Abstract
Vesicles and other carriers destined for the Golgi apparatus must be guided to the correct cisternae. Golgins, long coiled-coil proteins that localize to particular Golgi subdomains via their C termini, are candidate regulators of vesicle sorting. In this study, we report that the GRIP domain golgins, whose C termini bind the Arf-like 1 G protein on the trans-Golgi, can also bind four members of the Rab family of G proteins. The Rab2-, Rab6-, Rab19-, and Rab30-binding sites are within the coiled-coil regions that are not required for Golgi targeting. Binding sites for two of these Rabs are also present on two coiled-coil proteins of the cis-Golgi, the Drosophila melanogaster orthologues of GM130 and GMAP-210. We suggest an integrated model for a tentacular Golgi in which coiled-coil proteins surround the Golgi to capture and retain Rab-containing membranes, excluding other structures such as ribosomes. Binding sites for diverse Rabs could ensure that incoming carriers are captured on first contact and moved to their correct destination within the stack.
Introduction
The Golgi apparatus receives vesicles and other membrane-bound carriers from the ER and multiple compartments of the endocytic pathway (Bonifacino and Glick, 2004). In addition, carriers move between the cisternae within the Golgi stack, and in many organisms, tubular connections form between adjacent stacks to form a Golgi ribbon. These diverse carriers have to not only locate the Golgi stack, but also fuse with the correct cisternae within the stack. Electron micrographs of the Golgi show that cytosolic ribosomes are excluded from a zone surrounding the stack, which is sometimes referred to as a ribosome-excluding matrix (Lucocq et al., 1987; Mogelsvang et al., 2003; Donohoe et al., 2007). Because vesicular carriers have a diameter greater than the ∼25 nm of ribosomes, this implies that they must carry features that allow them to penetrate into and persist in this apparent zone of exclusion.
Actin and spectrin have been proposed to form a mesh or skeleton around the Golgi, which undergoes rearrangements to regulate vesicle arrival and departure (Lorra and Huttner, 1999; De Matteis and Morrow, 2000). Another group of proteins proposed to contribute to Golgi structure is a set of long coiled-coil proteins often referred to as golgins. At least 12 such proteins have been identified in mammalian cells, with many having orthologues in yeast, plants, and protozoa (Barr and Short, 2003; Gillingham and Munro, 2003; Lupashin and Sztul, 2005). Each is found on a particular part of the Golgi stack, and in those cases examined, they are attached to the Golgi by interactions at their C termini. Three of these proteins (golgin-84, giantin, and CASP) are anchored at the rims of Golgi cisternae by C-terminal transmembrane domains. However, most of the Golgi coiled-coil proteins are peripheral membrane proteins that bind to either the cis or the trans side of the Golgi. At the cis side, GM130 and GMAP-210 bind to GRASP65 and Arf1, respectively, through their C termini. On the trans side are several proteins, including four (golgin-97, golgin-245, GCC88, and GCC185) that share a C-terminal motif termed the GRIP domain. This domain binds to the small G protein Arf-like 1 (Arl1) to mediate Golgi recruitment (Panic et al., 2003; Wu et al., 2004). For GCC185, a second G protein, Rab6, binds next to the GRIP domain and has been suggested to aid Golgi targeting (Burguete et al., 2008). Most metazoans have four GRIP domain proteins, with a single GRIP domain protein present in lower eukaryotes. Several functions have been proposed for the Golgi coiled-coil proteins, including interacting with each other to tether transport vesicles before fusion, organizing the cisternae into stacks, and connecting these stacks into ribbons (Barr and Short, 2003; Gillingham and Munro, 2003; Lupashin and Sztul, 2005). In addition, some have been suggested to serve as scaffolds for Golgi-associated proteins such as kinases and motors.
We have investigated the metazoan GRIP domain proteins using Drosophila melanogaster as a model system. Drosophila have a Golgi apparatus similar to that of mammalian cells, although the stacks are not linked together in a ribbon near the microtubule-organizing center but are dispersed throughout the cytoplasm. In this study, we confirm that the Drosophila GRIP domain proteins are localized to the Golgi and interact with Arl1. By screening a panel of small G proteins, we identify new binding partners for the proteins from the Rab family of small G proteins. These include the well-characterized Golgi Rabs, Rab2 and Rab6, but also two further Rabs, Rab19 and Rab30, for which the GRIP domain proteins are the first reported effectors. For Rab2 and Rab30, we extend these observations by showing that they also bind to other Golgi coiled-coil proteins, including two on the cis-Golgi. Integrating this data with previous studies (Short et al., 2001; Diao et al., 2003; Rosing et al., 2007) produces a model in which coiled-coil proteins act like tentacles to entrap carriers arriving at the Golgi or moving through the stack and help guide them to the right part of the stack.
