Abstract
GnRH neurons are essential for the onset and maintenance of reproduction. Mutations in both fibroblast growth factor receptor (Fgfr1) and Fgf8 have been shown to cause Kallmann syndrome, a disease characterized by hypogonadotropic hypogonadism and anosmia, indicating that FGF signaling is indispensable for the formation of a functional GnRH system. Presently it is unclear which stage of GnRH neuronal development is most impacted by FGF signaling deficiency. GnRH neurons express both FGFR1 and -3; thus, it is also unclear whether FGFR1 or FGFR3 contributes directly to GnRH system development. In this study, we examined the developing GnRH system in mice deficient in FGF8, FGFR1, or FGFR3 to elucidate the individual contribution of these FGF signaling components. Our results show that the early emergence of GnRH neurons from the embryonic olfactory placode requires FGF8 signaling, which is mediated through FGFR1, not FGFR3. These data provide compelling evidence that the developing GnRH system is exquisitely sensitive to reduced levels of FGF signaling. Furthermore, Kallmann syndrome stemming from FGF signaling deficiency may be due primarily to defects in early GnRH neuronal development prior to their migration into the forebrain.
THE GNRH SYSTEM is crucial for reproduction (1,2,3). Studies in mouse embryos showed that GnRH mRNA and peptide-expressing neurons are first detected in the medial olfactory placode on embryonic day (E) 11.5 (2,3,4). Subsequent studies showed that these GnRH neurons migrate along peripherin-positive olfactovomeronasal fibers (5) through the cribriform plate into the preoptic region and hypothalamus, a process that is completed around E16.5 (2,3,4).
Aberrant embryonic development of the GnRH system results in severe clinical consequences. For example, abnormal GnRH system development is a hallmark of Kallmann syndrome (KS), a human neurological disorder characterized by hypogonadotropic hypogonadism associated with anosmia. Loss-of-function gene mutations have been shown to cause KS. These include mutations on anosmin-1, fibroblast growth factor receptor 1 (Fgfr1), prokineticin receptor-2, and prokineticin-2 genes (6,7,8,9).
Loss-of-function Fgfr1 mutations in KS is of specific interest because the targeted expression of a dominant-negative fibroblast growth factor receptor (FGFR) in mouse GnRH neurons resulted in a 30% decrease in the total number of GnRH neurons in the forebrain, late pubertal onset, and premature reproductive senescence (10). Nonetheless, several important mechanistic questions regarding the role of fibroblast growth factor (FGF) signaling during GnRH system development remain unanswered. First, developing GnRH neurons express both FGFR1 and FGFR3 (11). This and the existence of 22 FGF ligands (12) make it difficult to pinpoint the precise FGF ligand/receptor combination(s) mediating the development of the mouse GnRH system. Second, it is unclear which stage of GnRH system development is most severely disrupted by FGF signaling deficiency.
Recently loss-of-function mutations in the Fgf8 gene were found in KS patients (13), suggesting that FGF8 may be a crucial ligand for GnRH system development. Binding studies showed that FGF8 can readily bind to FGFR1 and FGFR3 with similar affinities, and the activation of both receptors induces mitogenic activity (14). These findings underscore the need to identify which specific FGFR mediates FGF8 signaling.
In this study, we investigated whether FGF8 plays a critical role in the development of the GnRH system and, if so, during which specific stage. We also examined whether FGFR1 or FGFR3 is important for the development of the GnRH system in mice. To elucidate these questions, we studied the mouse GnRH system development in transgenic mice that express significantly lower levels of functional FGF8 protein or FGFR1 protein or in mice in which FGFR3 expression is completely abolished. Our data suggest that FGF8 actions, primarily mediated through FGFR1, are critically required for the very early phase of GnRH neuronal development.
