) is visualized using this chromosome dot technique (Fig. 2 A; Hediger et al., 2002). In the strain background used, the nuclear envelope is also marked using a GFP-fused allele of the nuclear pore component Nup49 (Belgareh and Doye, 1997). Perinuclear telomere localization was then scored by means of the “three-zoning” method (Taddei et al., 2004). In brief, cells were scored only if the telomere “dot” was observed to be close to the equatorial region in a z stack of images (Fig. 2 B, left). If this criterion was met, localization of the telomere was then scored to one of three concentric zones with equal surface area (Fig. 2 B, right). If the telomere distributes randomly within the nucleus, localization to each zone will be 33.3%. Conversely, if the telomere is localized at the nuclear periphery, it will be scored predominantly in zone 1 (as for WT; Fig. 2, A and C). The cell cycle stage of individual cells was estimated according to bud size (see Materials and methods), and telomere localization in G1 and S phase cells was separately scored (Fig. 2 C). Cells in G2 and M phases were not counted because telomeres lose their nuclear peripheral tethering during these late cell cycle phases.
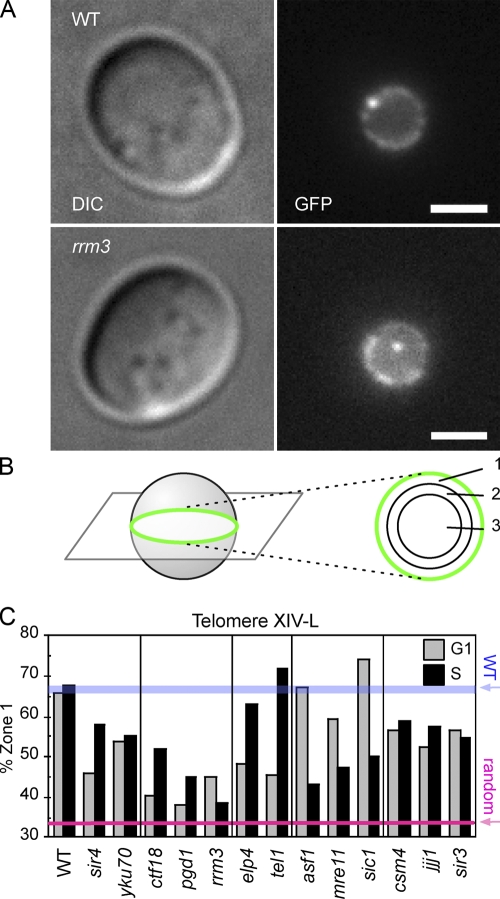
Figure 2.
Chromosome dot assay identifies a mutant with defective telomere localization to the nuclear periphery. (A) Typical images of WT and rrm3Δ strains with a chromosomal GFP tag 11 kb away from telomere XIV-L, seen as a bright dot. The strains also express Nup49-GFP to visualize nuclear membrane, seen as a dimmer ring. Differential interference contrast (DIC) images are shown on the left. Bars, 2 μm. (B) The zoning of images close to the equatorial plane is used to evaluate telomere localization. Localization of the GFP dot was scored against three concentric zones with equal surface area, as described in Materials and methods. (C) Telomere localization in the identified mutants, assessed separately for cells in G1 phase and S phase. The percentage of cells whose telomere XIV-L dot was peripheral (i.e., in zone 1) is plotted. The red line represents the distribution (33.3%) of a randomly positioned locus. WT values are also indicated (blue bar). For statistical details, see Table II.
Using this assay, 8 of the 23 mutants were observed to have a telomere localization defect; these were ctf18Δ, pgd1Δ, rrm3Δ, elp4Δ, tel1Δ, asf1Δ, mre11Δ, and sic1Δ. The results for these mutants are presented in Fig. 2 C, which shows the percentage of telomeres observed at the nuclear periphery (i.e., in zone 1) for the telomere localization-defective strains. Statistical analysis of these mutants is summarized in Table II. Data for all the mutants tested (including those that were telomere localization-proficient) is summarized in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1). sir4Δ and yku70Δ mutants were included in this analysis as controls because these mutants have previously been shown to display defective telomere localization (Hediger et al., 2002).
Table II. Summary of telomere localization mutants.
| Mutant | ORF name | Rap1 phenotype
|
% zone 1 (n,a p-valueb)
|
||
|---|---|---|---|---|---|
| Type | Score | G1 | S | ||
| Controls | |||||
| WT | 66.0 (295, 1.00) | 67.5 (336, 1.00) | |||
| sir4Δ | YDR227W | B | 3 | 46.0 (124, 1.5 × 10−5) | 58.2 (91, 0.01) |
| yku70Δ | YMR284W | B | 3 | 54.4 (184, 1.5 × 10−4) | 55.7 (87, 2.5 × 10−5) |
| G1 and S | |||||
| ctf18Δ | YMR078C | B | 3 | 40.2 (247, 5.3 × 10−12) | 51.9 (212, 0.002) |
| pgd1Δ | YGL025C | B | 3 | 37.9 (112, 3.5 × 10−11) | 44.8 (95, 6.2 × 10−7) |
| rrm3Δ | YHR031C | B | 3 | 45.2 (62, 0.002) | 38.4 (86, 2.0 × 10−8) |
| G1 specific | |||||
| elp4Δ | YPL101W | B/C | 2 | 45.7 (35, 0.01) | 71.9 (32, 0.68) |
| tel1Δ | YBL088C | B | 2 | 48.1 (54, 0.002) | 63.0 (46, 0.80) |
| S specific | |||||
| asf1Δ | YJL115W | B | 3 | 67.4 (46, 0.05) | 43.1 (51, 4.7 × 10−4) |
| mre11Δ | YMR224C | B | 2 | 59.4 (32, 0.66) | 47.3 (55, 5.9 × 10−4) |
| sic1Δ | YLR079W | A/B | 2 | 74.2 (31, 0.49) | 50.0 (42, 0.002) |
| Marginal | |||||
| csm4Δ | YPL200W | B | 2 | 56.8 (37, 0.31) | 59.0 (39, 0.26) |
| jjj1Δ | YNL227C | A/C | 2 | 52.3 (65, 0.03) | 57.6 (39, 0.26) |
| sir3Δ | YLR442C | B | 3 | 56.5 (46, 0.31) | 54.5 (44, 0.07) |
Localization of telomere XIV-L in each strain was measured as described in Materials and methods.