Results and discussion
Drosophila GRIP domain proteins are localized to the Golgi apparatus
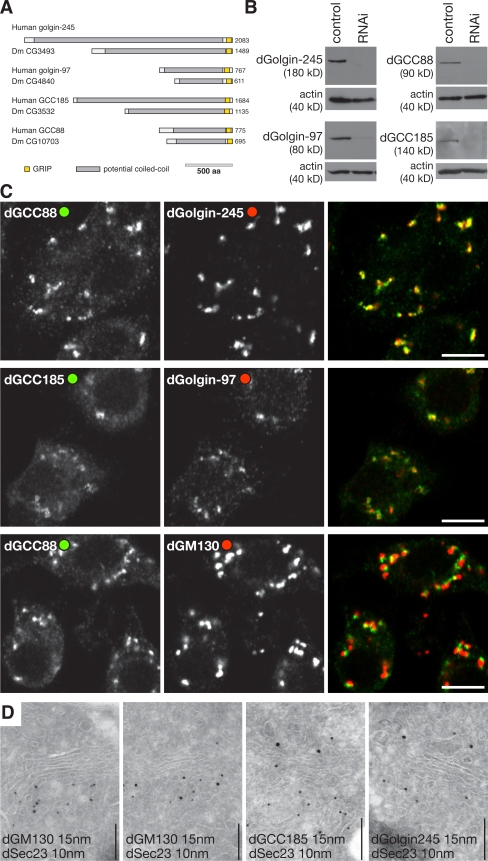
There is a single Drosophila orthologue of each of the four human GRIP domain proteins, and we will refer to the fly proteins by reference to their mammalian orthologues; i.e., dGolgin-245 (CG3493), dGolgin-97 (CG4840), dGCC88 (CG10703), and dGCC185 (CG3532; Fig. 1 A). Antisera raised against the N-terminal regions of the Drosophila GRIP domain proteins recognize a single protein in lysates from the Drosophila S2 cell line, with these being knocked down after treatment with specific double-stranded RNAs (dsRNAs; Fig. 1 B). Staining S2 cells with the antibodies yielded a punctate distribution typical of the Drosophila Golgi apparatus (Fig. 1 C). This staining was lost upon dsRNA treatment, but we could not detect any disruption of the Golgi or loss of cell viability upon such depletion either alone or in combination of all four proteins (unpublished data). All four proteins showed close colocalization with each other but only a partial overlap with the cis-Golgi coiled-coil protein dGM130, suggesting a trans-Golgi localization similar to the human GRIP domain proteins (Fig. 1 C). The cis side of the Drosophila Golgi is found adjacent to ER exit sites (Friggi-Grelin et al., 2006), and immunoelectron microcopy confirmed that GM130 was on the cis face and the GRIP proteins on the opposite trans face (Fig. 1 D and not depicted).
Figure 1.
Drosophila GRIP domain proteins localize to the Golgi apparatus. (A) Schematic representation of the human GRIP domain proteins and their Drosophila orthologues. Colors indicate regions predicted to be predominantly coiled-coil (gray) or the GRIP domains (yellow). (B) Protein blots of S2 lysates probed with antisera against the indicated GRIP domain proteins. The cells had been treated with dsRNA against GFP (control) or the corresponding GRIP domain proteins. Each antibody recognizes a single band, which is strongly reduced after RNAi. (C) Confocal micrographs of S2 cells labeled with the indicated antibodies. dGolgin-245 is coincident with dGCC88, and dGolgin-97 is coincident with dGCC185, but all GRIP domain proteins, here represented by dGCC88, are close to but slightly displaced from the cis-Golgi–specific dGM130. Bars, 5 μm. (D) Immunoelectron micrographs of frozen cryosections of S2 cells double labeled for dSec23 (15-nm gold), a marker of ER exit sites, and the indicated coiled-coil proteins (10-nm gold). dGM130 is present on the cis-Golgi cisternae that are adjacent to exit sites, whereas the GRIP proteins are on the opposite side of the stack. Sectioning and labeling were performed as described previously (Friggi-Grelin et al., 2006). Bars, 200 nm.
Identification of new binding partners for the Drosophila GRIP domain proteins
Interactions with labile determinants such as activated small G proteins are often found between peripheral membrane proteins and organelles or vesicles (Behnia and Munro, 2005). To test the possibility that the GRIP domain proteins bind to small G proteins in addition to the known interaction with Arl1, we used a yeast two-hybrid assay to test the proteins against a panel of 27 Drosophila G proteins (Table I). These represent the Drosophila orthologues of all the Arfs and Rabs found in mammals to be on the Golgi or endosomes or of unknown location (Gillingham and Munro, 2007; Zhang et al., 2007). For each small G protein, we generated forms with the Q→L mutation in the nucleotide-binding domain and for those that showed interactions with the S/T→N mutation, changes that in several members of these families result in constitutively active (GTP locked) or dominant-negative (GDP bound) forms, respectively (Zerial and McBride, 2001).