Materials and Methods
Transgenic animals
Fgf8 hypomorphs (129P2/OlaHsd* CD-1; Mouse Regional Resource Centers, Davis, CA), Fgfr1 hypomorphs (129sv/CD-1; Canadian Mutant Mouse Repository), and Fgfr3 knockout (KO) mice (Swiss-Webster/C57BL/6; Jackson Laboratories, Bar Harbor, ME) (15,16,17) were housed in our animal facility under a 12-h light, 12-h dark cycle and fed ad libitum. Embryos were generated through timed breeding of adult females with E0.5 being the morning copulatory plugs were detected. Day of birth was designated as postnatal day (PN) 0. All animal procedures complied with protocols approved by the Institutional Animal Care and Use Committee at the University of Colorado. Homozygous (HOMO) Fgf8 hypomorphic mice have been reported to exhibit about 54% decrease in normal Fgf8 mRNA (15), whereas HOMO Fgfr1 hypomorphic mice show a 66–80% decrease in normal Fgfr1 mRNA (17). HOMO Fgf8 and Fgfr1 hypomorphic mice die within 24 h after birth (15,17), whereas about 48% of HOMO Fgfr3 KO mice die between birth and 21 d of age (16). Comparisons between genotypes [wild-type (WT), heterozygous (HET), or HOMO] were made within the transgenic strain type.
Tissue collection
Embryos retrieved on E11.5 and E14.5 were immersion-fixed in 4% paraformaldehyde in 0.1 m phosphate buffer at room temperature for 2 h. Similarly, the heads of PN 0 pups were immersion fixed for 6 h at room temperature. Nonfixed tissue samples from each individual mouse embryo or pup were taken for genotyping purposes. Embryos and PN 0 tissues were stored in either 100% methanol at −20 C or 25% sucrose in phosphate buffer at 4 C before immunocytochemistry (ICC) (11) or in situ hybridization (ISH).
GnRH ICC
GnRH neurons in embryonic and neonatal heads were detected using the rabbit anti-GnRH polyclonal antibody LR5 (a gift from Dr. R. Benoit, Montréal General Hospital, Montréal, Québec, Canada) as described in detail previously (11). Briefly, whole E11.5 embryos or serial parasagittal sections of E14.5 embryos (20 μm) were incubated with LR-5 (1:5000) diluted in 0.1 m phosphate-buffered saline and 0.4% Triton X-100 containing 4% normal donkey serum and 10% normal horse serum for 5 d. Serial coronal sections (20 μm) from PN 0 animals were incubated with LR-5 (1:10,000) for 4 d at 4 C. Then E11.5 whole embryos were incubated in horseradish peroxidase-conjugated donkey antirabbit (Jackson ImmunoResearch Laboratories, West Grove, PA) for 2 d, reacted with 0.05% 3, 3-diaminobenzidine, and embedded in paraffin. Serial ribbons of parasagittal sections (12 μm) were made with a rotary microtome (Leica, Bannockburn, IL), mounted on gelatin-coated glass slides and deparaffinized. Sections from E14.5 embryos and PN 0 pups were incubated with a biotinylated-donkey antirabbit antibody (1:500), followed by avidin and biotin-coupled horseradish peroxidase (ABC kit; Vector Labs, Burlingame, CA), and reacted with DAB. All sections were dehydrated and coverslipped with Permount (Fisher, Pittsburgh, PA).
Nonradioactive GnRH ISH
Coronal sections (20 μm) through the PN 0 preoptic region of WT and HOMO Fgf8 hypomorphs were processed for ISH using a standard nonradioactive protocol (10). Sections were hybridized with a 270-nt digoxigenin-label riboprobe, which encompasses the entire open reading frame of the GnRH cDNA, from the start to the stop codon (NM_008145), washed extensively, and incubated with an antidigoxigenin antibody conjugated to alkaline phosphatase (Roche, Indianapolis, IN). Following, GnRH mRNA was detected using 5-bromo-4-chloro-3-indoyl-phosphate, 4-toluidine salt/4-nitro blue tetrazolium chloride as chromagens. The distribution of GnRH neurons detected by this probe is identical with that detected by the specific GnRH antibody LR5.