Total number of cells scored in each cell cycle phase.
Calculated by χ2 test against WT value.
Three of the mutants, ctf18Δ, pgd1Δ, and rrm3Δ, were found to be defective in telomere localization during both G1 and S phases. Each of the three mutations did, however, have slightly different impact on telomere localization at each cell cycle stage. Two mutants, elp4Δ and tel1Δ, are defective in G1 phase but not in S phase. Three mutations, asf1Δ, mre11Δ, and sic1Δ, have a distinct telomere localization defect in S phase, but no obvious defect in G1 phase. These cell cycle–specific defects of the various mutants suggest that different mechanisms are responsible for telomere localization in G1 and S phases.
3 of the 23 mutants, csm4Δ, jjj1Δ, and sir3Δ, displayed slight defects in telomere XIV-L peripheral localization in both in G1 and S phases. However, statistical analysis failed to demonstrate that these effects are significant (Table II).
Of the 23 mutants carried forward, 18 mutants were successfully examined for localization of telomere XIV-L by using the chromosomal GFP dot technique. Five of the mutants were left untested for technical reasons (see Table S2), usually because of difficulties in obtaining the required chromosome dot strains.
No correlation between telomere localization and telomere length
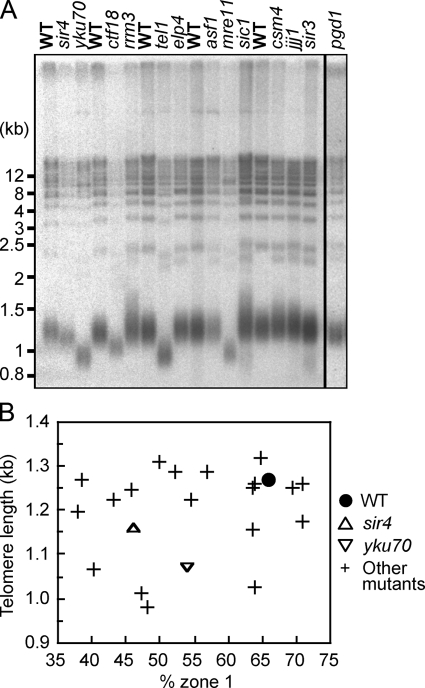
Some of the genes identified are implicated in telomere length regulation (Askree et al., 2004; Gatbonton et al., 2006). To investigate whether telomere length directly regulates telomere localization, we measured telomere length in the localization mutants (Fig. 3 A). Telomeres in mre11Δ, tel1Δ, and yku70Δ mutants are significantly shorter than in WT. Although the effect is smaller, ctf18Δ, pgd1Δ, and sir4Δ mutants also have short telomeres. Subpopulations of rrm3Δ and sic1Δ mutants have abnormally long telomeres. No distinct effect on telomere length was observed in asf1Δ and elp4Δ mutants. These results are consistent with previously published data (Askree et al., 2004; Gatbonton et al., 2006). To investigate any relationship between telomere localization and telomere length, telomere length was plotted against the “% zone 1” value for all the mutants tested for telomere localization (Fig. 3 B). For each mutant, the lower of the G1 and S phase localization values was used in this assessment. As shown in Fig. 3, no clear correlation between telomere length and localization proficiency was observed. Telomere length was also plotted against G1 and S phase localization values separately (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1). Again, no direct correlation between telomere length and telomere localization defect was observed. Because some mutants display short but well-localized telomeres, whereas others contain mislocalized telomeres of normal length, we conclude that telomere shortening is neither necessary nor sufficient to cause a telomere localization defect.
Figure 3.
Telomere delocalization does not cause shortened telomeres, and short telomeres do not preclude peripheral localization. (A) Telomere lengths of telomere localization-defective mutant strains were tested as described in Materials and methods. The result for pgd1Δ is from a different gel performed under identical condition to the others. Positions of molecular weight markers are shown on the left. The black line indicates that lanes have been spliced out. (B) Mean length of the terminal fragment derived from Y′ telomeres plotted against “% zone 1” value of each mutant. For all mutants, the “% zone 1” values differ in G1 and S phases; the lower value is shown. (e.g., for the asf1Δ mutant, “% zone 1” values in G1 and S phase are 67.4% and 43.1%, respectively; the value plotted is 43.1%.)
Histone acetyltransferase Rtt109 is required for telomere localization
The asf1Δ mutant displayed a severe telomere localization defect during S phase but had telomeres of normal length. Asf1 is a histone chaperone involved in assembling the histone H3–H4 subcomplex into nucleosomes, playing a role in histone deposition during DNA replication as well as transcription-coupled chromatin remodeling. Asf1 binding to H3–H4 also stimulates acetylation of H3K56 in vitro and is required for efficient acetylation of H3K56 in vivo.
We tested which activity of Asf1 is responsible for telomere localization. Asf1 is implicated in nucleosome loading/unloading and chromatin remodeling through its cooperation with other histone chaperones, Hir and CAF-1. If this function of Asf1 is important for telomere localization, disruption of Hir or CAF-1 components should also affect telomere localization. However, disruption of the genes encoding these histone chaperones (HIR1, HIR2, RLF2, CAC2, or MSI1) had little or no effect on Rap1 organization (Table S1), which suggests that neither Hir nor CAF-1 is crucial for telomere localization. We cannot, however, exclude the possibility that redundant function of CAF-1 and Rtt106 in loading H3–H4 into nucleosomes (Li et al., 2008) masked an involvement of these histone chaperones in Rap1 organization.