Table I. Drosophila Arf and Rab proteins used in the Y2H panel.
| Drosophila name | Human orthologues | Localization in mammals |
|---|---|---|
| Sar1, CG7073 | Sar1a/b | ER |
| Arf72A, CG6025 | Arl1 | Golgi |
| Arf84F, CG7039 | ARFRP1 | Golgi |
| Arf79F, CG8385 | Arf1/2 | Golgi |
| Arf102F, CG11027 | Arf4/5 | Golgi |
| CG7197 | Arl5A/B/C | Golgi |
| Rab1, CG3320 | Rab1A/B | Golgi |
| Rab2, CG3269 | Rab2A/B | Golgi |
| Rab6, CG6601 | Rab6A/B/C | Golgi |
| Rab10, CG17060 | Rab10 | Golgi |
| Rab19, Rab-RP3, CG7062 | Rab19B/Rab43 | Golgi |
| Rab30, CG9100 | Rab30 | Golgi |
| Rab39, CG12156 | Rab39A/B/Rab42 | Golgi |
| Rab5, CG3664 | Rab5A/B/C | early endosome |
| Rab7, CG5915 | Rab7 | late endosome |
| Rab9, CG9994 | Rab9A/B | late endosomes |
| Rab26, CG34410 | Rab26/Rab37 | late endosome |
| Rab27, CG14791 | Rab27A/B | late endosome |
| Rab11, CG5771 | Rab11A/B/Rab25 | recycling endosome |
| Rab4, CG4921 | Rab4A/B | endosomes |
| Rab8, CG8287 | Rab8A/B | endosomes |
| Rab14, CG4212 | Rab14 | endosomes |
| Rab21, CG17515 | Rab21 | endosome |
| Rab23, CG2108 | Rab23 | endosomes |
| Rab35, CG9575 | Rab35 | endosomes |
| Rab40, CG1900 | Rab40A/B/C | endosomes |
| Rab18, Rab-RP4, CG3129 | Rab18 | lipid droplets |
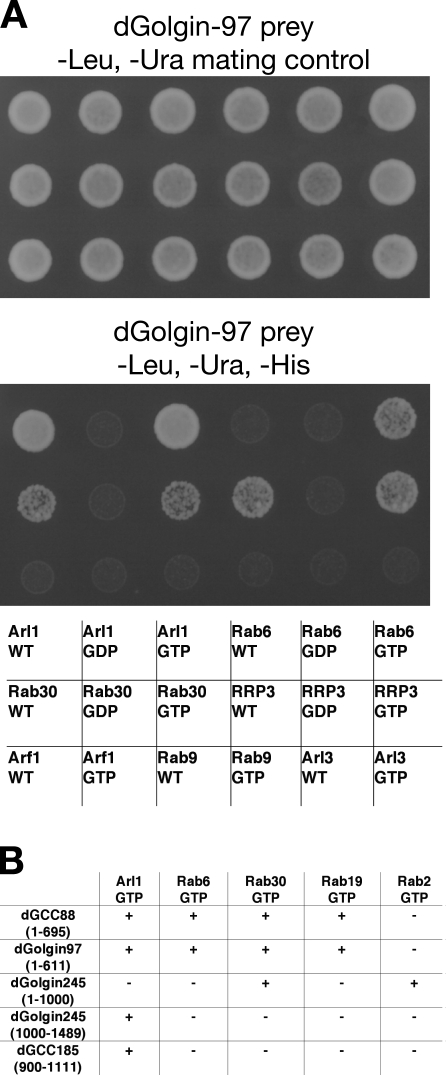
These small G proteins were tested against full-length forms of dGolgin-245, dGolgin-97, and dGCC88. We were unable to recover a full-length cDNA for dGCC185 but were at least able to test the C-terminal part of the protein. Fig. 2 shows an example of the data from the screen along with a summary of the overall results. Of the Arf family, only Arl1 showed an interaction, interacting with all four GRIP domain proteins. In contrast, we found that several of the Rabs interacted with the proteins. dGCC88 and dGolgin-97 both showed an interaction with the GTP-locked but not the GDP-bound forms of Rab6, Rab30, and Rab19. In the case of dGolgin-245, we could not detect any interactions with the full-length protein. However, dGolgin-245 is the longest GRIP domain protein, and when we split it into two parts, we found Rab30 and Rab2 binding to the N-terminal region and Arl1 binding to the C-terminal region of the protein. In summary, these results indicate that Arl1 can bind to all four GRIP proteins as expected but that in addition, there are several Rab-binding sites on these proteins (Fig. 2 B).
Figure 2.
Yeast two-hybrid assay identifies Rab-binding partners of the Drosophila GRIP domain proteins. (A) A representative yeast two-hybrid assay with part of the panel of Arf and Rab proteins. A full-length dGolgin-97 prey was tested against bait constructs for the indicated Arf and Rab proteins, which were either native (WT, wild type) or mutants expected to alter the nucleotide state T→N (GDP) or Q→L (GTP). The top panel shows the mating control, and the bottom panel shows the growth under selection for the HIS3 reporter gene. (B) Summary of the interactions found between the GRIP domain proteins and all of the small G proteins that showed a reaction with any of them. G proteins or their mutants that gave no reaction with any prey were omitted.