Olf-1 and peripherin ICC
ICC for Olf-1 [a marker for olfactory receptor neurons (ORN)] and peripherin (a marker for intermediate filaments in olfactovomeronasal fibers) were conducted to assess whether Fgf8 hypomorphs suffer disruption in ORN development and the GnRH migratory pathway. Serial parasagittal sections (20 μm) were mounted on gelatin-coated glass slides and incubated with a rabbit anti-Olf-1 polyclonal antibody (for E11.5 embryos, 1:2000, a gift from Dr. R. R. Reed, Johns Hopkins University, School of Medicine, Baltimore, MD) or a rabbit antiperipherin polyclonal antibody (for E14.5 embryos, 1:3000, AB1530. Millipore, Bedford, MA) for 4 d at 4 C, incubated with a Cy3-conjugated donkey antirabbit antibody (1:500), and coverslipped with an antifade medium containing the nuclear stain, 4′,6′-diamino-2-phenylindole (5 μm).
Apoptosis
To compare the incidence of apoptosis in the nasal region of WT, HET, and HOMO Fgf8 hypomorphic E11.5 embryos, we stained corresponding parasagittal sections with the histological stain cresyl violet, which reveals the pyknotic cell nuclei of apoptotic cells (18). Comparisons were made in the Olf-1 positive region of the olfactory epithelium.
GnRH neuronal counts
The total number of GnRH neurons was counted in coded serial sections (20 μm) through the whole neonatal head, from the tip of nose to past the median eminence.
Statistical analysis
One-way ANOVA or Student’s t tests were used to test for significant differences (P < 0.05). Student-Newman-Keuls test was used for post hoc testing.
Results
GnRH neurons in Fgf8 hypomorphs
On E11.5, both WT and HET Fgf8 hypomorphs possessed morphologically distinct thickening and invagination of the medial-ventral olfactory epithelium, an elaboration of the olfactory placode (4) (see Figs. 1A and 3, A and B). The medial-ventral olfactory epithelium in HOMO Fgf8 hypomorphs also showed invagination, albeit to an extent much less than WT and HET Fgf8 hypomorphs (Figs. 1B and 3C).
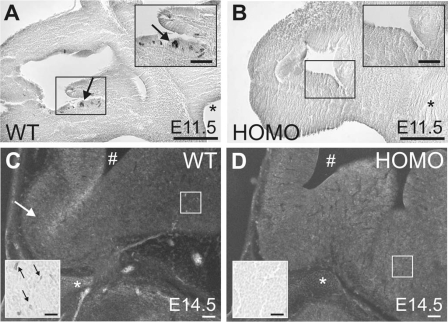
Figure 1.
Representative bright-field photomicrographs of GnRH ICC in parasagittal sections of Fgf8 hypomorphs. WT (A) and homozygous HOMO (B) E11.5 embryos are shown. Note the presence of GnRH-IR neurons (black arrows) in the invaginated medial-ventral olfactory epithelium (boxed) of WT but not in HOMO embryos. Black asterisk indicates preoptic recess. Black scale bar, 200 μm or 100 μm (inset). C and D, Inverted bright-field photomicrographs of WT and HOMO E14.5 embryos. Note the presence of GnRH-IR neurons in the preoptic area of WT but not in HOMO embryos. White arrow indicates the olfactory bulb. White asterisk indicates the sphenoid cartilage. #, Lateral ventricle. White scale bar, 500 μm or 50 μm (inset). Insets are higher-magnification photomicrographs of the boxed areas.
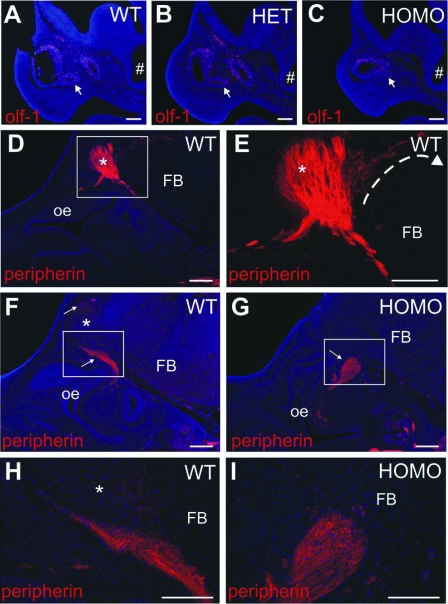
Figure 3.