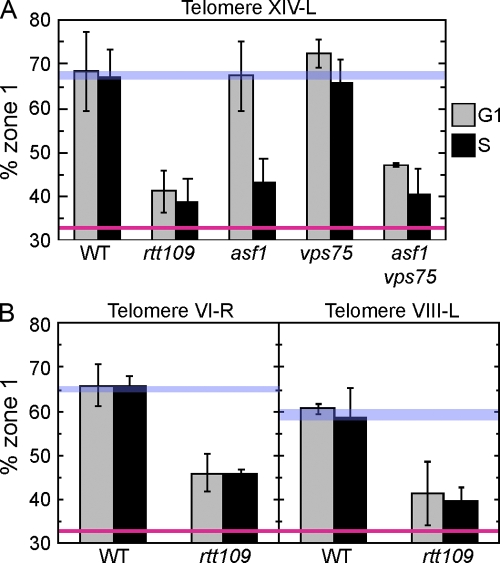
We next tested the alternative possibility that acetylation of H3K56 is required for telomere localization. The RTT109 gene encodes a histone acetyltransferase responsible for acetylation of H3K56 in vivo (Schneider et al., 2006; Driscoll et al., 2007; Han et al., 2007a; Tsubota et al., 2007). Both in vivo and in vitro studies confirmed that Asf1 dramatically stimulates the catalytic activity of Rtt109. If H3K56 acetylation is important for telomere localization, telomere localization will be compromised in the rtt109Δ mutant, which is deficient for H3K56 acetylation. As shown in Fig. 4, we found using the chromosome dot assay that the rtt109Δ mutant exhibits a severe telomere localization defect for all telomeres tested (telomeres XIV-L, VI-R, and VIII-L; Fig. 4, A and B). Interestingly, the defect of rtt109Δ was cell cycle–independent, in contrast to the S phase–specific defect of the asf1Δ mutant. This discrepancy suggests the possibility that Asf1-dependent acetylation of H3K56 is responsible for telomere localization only in S phase, whereas another protein with similar activity could play a role in localization during G1 phase. The VPS75 gene encodes a recently identified histone H3–H4 chaperone (Selth and Svejstrup, 2007). In vitro studies demonstrated that, like Asf1, Vps75 can stimulate acetylation of H3K56 by Rtt109 (Han et al., 2007b; Tsubota et al., 2007). Hence, Vps75 is a candidate for a G1 phase–specific counterpart of Asf1. We tested whether Vps75 is involved in telomere localization. A vps75Δ single mutant showed no detectable telomere localization defect, which demonstrates that VPS75 is dispensable for telomere localization in the presence of ASF1 (Fig. 4 A). We investigated whether Asf1 and Vps75 might play redundant roles in telomere localization during G1 phase. Consistent with this idea, an asf1Δ vps75Δ double mutant showed a severe telomere localization defect in both G1 and S phases, essentially recapitulating the phenotype of rtt109Δ (Fig. 4 A). To summarize, we found that Asf1 and Vps75 function redundantly in telomere localization during G1 phase, whereas only Asf1 can mediate correct telomere localization during S phase.
Figure 4.
Histone acetyltransferase Rtt109 and histone chaperones Asf1 and Vps75 are crucial for telomere localization. (A) Localization of telomere XIV-L was tested as in Fig. 2. Strains used were GA-1985 (WT), SHY196 (rtt109Δ), SHY173 (asf1Δ), SHY227 (vps75Δ), and SHY229 (asf1Δ vps75Δ). (B) Localization of telomeres VI-R and VIII-L was tested as in Fig. 2. Strains used were GA-1459 (WT) and SHY298 (rtt109Δ) for telomere VI-R, and GA-1986 (WT) and SHY299 (rtt109Δ) for telomere VIII-L. Error bars represent SD of values obtained from independent strain isolates (n = 4 for asf1Δ vps75Δ strain, n = 3 for rtt109Δ strain in A, n = 2 for others). WT and random localization values are indicated by blue and red lines, respectively.
Acetylation of H3K56 is crucial for telomere localization
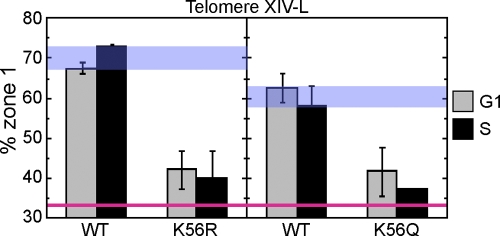
To assess the importance of H3K56 in telomere localization directly, we examined the competence for telomere localization of mutants where H3K56 is substituted by the nonacetylatable residue arginine (K56R) or glutamine (K56Q), which may mimic constitutive acetylation (Hyland et al., 2005; Masumoto et al., 2005; Ozdemir et al., 2005; Xu et al., 2005). We found that telomere localization was almost completely disrupted in either mutant in both G1 and S phases, with telomere XIV-L showing virtually random localization within the nuclear space (Fig. 5). This defect is very similar to the telomere localization defects observed for the rtt109Δ mutant and asf1Δ vps75Δ mutants (Fig. 4). The fact that the K56Q shows a very similar defect as the K56R mutant (Fig. 5) suggests that constitutively acetylated or deacetylated H3K56 would not support telomere localization. Together, these results demonstrate that both acetylation and deacetylation of H3K56 are crucial for telomere localization to the nuclear periphery. The effect of H3K56 acetylation in telomere localization appears to be rather specific because deleting components of other histone-modifying enzymes (including components of the ADA and NuA3 histone acetyltransferase complexes, the SAS complexes, and the SET3 histone deacetylase complex) caused little or no Rap1 organization defect (Table S1).
Figure 5.
Acetylation of H3K56 is crucial for telomere localization. Localization of telomere XIV-L (GFP-tagged 19 kb from telomere) in WT, K56R, and K56Q strains was tested as in Fig. 2. (left) Telomere localization in strains carrying either WT histone H3 allele (SHY248) or K56R allele (SHY247). (right) Telomere localization in strains with either a WT histone H3 allele (SHY310) or K56Q allele (SHY304). Histones H3 and H4 are encoded by only one gene in each strain. Error bars represent SD of values obtained from independent strain isolates (n = 2). Note that the “% zone 1” values shown here for telomere XIV-L are not directly comparable with the other figures because the GFP-tagged locus is different. WT and random localization values are indicated by blue and red lines, respectively. The difference in WT values between left and right panels is presumably caused by the different strain constructs used.