GRIP domain proteins can bind to different Rabs in vitro
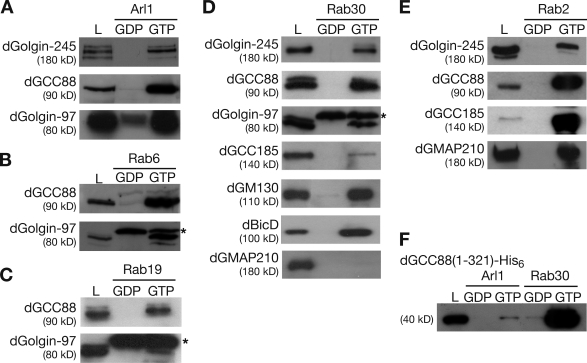
To obtain biochemical confirmation of the results from the aforementioned yeast two-hybrid assays, we investigated whether the GRIP domain proteins can interact in vitro with Arl1 and the various Rab proteins. GST fusions to dominant-negative and constitutively active forms of the small G proteins were expressed in bacteria and immobilized on glutathione beads. Cytosol from Drosophila S2 cells was incubated with the G protein–coated beads, and the amounts of GRIP proteins retained by the beads were analyzed by immunoblotting. We initially confirmed that Arl1 was able to bind to dGolgin-245, dGolgin-97, and dGCC88 in a GTP-dependent manner (Fig. 3 A). In addition, we found that dGCC88 and dGolgin-97 showed a GTP-specific binding to Rab6, Rab19, and Rab30 (Fig. 3, B–D). Likewise, dGolgin-245 binding to Rab2 and Rab30 could be confirmed (Fig. 3, D and E). We were not able to detect an interaction between dGCC185 and Arl1, suggesting that the interaction is either transient or needs other factors to help to stabilize it. It has been recently reported that in mammalian cells, Rab6 stabilizes the Arl1-dependent interaction of GCC185 with Golgi membranes (Burguete et al., 2008), although we could not detect binding of dGCC185 to Drosophila Rab6 (unpublished data). We did observe a robust binding of dGCC185 to Rab2-GTP (Fig. 3 E) and some binding to Rab30, although this was weak compared with the interaction of Rab30 with the other GRIP proteins (Fig. 3 D).
Figure 3.
Testing of the putative effectors of Arl1 and Rab using affinity chromatography. (A–E) Immunoblots of proteins from S2 cell lysates that bound to beads coated in GST fusions to the indicated small G proteins. The G proteins carried mutants of the type used in Fig. 2 (T→N, GDP and Q→L, and GTP). In addition, 5% of the lysates loaded on the columns was run in parallel (L). Anti–dGolgin-97 recognizes a band that is nonspecific, as it is still seen in eluates of lysates from RNAi-treated S2 cells and may be a bacterial protein (*). (F) Anti-His6 immunoblots of proteins eluted from Arl1 and Rab30 beads as in A–E but were instead incubated with a whole cell lysate from E. coli expressing a His6-tagged form of the N-terminal 321 residues of dGCC88.
Mapping of the Rab-binding sites on the GRIP domain proteins
The aforementioned results indicate that a single Rab can bind to multiple GRIP domain proteins and that a single GRIP domain protein can bind to more than one Rab. We next investigated how the binding sites for these G proteins were distributed along these coiled-coil proteins by using yeast two-hybrid assays with fragments of the proteins. The mapping data indicate that for all four proteins, the C-terminal GRIP domain binds to Arl1 but not to any of the interacting Rabs (Fig. 4). In contrast, the Rab-binding sites are found distributed at sites along the length of the proteins, in some cases far from the GRIP domain. In golgin-97, there appear to be distinct binding sites for the three Rabs that it interacts with (Rab6, Rab19, and Rab30), and for dGCC88, there are two binding sites for the same Rab, Rab30. The C-terminal coiled-coil part of dGCC88 (residues 550–600) is required for the second Rab30 site and also for the Rab6- and Rab19-binding sites. These Rab-binding sites mapped upstream of the GRIP domain but depend on the presence of the GRIP domain. Several Rab-binding sites on coiled-coil effectors are known to be comprised of residues from both helices of the coiled coil (Kawasaki et al., 2005; Wu et al., 2005). Thus, it is possible that the GRIP domain is stabilizing the adjacent regions of coiled coil in the tested fragments. In the yeast two-hybrid assays, we observed that Arl1 showed an interaction with a region of GCC88 that is distal to the GRIP domain and also binds Rab30 (Fig. 4 A). However, the in vitro binding of this N-terminal region of GCC88 (1–321) was much less effective for Arl1 than for Rab30, and so the significance of this Arl1 interaction is questionable (Fig. 3 F).
Figure 4.
Mapping of the Rab-binding sites on the GRIP domain proteins. (A) Yeast two-hybrid interactions between the indicated small G proteins (all Q-L GTP forms) and fragments of the GRIP domain proteins dGCC88, dGolgin-245, and dGolgin-97 and the cis-Golgi coiled-coil proteins dGMAP (CG33206; Friggi-Grelin et al., 2006) and dGM130 (CG11061; Kondylis et al., 2005). The prey fragments were fused to the C terminus of the Gal4 activation domain and tested for robust growth on selective plates (+/−). (B) Summary of the interactions shown in A. Rab2 also binds GCC88 and GC185, but the former interaction could not be detected by two hybrid, and the latter was not mapped because of the lack of a full-length cDNA clone. Colors indicate regions predicted to be predominantly coiled-coil (gray) or the GRIP domains (yellow). Orange indicates a GRAB (GRIP-related Arf-binding) domain.
Localization of Rab19 and Rab30 and binding partners of Rab30
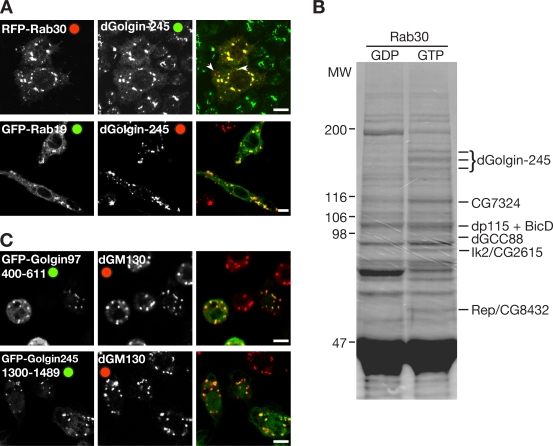
Among the four Rabs found to bind to the GRIP domain proteins, Rab2 and Rab6 are well characterized in mammalian cells as acting in membrane traffic at the Golgi apparatus and having established effectors (Zerial and McBride, 2001; Barr and Short, 2003). In contrast, little is known about the role of Rab19 and Rab30. There is a single mammalian orthologue of Drosophila Rab30 that has been shown to localize to the Golgi apparatus, but there are no reported functions or binding partners (de Leeuw et al., 1998). Drosophila Rab19 corresponds to two closely related paralogues in vertebrates Rab19B and Rab43. Rab43 localizes to the Golgi and has a function in the retrograde transport of Shiga toxin from endosomes to Golgi (Fuchs et al., 2007), whereas Rab19B is uncharacterized. To examine the distribution of the Drosophila proteins, GFP-Rab19 and RFP-Rab30 were expressed in S2 cells, and both were found to be primarily Golgi localized (Fig. 5 A).
Figure 5.
Rab30 and Rab19 in Drosophila S2 cells. (A) Confocal micrographs of S2 cells expressing RFP-Rab30 or GFP-Rab19 and labeled for dGolgin-245. Although primarily Golgi localized, RFP-Rab30 also labeled some additional structures (arrowheads), which are apparently endosomes, as they label with fluorescent dextran after 10 min of uptake (not depicted). (B) Coomassie-stained protein gel resulting from large-scale affinity chromatography of S2 cell lysates using GST-Rab30 T-N (GDP) or Q-L (GTP) forms. Bands enriched in the GTP lane were excised and digested in gel with trypsin (Roche), and peptides were identified by matrix-assisted laser desorption/ionization mass spectrometry as indicated. MW, molecular weight (in kilodaltons). (C) Confocal micrographs of S2 cells expressing the indicated C-terminal fragments of GRIP domain proteins fused to GFP and stained for the Golgi marker dGM130. Bars, 5 μM.
Because no effectors of either Rab19 or Rab30 have been reported, we next investigated for one of these Rabs whether the GRIP domain proteins were major effectors or whether the Rab has other interaction partners. Thus, we performed a large-scale affinity purification from S2 cell lysate using GST-Rab30(T-N) and GST-Rab30(Q-L) fusion proteins bound to glutathione–Sepharose beads (Fig. 5 B). Although a set of background proteins bound to both forms, there were several proteins that were specific to the GTP-locked form of Rab30, and these were identified by mass spectrometry. This revealed that the abundant binding partners for Rab30 included dGolgin-245 and dGCC88 and two further Golgi-localized coiled-coil proteins, BicaudalD (BicD) and dp115. There were only three other interacting proteins that we could detect using this approach. These were Rep (CG8432), the Drosophila orthologue of Rab escort protein that delivers unprenylated Rabs to the prenylation machinery (Wu et al., 2007); CG7324, an uncharacterized protein with a TBC (Tre-2/Bub2/Cdc16) domain that in all cases so far investigated has GTPase-activating protein activity on Rab proteins (Fuchs et al., 2007); and Ik2 (DmIKKε; CG2615), one of two IkB kinases in Drosophila. Further experiments will be necessary to establish the significance of the interactions with CG7324 and Ik2, but the interaction between Rab30 and BicD was confirmed using a specific antibody (Fig. 3 D). Collectively, these data indicate that Drosophila Rab30 and Rab19 both localize to the Golgi and that the principal effectors of Rab30, at least detectable in vitro, include GRIP domain proteins and two further coiled-coil proteins, BicD and dp115.