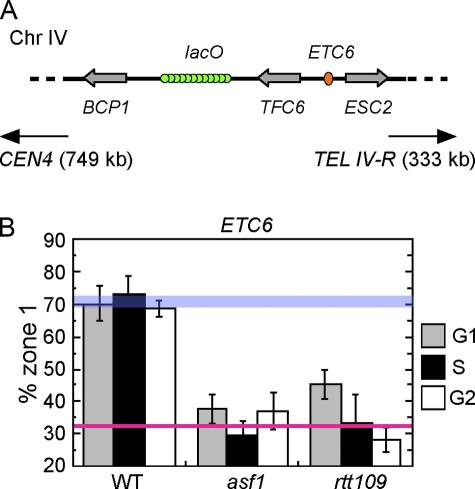
Peripheral positioning of the ETC6 chromosome locus
We wished to test whether the effect of H3K56 acetylation on chromatin intranuclear positioning is limited to telomeres, or whether instead other chromosome loci require the same histone modification for peripheral positioning. In the fission yeast S. pombe, COC sites have recently been identified that associate with the nuclear periphery. COC sites were discovered via a genome-wide chromatin immunoprecipitation analysis of locations bound by transcription factor IIIC (TFIIIC; Noma et al., 2006). Several sites were identified that bind TFIIIC but not its cognate polymerase (RNA Pol III), and these TFIIIC-bound loci were shown to localize to the nuclear periphery. Their perinuclear localization led to the naming of these sequences as COCs. Interestingly, at least one such site serves as a heterochromatin boundary.
Chromosomal sites were previously identified within the budding yeast genome that bind TFIIIC but not RNA Pol III (Moqtaderi and Struhl, 2004). These sites are good candidates for S. cerevisiae counterparts of the S. pombe COC sites. These loci were named extra TFIIIC (ETC) sites. We tested whether one of the ETC sites, ETC6, localizes to the nuclear periphery. ETC6 lies in a divergent intergene on the right arm of chromosome IV (genome coordinate: IV, 1198690–1199174), more than 300 kb from telomere IV-R (Fig. 6 A; Moqtaderi and Struhl, 2004). We tagged the intergene neighboring ETC6 using the chromosome dot system and examined its subnuclear localization. As shown in Fig. 6 B, we found that in a WT strain background, the ETC6 locus is positioned predominantly at the nuclear periphery. The ETC6 locus is preferentially localized to the nuclear periphery in G1 and S phases, and, indeed, its peripheral positioning persists in G2 phase, showing that the cell cycle regulation of ETC6 positioning differs from that of telomeres. The S. cerevisiae genome therefore contains at least one peripherally positioned chromosome locus equivalent to an S. pombe COC. More detailed analysis of the localization of S. cerevisiae COC sites will be described elsewhere.
Figure 6.
Acetylation of H3K56 is crucial for perinuclear localization of ETC6. (A) Strain construct used to test the intranuclear position of ETC6 (located within TFC6-ESC2 intergene on chromosome IV). The neighboring intergene (BCP1-TFC6) was GFP-tagged in WT, asf1Δ, and rtt109Δ mutant strains. The center of the lacO array is 11 kb from the ETC6 locus. (B) Peripheral localization was tested as in Fig. 2. Error bars represent SD of values obtained from independent strain isolates (n = 3, 2, and 4 for the WT, asf1Δ, and rtt109Δ strain, respectively). WT and random localization values are indicated by blue and red lines, respectively.
Peripheral positioning of ETC6 requires H3K56 acetylation
We tested whether Asf1 and H3K56 acetylation are important for perinuclear localization of the ETC6 locus. We deleted the ASF1 and RTT109 genes from the strain illustrated in Fig. 6 A and examined the effect on ETC6 localization. Deleting ASF1 resulted in random positioning of the ETC6 locus throughout interphase (Fig. 6 B), showing that Asf1 is required for ETC6 peripheral localization. This result contrasts with the S phase–specific defect of asf1Δ in telomere localization, and suggests that VPS75 cannot substitute for ASF1 in perinuclear localization of ETC6 in any cell cycle phase. In the rtt109Δ mutant, peripheral positioning of ETC6 was also largely ablated, with a slight preference for localization to zone 1 remaining only in G1 phase of the cell cycle. Deletion of VPS75, SIR4, and YKU70 had only minor effects on perinuclear localization of the ETC6 locus (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1), which suggests that the function of these genes is not critical for ETC6 localization, although they may contribute to some extent. Although believed to bind primarily to telomeres and other silenced loci, a role for Ku and Sir in organizing other chromatin domains has been described previously (Thrower and Bloom, 2001), and both proteins have been implicated in confining chromatin movement (Gartenberg et al., 2004; Bystricky et al., 2005).
To summarize, these mutant phenotypes demonstrate that H3K56 acetylation plays a critical role in perinuclear localization of the COC ETC6. H3K56 acetylation is therefore crucial to mediate the correct intranuclear positioning of another peripherally localized chromatin domain, in addition to telomeres.
Discussion
We describe here the identification of seven genes that are required for telomere localization to the nuclear periphery. Overall, the components identified fall in three general categories: first, gene products implicated in chromatin structure or assembly, especially during DNA replication (Ctf18, Asf1, and Rrm3); second, gene products already known to localize to telomeres, whose role in telomere localization highlights a previously unrecognized function (Mre11 and Tel1); and third, gene products thought be involved in unrelated process, whose effect on telomere localization may be indirect (Elp4, Pgd1, and Sic1).