Some Rabs that bind GRIP domain proteins also bind cis-Golgi coiled-coil proteins
We next examined the eluates from the Rab30 beads for dGM130 and Drosophila Golgi microtubule-associated protein (dGMAP), two cis-Golgi–localized coiled-coil proteins for which antibodies are available, and found that dGM130 also binds to Rab30 but that dGMAP does not (Fig. 3 D). Thus, we tested whether the latter coiled-coil protein could bind to any Rab by using the yeast two-hybrid panel and found an interaction with Rab2 that could be readily confirmed using affinity chromatography with GST-Rab2 beads (Fig. 3 E). These interactions were mapped by two-hybrid assay and again were found to be along the coiled-coil regions of the proteins (Fig. 4, A and B). Human GM130 has been previously reported to bind to Rab1, 2, and 33b, although the regions of binding were not mapped (Short et al., 2001; Valsdottir et al., 2001). Rab33b has no Drosophila orthologue (although its closest relative is Rab30), but we were able to detect dGM130 bound to the Rab2 column, and for Rab1 and Rab30, we were able to map the binding sites by yeast two-hybrid assay (Fig. 4, A and B).
It is possible that there are further Rab-binding sites on these coiled-coil proteins whose affinity is too weak to detect by either yeast two hybrid or affinity chromatography. Weak binding has been seen for other membrane traffic proteins that make multiple interactions such as coat adaptors, presumably to allow reversibility. This may provide an explanation for our inability to find Rab6 binding to dGCC185. Likewise, Rab2 was found to bind to dGCC88 by affinity chromatography even though we could not detect the interaction by yeast two hybrid (Fig. 3 E). However, even with the set of interactions identified so far, some interesting patterns are apparent. The first is that the Rab-binding sites are distributed along the length of the coiled-coil proteins, which implies that Rab binding is not just involved in strengthening the interaction of the C terminus with Golgi membranes, as has recently been suggested for a Rab6-binding site next to the GRIP domain in human GCC185 (Burguete et al., 2008). Indeed, C-terminal fragments of dGolgin-97 and dGolgin-245 that contain the GRIP domain but lack all of the aforementioned Rab-binding sites are still targeted to the Golgi (Fig. 5 C). Thus, at least some of the Rab-binding sites would allow the GRIP domain proteins and other coiled-coil proteins to reach more distant G proteins, potentially on incoming transport carriers or other parts of the Golgi stack. Interestingly, in mammalian cells, the transport of Shiga toxin from endosomes to the Golgi requires golgin-97 and Rab43, one of the mammalian orthologues of Drosophila Rab19 (Lu et al., 2004; Fuchs et al., 2007). A second implication of our results is that for some Rabs, any membrane bearing an activated form of the Rab would have the potential to bind to several different coiled-coil proteins on both the trans- and cis-Golgi.
A model for the function of Golgi coiled-coil proteins
These observations, combined with previous reports of Rabs binding to coiled-coil proteins from the Golgi rim (Diao et al., 2003; Rosing et al., 2007), point to a model in which the coiled-coil proteins of the Golgi act collectively to form an array that surrounds the organelle. In the model, most, if not all, coiled-coil proteins are anchored to the Golgi via their C termini, with the rest of the molecules projecting into the cytoplasm like tentacles. These tentacles are studded along their length with Rab-binding sites that are shared by subsets of the coiled-coil proteins. Thus, the proteins could capture membranes bearing Rabs, including arriving carriers, intra-Golgi vesicles and tubes, and even other cisternae, and rounds of binding and release would allow the captured membranes to move between tentacles. In contrast, large structures that lack Rabs such as ribosomes would be excluded. This would be analogous to the proposed movement of importins through the nuclear pore by binding to a gel of phenylalanine-glycine repeats formed by several nuclear porins (Patel et al., 2007) and would be consistent with apparent stringlike structures observed around Golgi-associated vesicles by electron microscopy (Orci et al., 1998). Binding sites for different Rabs could conceivably allow for a hierarchy of interactions in which initial capture of a carrier anywhere in the stack via Rabs with binding sites broadly distributed over the stack such as Rab2 or Rab30 would be followed by binding by a different set of Rabs such as Rab1 or Rab6, whose binding sites appear to be concentrated toward the cis or trans face, respectively.
If correct, this model implies a degree of redundancy between the different Golgi coiled-coil proteins and Rabs, which could provide an explanation for the surprisingly mild phenotypes observed when some Golgi Rabs and coiled-coil proteins are knocked down or deleted (Kondylis et al., 2005; Friggi-Grelin et al., 2006; Fuchs et al., 2007). Indeed, we have recently found that dGolgin-245 is not essential for Drosophila viability or fertility (unpublished data). Investigating this and other models will require considerable further study, and it should be stressed that even if correct, our model does not exclude additional roles for the Rab-binding sites on the coiled-coil proteins, or indeed for the rest of the coiled-coil domain, such as contributing to Golgi recruitment or mediating interactions with other binding partners (Lupashin and Sztul, 2005; Burguete et al., 2008). Nonetheless, the finding that several Rabs can bind to the GRIP proteins and that at least Rab2 and Rab30 have additional coiled-coil effectors on the cis-Golgi suggests that a tentacular model for the Golgi is worth exploring as a route to understanding the Golgi and its coiled-coil proteins.