Elp4, Pgd1, and Sic1
Elp4 is part of the multifunctional Elongator complex, which is a major histone acetyltransferase component of the RNA polymerase II holoenzyme, and acts as well in other processes including tRNA modification (Svejstrup, 2007). PGD1 (also called MED3) encodes a subunit of the Mediator global transcriptional cofactor complex, part of the RNA polymerase II holoenzyme that plays an essential role in basal and activated transcription (Myers et al., 1998). Elp4 and Pgd1 may affect telomere localization indirectly, possibly by altering the expression of other genes required for telomere localization, although we cannot exclude the possibility that histone acetylation activity of Elp4 plays a similar role to Rtt109 (see the following paragraphs).
SIC1 encodes a cyclin-dependent kinase inhibitor that binds B-cyclin–Cdk complexes during G1 phase to restrain Cdk activity (Lengronne and Schwob, 2002). Once cells enter S phase, Sic1 is rapidly degraded by the proteasome. This previously described function of Sic1 occurs in G1 phase. SIC1 is, however, required for telomere localization during S phase and not in G1 phase (Fig. 2 and Table II). Origin initiation and the DNA replication program are disrupted in a sic1Δ mutant, as premature activation of Cdk activity driven by B-type cyclin leads to deregulated entry into S phase (Lengronne and Schwob, 2002). Disengagement of telomeres from the nuclear periphery is caused by telomere replication, which normally occurs late during S phase (Ebrahimi and Donaldson, 2008). The S phase–specific telomere localization of the sic1Δ mutant may therefore result from a derailed replication program, in which telomeres replicate throughout S phase.
Mre11 and Tel1
Mre11 and Tel1 are both telomere-binding proteins that are required for telomere length regulation. TEL1 encodes an ATM/ATR-like kinase, whereas Mre11 is a member of heterotrimeric MRX complex (consisting of Mre11, Rad50, and Xrs2) that is involved in telomere protection and length regulation (Askree et al., 2004; Gatbonton et al., 2006). Recent studies demonstrated that Tel1 preferentially binds shorter telomeres in an Mre11-dependent manner (Hector et al., 2007; Sabourin et al., 2007). These data suggest that they may have coordinated function in recruitment of telomerase to short telomeres. However, the telomere localization phenotypes of mre11Δ and tel1Δ mutants showed differing cell cycle specificity (Fig. 2 and Table II), which suggests that their role in telomere localization may be independent of their function in telomere length regulation. Consistently, no direct correlation between telomere length and localization was observed (Fig. 3).
Ctf18, Rrm3, and Asf1
A group of gene products that are implicated in DNA replication were identified as important for telomere positioning. The group consists of Rrm3, Asf1, and Ctf18. Ctf18 is the largest of three subunits specific to the Ctf18–replication factor C–like complex (RLC). As we previously described, all three subunits specific to the Ctf18-RLC (Ctf18, Ctf8, and Dcc1) are required for telomere positioning (Hiraga et al., 2006). The molecular target of the Ctf18-RLC remains unclear, although Ctf18-RLC is thought to load or unload proteins at replication forks. Rrm3 is also a replication fork component; RRM3 encodes a DNA helicase that interacts with proliferating cell nuclear antigen (Schmidt et al., 2002) and promotes passage of replication forks through protein–DNA complexes, including the telomeric nucleoprotein structure (Ivessa et al., 2003; Makovets et al., 2004). Both ctf18Δ and rrm3Δ mutants show telomere localization defects during G1 phase, despite the fact that their primary function is believed to occur during S phase. Ctf18 and Rrm3 are similar to Asf1 in being mainly implicated in replication-coupled events, and the identification of all three proteins in our telomere-positioning screen suggests that an S phase event is required for proper telomere localization in the subsequent G1 phase.
Asf1 is of particular interest when considering the possibility that the telomere localization mechanism is coupled to DNA replication. Asf1 physically interacts with components of the replication machinery and mediates replication-coupled nucleosome assembly (Mousson et al., 2007). Acetylation of chromatin-associated H3K56 is detectable throughout the cell cycle but dramatically increases in S phase because of incorporation of K56-acetylated H3 into chromatin by Asf1 (Masumoto et al., 2005; Maas et al., 2006; Ozdemir et al., 2006). This “wave” of H3K56 acetylation on chromatin in S phase is reversed by the histone deacetylases Hst3 and Hst4 (Celic et al., 2006; Maas et al., 2006) and at silenced loci by Sir2 (Xu et al., 2007).
We found that the histone chaperones Asf1 and Vps75 play redundant roles in telomere localization during G1 phase. Vps75 can stimulate acetylation of H3K56 efficiently in vitro (Han et al., 2007b; Selth and Svejstrup, 2007; Tsubota et al., 2007). Deletion of VPS75 does not however detectably decrease bulk cellular levels of acetylated H3K56. One possibility is that Vps75 regulates chromatin structure at a limited number of genomic loci, including telomeric regions. Short telomeres in the vps75Δ mutant (Askree et al., 2004) but not in asf1Δ (Fig. 3) are consistent with a telomeric function for Vps75. The observation that Vps75 does not substitute for Asf1 in positioning the ETC6 locus (Fig. 6 B and Fig. S2) also suggests a telomere-specific role for Vps75 in chromosome organization. It should be noted, however, that although our results clearly show that the H3K56 acetylation cycle is important for telomere positioning, we do not know at which site acetylation matters. Modulation of H3K56 acetylation in telomeric chromatin may be the critical factor, but an alternative possibility is that the acetylation state of a distant chromosomal site affects telomere localization, perhaps indirectly.
Our identification of the role of Asf1 in telomere positioning led to our observation that H3K56 acetylation by Rtt109 is crucial for telomere localization. Lack of H3K56 acetylation resulted in loss of telomere localization both in G1 and S phases (Fig. 5), despite the fact that H3K56 acetylation is most pronounced after DNA replication. This observation again implies that an event during replication is required for subsequent telomere localization. The histone H3 K56Q and K56R mutants are equally defective for telomere localization, which suggests that the acetylation/deacetylation cycle that occurs after DNA replication is important for positioning (rather than simply the presence of acetylated H3K56). A role for the acetylation/deacetylation cycle is consistent with the observation that H3K56 is in fact hypoacetylated at subtelomeric regions (Xu et al., 2007).
Telomeres are packaged in a heritable heterochromatin structure (Gottschling, 1992; Wright et al., 1992). Like transcriptional silencing, telomere localization status is subject to epigenetic inheritance (Maillet et al., 2001). A role for H3K56 acetylation in the epigenetic inheritance of chromatin structure might explain the effect of H3K56 acetylation on telomere and COC site localization. For example, one possibility is that H3K56 acetylation distinguishes “newly synthesized” nucleosomes from “mature” nucleosomes inherited as histone tetrasomes from parental DNA (Annunziato, 2005). New histone H3 is subject to K56 acetylation at the H3–H4 dimer stage of nucleosome assembly. Acetylated H3K56 could be required to ensure correct transmission of other histone modifications (such as N-terminal tail methylations and acetylations) to nucleosomes containing newly synthesized histones. In this model, the inherited nucleosomes, distinguished by absence of H3K56 acetylation, could be used as a template for copying the required modifications. H3K56 is positioned at the entry/exit point of DNA wound around nucleosomes, and acetylation of the residue is suggested to loosen histone–DNA interactions (Masumoto et al., 2005). Therefore, nucleosomes acetylated at H3K56 may be easily repositioned to allow reorganization of chromatin to achieve a preferred local chromatin structure (e.g., the compact heterochromatin structure close to telomeres), after which deacetylation of H3K56 might “fix” the chromatin status. In this way, the acetylation/deacetylation cycle of H3K56 could be envisaged to be critical for setting up a particular chromatin status (at telomeres [Xu et al., 2007] and other chromosome loci including COC sites) that allows correct localization to the nuclear periphery in the ensuing cell cycle. An early study found that deacetylation of histone is required for maturation of newly replicated chromatin in HeLa cells (Annunziato and Seale, 1983), which suggests that such an acetylation/deacetylation cycle may establish chromatin status in mammalian cells.
In this model, the ctf18Δ and rrm3Δ mutations could be envisaged to affect telomere positioning by affecting replication fork progression in a way that interferes with chromatin establishment, for example, by disrupting the balance between K56-acetylated and nonacetylated H3 on newly synthesized DNA.
The model we outline represents one possible route by which Asf1-stimulated H3K56 acetylation might be important for positioning chromosome domains. Further investigations will of course be necessary to understand how Asf1 and other components identified here regulate the correct organization of chromosomes within the nuclear interior.
Materials and methods
Yeast strains and plasmids
For screening of Rap1 organization mutants, deletion mutants were picked from the Saccharomyces Genome Deletion Project (Giaever et al., 2002) and transformed with plasmid YCp-GFP-RAP1 (Hiraga et al., 2006). For the genes tested, refer to Table S1. Strains used only for screening are not listed individually; all other strains used are listed in Table S3 (available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1).
The VPS75 open reading frame (ORF) overlaps with the flanking CWC25 ORF by one base pair. To avoid interfering with CWC25 expression, we removed the N-terminal 825 bases of the 890-bp VPS75 ORF, rather than removing entire coding sequence. To construct strain SHY227, the 826 bases of VPS75 gene in strain GA-1985 were replaced with the natMX4 cassette of the plasmid pAG25 (Goldstein and McCusker, 1999) by one-step gene replacement. The ASF1 gene of SHY227 was further replaced by kanMX4 to construct SHY229.
To tag the ETC6 locus with GFP, the lacO array of plasmid pAFS52 (Straight et al., 1996) was integrated into the intergene between BCP1 and TFC6 in the strain GA-1320 (Heun et al., 2001) to create SBY1, SBY2, and SBY6.
To construct strain SHY234, the mating type of HMY57 (Masumoto et al., 2005) was converted to MATα by expressing HO endonuclease transiently from the plasmid pJH132 (Connolly et al., 1988), followed by integration of pSH114 (carrying a lacO array) 19 kb from the left end of chromosome XIV (identical to the insertion locus used in Bystricky et al., 2005). GFP-lacI (pAFS135) was integrated at the HIS3 locus. Strains SHY247 and SHY248 were made by crossing SHY234 to HMY140 or HMY152 (Masumoto et al., 2005), respectively, followed by transformation with pSH115, which carries the GFP-NUP49 fusion gene.
HMY133 and HMY135 (Masumoto et al., 2005) were crossed to SHY296.2 to construct SHY310 and SHY304, respectively.
Other deletion mutant strains were derived from GA-1459 (GFP tagged at 14 kb away from telomere VI-R), GA-1985 (19 kb from VIII-L), GA-1986 (11 kb from telomere XIV-L) (Hediger et al., 2002), and SBY1 by replacing the genes of interest with kanMX4-disrupted alleles unless otherwise noted. kanMX4-disrupted alleles were obtained by PCR amplification of a suitable DNA fragment from the relevant mutant in the S. cerevisiae deletion collection (Giaever et al., 2002).
Plasmids used are listed in Table S4 (available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1). Plasmid pSH114 was constructed by cloning a PCR-amplified genomic fragment (coordinate XIV: 18,831–19,853) into pAFS59, followed by replacing the LEU2 marker with the ADE2 gene from plasmid pGVH30.
The plasmid pSH115 was constructed by recloning the GFP-NUP49 fusion gene of the plasmid pUN100-GFP-NUP49 (Belgareh and Doye, 1997) into YCplac33 (Gietz and Sugino, 1988).
Cytological techniques
Microscopic images were acquired by using a microscope (Deltavision RT; Applied Precision, LLC) with a UPlan-Apo 100× objective (1.35 NA; Olympus) and a cooled charge-coupled device camera (CoolSnap HQ monochrome; Photometrics). The acquisition software used was SoftWoRx (Applied Precision, LLC). The pixel size of acquired images is 66.3 nm under these conditions. Living yeast cells (grown at 30°C) were mounted on 1.7% agarose pad containing appropriate synthetic medium after brief sonication and observed at RT (23°C). For observation of GFP(S65T) fluorescence, an FITC filter set was used to capture z stacks containing 20 images at 250-nm intervals. Differential interference contrast images acquired at the same z intervals were used for determination of cell cycle stage by bud size (see the following paragraph). SoftWoRx explorer (Applied Precision, LLC) was used for subsequent measurements.