Materials and methods
Cell culture and antibodies
Polyclonal antibodies were raised in rabbits (dGolgin-245 and dGolgin-97), rats (dGCC185), or guinea pigs (GCC88) with GST fusion proteins containing the N-terminal 200 residues of each protein expressed from the pGEX-6P-2 vector (GE Healthcare). After affinity purification, anti–dGolgin-245 and anti–Golgin-97 were used at 1:4,000/1:400, and anti-dGCC88 and anti-GCC185 were used at 1:2,000/1:200 for blotting and immunofluorescence, respectively. Monoclonal mouse anti–BicD-1B11 (1:500; Developmental Studies Hybridoma Bank), polyclonal rabbit anti-dGMAP (1:5,000; Friggi-Grelin et al., 2006), and polyclonal rabbit anti-GM130 (1:3,000; Abcam) were used.
Drosophila S2 cells were grown according to the manufacturer's protocol (Dmel; Invitrogen). Cells were fixed (4% formaldehyde in PBS for 15 min) and blocked for 1 h in PBTB (PBS, 0.1% Triton X-100, and 1% BSA). Primary and secondary (Alexa Fluor; Invitrogen) antibodies in PBTB were applied for 1 h, and cells were washed three times with PBTB, mounted in Fluoromount-G (SouthernBiotech), and imaged on a confocal microscope with a 63× 1.4 NA objective (LSM510; Carl Zeiss, Inc.).
dsRNAs were amplified using T7 Ribomax Express (Promega) against dGolgin-245 (4,266–4,447 bp), dGolgin-97 (587–838 bp), dGCC88 (1,601–1,886 bp), and dGCC185 (1,405–1,604 bp). Dmel cells were transfected with 20 μg dsRNA and 15 μl of Transfast (Promega) in 6-well plates as described previously (Bettencourt-Dias et al., 2005) and were analyzed 4 d later.
Plasmids for expression and yeast two-hybrid assays
Drosophila Arf family proteins without the first 14 residues comprising the amphipathic helix and full-length Rabs were inserted into the bait vector pGBDUC1 for yeast two-hybrid assays and into pGEX-6P-2 for bacterial expression. Full-length and truncated versions of GRIP proteins were PCR amplified (EST clones: SD05887 for dGolgin-245, LD35238 for dGolgin-97, LD06167 for dGCC88, and total cDNA for CG3532) and cloned into pGAD424 (Clontech Laboratories, Inc.). Point mutations to create dominant-negative (T-N or S-N for Rab2 and Rab9) or constitutively active (Q-L) versions of the G proteins were performed by the Quickchange method (Agilent Technologies), and yeast two-hybrid assays were performed as described previously (James et al., 1996).
Affinity chromatography and mass spectrometry
For affinity chromatography, Arl1 and Rab-GST fusion proteins were expressed in Escherichia coli BL21-GOLD (DE3; Agilent Technologies). Cells grown to OD600 = 0.8 at 37°C were induced with 0.25 mM IPTG at 16°C overnight. E. coli lysates were prepared from pellets by sonication in lysis buffer (20 mM Tris-HCl, pH 7.4, 110 mM KCl, 5 mM MgCl2, 1% Triton X-100, 5 mM MgCl2, 5 mM β-mercaptoethanol, protease inhibitors, and 200 μM GDP or nonhydrolyzable GTP analogue [GppNHp]; Sigma-Aldrich). The lysates were clarified by centrifugation at 12,000 g for 20 min, incubated with glutathione–Sepharose beads (GE Healthcare) at 4°C for 30 min, and GST fusion–coated beads were washed in lysis buffer. Dmel cell lysates were prepared from 5 × 107 cells for small scale or 5 × 108 cells for large scale by using 5 ml/10 ml of lysis buffer, Dounce homogenizing, and passage through a 30-G needle and were clarified by centrifugation at 50,000 g for 30 min at 4°C. Supernatants were incubated with 50 μl/150 μl of GST fusion–coated beads in the presence of 100 μM GDP or GppNHp for 2 h at 4°C. The beads were washed three times with lysis buffer and eluted with 100 μl of SDS sample buffer (small scale) or elution buffer (20 mM Tris-HCl, pH 7.4, 1.5 M KCl, 20 mM EDTA, 5 mM β-mercaptoethanol, and 5 mM GDP or GppNHp) followed by methanol/chloroform precipitation and resuspension in 40 μl SDS-PAGE buffer (large scale).
Acknowledgments
We thank F. Begum for expert mass spectrometric analysis, C. Rabouille for advice on electron microscopy, and G. Warren for valuable discussions about Golgi coiled-coil proteins.
Abbreviations used in this paper: Arl1, Arf-like 1; BicD, BicaudalD; dGMAP, Drosophila Golgi microtubule-associated protein; dsRNA, double-stranded RNA.