The cell cycle stage of individual cells was estimated from bud size: G1 phase, unbudded; S phase, cells with bud smaller than or equal to 2 μm; G2 phase, cells with bud larger than 2 μm and a spherical (i.e., nonmitotic) nucleus.
Quantitative evaluation of telomere localization was performed as described previously (Taddei et al., 2004). In brief, yeast cells whose GFP dot is in any of three equatorial sections of the nucleus were chosen for counting. Localization of the GFP dot was scored in two dimensions against three imaginary concentric zones at the equatorial plane, as shown in Fig. 2 B. At least 100 cells were scored in each culture unless otherwise noted. P-values were calculated by χ2 test against either random distribution or WT values.
Telomere length assay
XhoI-digested genomic DNA was separated by 1.5% agarose gel electrophoresis in 1× Tris/Borate/EDTA buffer and blotted to neutral nylon membrane (MP Biomedicals) by capillary transfer. The telomeric TG1-3 sequence was probed with a radioactively labeled 0.1-kb BamHI–EcoRI fragment from the plasmid pVII-L-URA3-TEL (Gottschling et al., 1990). Images were acquired by using a phosphorimager (FLA-3000) and analyzed by using ImageGauge software (both from Fujifilm).
Online supplemental material
Table S1 shows the results of the primary screening using the GFP-Rap1 fusion construct. Table S2 shows the result of the secondary screen testing telomere localization by using chromosomal dot strains. Table S3 shows a list of yeast strains. Table S4 shows plasmids used in this study. Fig. S1 shows the telomere length study that is supplementary to Fig. 3. Fig. S2 shows the analysis of the perinuclear localization of ETC6 in additional mutant strains. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806065/DC1.
Supplementary Material
Acknowledgments
We thank Valérie Doye, Marc R. Gartenberg, Susan M. Gasser, Hiroshi Masumoto, and Alain Verreault for strains and plasmids. We are grateful to Conrad Nieduszynski and Ian Stansfield for critical reading of the manuscript. Thanks to Rhiannon Grant, Amina Harb, and Yvonne Knox for technical assistance.
This research was supported by Wellcome Trust grant No. 082377/Z/07/Z.
Abbreviations used in this paper: COC, chromosome-organizing clamp; ETC, extra TFIIIC; H3–H4, histone H3–H4 complex; H3K56, histone H3 lysine 56; NPC, nuclear pore complex; ORF, open reading frame; RLC, replication factor C–like complex; TFIIIC, transcription factor IIIC; WT, wild-type; XIV-L, XIV-left.
References
- Annunziato, A.T. 2005. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 280:12065–12068. [DOI] [PubMed] [Google Scholar]
- Annunziato, A.T., and R.L. Seale. 1983. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 258:12675–12684. [PubMed] [Google Scholar]
- Askree, S.H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk, C. Coker, A. Krauskopf, M. Kupiec, and M.J. McEachern. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA. 101:8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., and V. Doye. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, J.H., and P. Walter. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp, J.M., A.E. Martin, E.S. Stensrud, and S.L. Jaspersen. 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky, K., T. Laroche, G. van Houwe, M. Blaszczyk, and S.M. Gasser. 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari, J.M., C.R. Brown, S. Komili, J. West, H. Hieronymus, and P.A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. [DOI] [PubMed] [Google Scholar]
- Celic, I., H. Masumoto, W. Griffith, P. Meluh, R. Cotter, J. Boeke, and A. Verreault. 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 16:1280–1289. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., and W.A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, B., C.I. White, and J.E. Haber. 1988. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2342–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, A.J., C.A. Nieduszynski, and A.D. Donaldson. 2002. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 16:2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, R., A. Hudson, and S.P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 315:649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi, H., and A.D. Donaldson. 2008. Release of yeast telomeres from the nuclear periphery is triggered by replication and maintained by suppression of Ku-mediated anchoring. Genes Dev. In press. [DOI] [PMC free article] [PubMed]
- Franco, A.A., W.M. Lam, P.M. Burgers, and P.D. Kaufman. 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19:1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg, M.R., F.R. Neumann, T. Laroche, M. Blaszczyk, and S.M. Gasser. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119:955–967. [DOI] [PubMed] [Google Scholar]
- Gasser, S.M. 2002. Visualizing chromatin dynamics in interphase nuclei. Science. 296:1412–1416. [DOI] [PubMed] [Google Scholar]
- Gatbonton, T., M. Imbesi, M. Nelson, J.M. Akey, D.M. Ruderfer, L. Kruglyak, J.A. Simon, and A. Bedalov. 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A.M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.L., and J.H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S.M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling, D.E. 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA. 89:4062–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling, D.E., O.M. Aparicio, B.L. Billington, and V.A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 63:751–762. [DOI] [PubMed] [Google Scholar]
- Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R.M. Xu, and Z. Zhang. 2007. a. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 315:653–655. [DOI] [PubMed] [Google Scholar]
- Han, J., H. Zhou, Z. Li, R.M. Xu, and Z. Zhang. 2007. b. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 282:14158–14164. [DOI] [PubMed] [Google Scholar]
- Hayashi, A., H. Ogawa, K. Kohno, S.M. Gasser, and Y. Hiraoka. 1998. Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells. 3:587–601. [DOI] [PubMed] [Google Scholar]
- Hector, R.E., R.L. Shtofman, A. Ray, B.R. Chen, T. Nyun, K.L. Berkner, and K.W. Runge. 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell. 27:851–858. [DOI] [PubMed] [Google Scholar]
- Hediger, F., F.R. Neumann, G. Van Houwe, K. Dubrana, and S.M. Gasser. 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12:2076–2089. [DOI] [PubMed] [Google Scholar]
- Heun, P., T. Laroche, M.K. Raghuraman, and S.M. Gasser. 2001. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 152:385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga, S., E.D. Robertson, and A.D. Donaldson. 2006. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J. 25:1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland, E.M., M.S. Cosgrove, H. Molina, D. Wang, A. Pandey, R.J. Cottee, and J.D. Boeke. 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:10060–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa, A.S., B.A. Lenzmeier, J.B. Bessler, L.K. Goudsouzian, S.L. Schnakenberg, and V.A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 12:1525–1536. [DOI] [PubMed] [Google Scholar]
- Kanoh, J., and F. Ishikawa. 2003. Composition and conservation of the telomeric complex. Cell. Mol. Life Sci. 60:2295–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne, A., and E. Schwob. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell. 9:1067–1078. [DOI] [PubMed] [Google Scholar]
- Li, Q., H. Zhou, H. Wurtele, B. Davies, B. Horazdovsky, A. Verreault, and Z. Zhang. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 134:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas, N.L., K.M. Miller, L.G. DeFazio, and D.P. Toczyski. 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell. 23:109–119. [DOI] [PubMed] [Google Scholar]
- Maillet, L., F. Gaden, V. Brevet, G. Fourel, S.G. Martin, K. Dubrana, S.M. Gasser, and E. Gilson. 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets, S., I. Herskowitz, and E.H. Blackburn. 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 24:4019–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 436:294–298. [DOI] [PubMed] [Google Scholar]
- Moqtaderi, Z., and K. Struhl. 2004. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 24:4118–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousson, F., F. Ochsenbein, and C. Mann. 2007. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 116:79–93. [DOI] [PubMed] [Google Scholar]
- Myers, L.C., C.M. Gustafsson, D.A. Bushnell, M. Lui, H. Erdjument-Bromage, P. Tempst, and R.D. Kornberg. 1998. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K., H.P. Cam, R.J. Maraia, and S.I. Grewal. 2006. A role for TFIIIC transcription factor complex in genome organization. Cell. 125:859–872. [DOI] [PubMed] [Google Scholar]
- Ozdemir, A., S. Spicuglia, E. Lasonder, M. Vermeulen, C. Campsteijn, H.G. Stunnenberg, and C. Logie. 2005. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 280:25949–25952. [DOI] [PubMed] [Google Scholar]
- Ozdemir, A., H. Masumoto, P. Fitzjohn, A. Verreault, and C. Logie. 2006. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle. 5:2602–2608. [DOI] [PubMed] [Google Scholar]
- Porter, S.E., P.W. Greenwell, K.B. Ritchie, and T.D. Petes. 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24:582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde, F.E., H.C. Gorham, and E.J. Louis. 1997. Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7:822–828. [DOI] [PubMed] [Google Scholar]
- Raghuraman, M.K., E.A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D.J. Lockhart, R.W. Davis, B.J. Brewer, and W.L. Fangman. 2001. Replication dynamics of the yeast genome. Science. 294:115–121. [DOI] [PubMed] [Google Scholar]
- Recht, J., T. Tsubota, J.C. Tanny, R.L. Diaz, J.M. Berger, X. Zhang, B.A. Garcia, J. Shabanowitz, A.L. Burlingame, D.F. Hunt, et al. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA. 103:6988–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin, M., C.T. Tuzon, and V.A. Zakian. 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 27:550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., G. Arib, C. Laemmli, J. Nishikawa, T. Durussel, and U.K. Laemmli. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21:379–391. [DOI] [PubMed] [Google Scholar]
- Schmidt, K.H., K.L. Derry, and R.D. Kolodner. 2002. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 277:45331–45337. [DOI] [PubMed] [Google Scholar]
- Schneider, J., P. Bajwa, F.C. Johnson, S.R. Bhaumik, and A. Shilatifard. 2006. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281:37270–37274. [DOI] [PubMed] [Google Scholar]
- Selth, L., and J.Q. Svejstrup. 2007. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 282:12358–12362. [DOI] [PubMed] [Google Scholar]
- Singer, M.S., A. Kahana, A.J. Wolf, L.L. Meisinger, S.E. Peterson, C. Goggin, M. Mahowald, and D.E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 150:613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A.F., A.S. Belmont, C.C. Robinett, and A.W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608. [DOI] [PubMed] [Google Scholar]
- Svejstrup, J.Q. 2007. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19:331–336. [DOI] [PubMed] [Google Scholar]
- Taddei, A., F. Hediger, F.R. Neumann, C. Bauer, and S.M. Gasser. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, H., F.A. Habermann, I. Solovei, M. Cremer, and T. Cremer. 2002. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat. Res. 504:37–45. [DOI] [PubMed] [Google Scholar]
- Thrower, D.A., and K. Bloom. 2001. Dicentric chromosome stretching during anaphase reveals roles of Sir2/Ku in chromatin compaction in budding yeast. Mol. Biol. Cell. 12:2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota, T., C. Berndsen, J. Erkmann, C. Smith, L. Yang, M. Freitas, J. Denu, and P. Kaufman. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 25:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.R.E., V. Azuara, P. Perry, S. Sauer, M. Dvorkina, H. Jorgensen, J. Roix, P. McQueen, T. Misteli, M. Merkenschlager, and A.G. Fisher. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119:132–140. [DOI] [PubMed] [Google Scholar]
- Wright, J.H., D.E. Gottschling, and V.A. Zakian. 1992. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 6:197–210. [DOI] [PubMed] [Google Scholar]
- Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 121:375–385. [DOI] [PubMed] [Google Scholar]
- Xu, F., Q. Zhang, K. Zhang, W. Xie, and M. Grunstein. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell. 27:890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink, D., M.D. Amaral, A. Englmann, S. Lang, L.A. Clarke, C. Rudolph, F. Alt, K. Luther, C. Braz, N. Sadoni, et al. 2004. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.