References
- Barr, F.A., and B. Short. 2003. Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 15:405–413. [DOI] [PubMed] [Google Scholar]
- Behnia, R., and S. Munro. 2005. Organelle identity and the signposts for membrane traffic. Nature. 438:597–604. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., R. Sinka, L. Frenz, and D.M. Glover. 2005. RNAi in Drosophila cell cultures. In Gene Silencing by RNA Interference: Technology and Application. M. Sohail, editor. CRC Press, Boca Raton, FL. 147–166
- Bonifacino, J.S., and B.S. Glick. 2004. The mechanisms of vesicle budding and fusion. Cell. 116:153–166. [DOI] [PubMed] [Google Scholar]
- Burguete, A.S., T.D. Fenn, A.T. Brunger, and S.R. Pfeffer. 2008. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 132:286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw, H.P., P.M. Koster, J. Calafat, H. Janssen, A.J. van Zonneveld, J.A. van Mourik, and J. Voorberg. 1998. Small GTP-binding proteins in human endothelial cells. Br. J. Haematol. 103:15–19. [DOI] [PubMed] [Google Scholar]
- De Matteis, M.A., and J.S. Morrow. 2000. Spectrin tethers and mesh in the biosynthetic pathway. J. Cell Sci. 113:2331–2343. [DOI] [PubMed] [Google Scholar]
- Diao, A., D. Rahman, D.J. Pappin, J. Lucocq, and M. Lowe. 2003. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 160:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe, B.S., B.H. Kang, and L.A. Staehelin. 2007. Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc. Natl. Acad. Sci. USA. 104:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin, F., C. Rabouille, and P. Therond. 2006. The cis-Golgi Drosophila GMAP has a role in anterograde transport and Golgi organization in vivo, similar to its mammalian ortholog in tissue culture cells. Eur. J. Cell Biol. 85:1155–1166. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., A.K. Haas, R.A. Spooner, S. Yoshimura, J.M. Lord, and F.A. Barr. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and S. Munro. 2003. Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta. 1641:71–85. [DOI] [PubMed] [Google Scholar]
- Gillingham, A.K., and S. Munro. 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23:579–611. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, M., K. Nakayama, and S. Wakatsuki. 2005. Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr. Opin. Struct. Biol. 15:681–689. [DOI] [PubMed] [Google Scholar]
- Kondylis, V., K.M. Spoorendonk, and C. Rabouille. 2005. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell. 16:4061–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorra, C., and W.B. Huttner. 1999. The mesh hypothesis of Golgi dynamics. Nat. Cell Biol. 1:E113–E115. [DOI] [PubMed] [Google Scholar]
- Lu, L., G. Tai, and W. Hong. 2004. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol. Biol. Cell. 15:4426–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq, J.M., J.G. Pryde, E.G. Berger, and G. Warren. 1987. A mitotic form of the Golgi apparatus in HeLa cells. J. Cell Biol. 104:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin, V., and E. Sztul. 2005. Golgi tethering factors. Biochim. Biophys. Acta. 1744:325–339. [DOI] [PubMed] [Google Scholar]
- Mogelsvang, S., N. Gomez-Ospina, J. Soderholm, B.S. Glick, and L.A. Staehelin. 2003. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol. Biol. Cell. 14:2277–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci, L., A. Perrelet, and J.E. Rothman. 1998. Vesicles on strings: morphological evidence for processive transport within the Golgi stack. Proc. Natl. Acad. Sci. USA. 95:2279–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic, B., O. Perisic, D.B. Veprintsev, R.L. Williams, and S. Munro. 2003. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell. 12:863–874. [DOI] [PubMed] [Google Scholar]
- Patel, S.S., B.J. Belmont, J.M. Sante, and M.F. Rexach. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 129:83–96. [DOI] [PubMed] [Google Scholar]
- Rosing, M., E. Ossendorf, A. Rak, and A. Barnekow. 2007. Giantin interacts with both the small GTPase Rab6 and Rab1. Exp. Cell Res. 313:2318–2325. [DOI] [PubMed] [Google Scholar]
- Short, B., C. Preisinger, R. Korner, R. Kopajtich, O. Byron, and F.A. Barr. 2001. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsdottir, R., H. Hashimoto, K. Ashman, T. Koda, B. Storrie, and T. Nilsson. 2001. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 508:201–209. [DOI] [PubMed] [Google Scholar]
- Wu, M., L. Lu, W. Hong, and H. Song. 2004. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 11:86–94. [DOI] [PubMed] [Google Scholar]
- Wu, M., T. Wang, E. Loh, W. Hong, and H. Song. 2005. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J. 24:1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y.W., K.T. Tan, H. Waldmann, R.S. Goody, and K. Alexandrov. 2007. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc. Natl. Acad. Sci. USA. 104:12294–12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117. [DOI] [PubMed] [Google Scholar]
- Zhang, J., K.L. Schulze, P.R. Hiesinger, K. Suyama, S. Wang, M. Fish, M. Acar, R.A. Hoskins, H.J. Bellen, and M.P. Scott. 2007. Thirty-one flavors of Drosophila Rab proteins. Genetics. 176:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]