Representative epifluorescence photomicrographs of Olf-1-IR olfactory neurons (A–C) and peripherin-IR olfactovomeronasal fibers (D–I) in parasagittal sections of Fgf8 hypomorphs. Olf-1-positive staining (red) WT (A), HET (B), and HOMO (C) E11.5 embryos. Note Olf-1-positive staining in the medial-ventral olfactory epithelium (arrows). #, Preoptic recess. Scale bar, 200 μm. D–I, Peripherin-positive staining (red, arrows) in WT (D–F) and HOMO (G) E14.5 embryos. Asterisk indicates the olfactory bulb. Boxed areas are magnified in WT (H) and HOMO (I) E14.5 embryos. Scale bar, 200 μm. oe, Olfactory epithelium; FB, forebrain. Blue is 4′,6′-diamino-2-phenylindole nuclear counterstain.
GnRH ICC detected GnRH-immunoreactive (IR) neurons in the invaginated zone of the medial-ventral olfactory epithelium in WT but not in HOMO Fgf8 hypomorphs on E11.5 (Fig. 1, A and B). Similarly, GnRH-IR neurons were found in the preoptic region of WT but not in HOMO Fgf8 hypomorphs on E14.5 (Fig. 1, C and D).
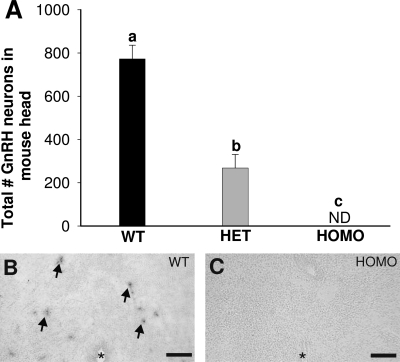
Consistent with the absence of GnRH neurons in HOMO Fgf8 hypomorphic embryos, no GnRH-IR neurons were detected in HOMO Fgf8 hypomorphs on PN 0 (Fig. 2A). The total number of GnRH-IR neurons in the whole head on PN 0 was significantly higher in WT (n = 4; 773 ± 64) than HET (n = 4; 268 ± 63) or HOMO (n = 4; none detected) Fgf8 hypomorphs on PN 0 (P < 0.01). Moreover, HET Fgf8 hypomorphs had less than 50% of the total number of GnRH-IR neurons, compared with WT Fgf8 hypomorphs (Fig. 2A: P < 0.01).
Figure 2.
GnRH neurons in Fgf8 hypomorphs on PN 0. A, Total number of GnRH-IR neurons in the whole head of WT (n = 4), HET (n = 4), and HOMO (n = 4) Fgf8 hypomorphs. Different letters indicate significant differences (P < 0.01). Data are presented as mean ± sem. ND, Not detectable. GnRH mRNA-positive neurons (arrows) are present in coronal sections of the preoptic area in WT (B) but not HOMO animals (C). Asterisk indicates third ventricle. Scale bar, 100 μm.
GnRH ISH was conducted to verify the lack GnRH-IR resulted from the lack of GnRH mRNA, not from defects in GnRH prohormone processing. GnRH mRNA-containing neurons are present in the preoptic area of WT but not in HOMO Fgf8 hypomorphs on PN 0 (Fig. 2, B and C).
Olf-1 neurons and peripherin fibers in Fgf8 hypomorphs
On E11.5, Olf-1-IR sensory neurons were detected in the olfactory epithelium of WT (n = 3), HET (n = 3) and HOMO (n = 3) Fgf8 hypomorphs (Fig. 3, A–C). Specifically, Olf-1-IR cells were found in the dorsal and medial-ventral invagination of olfactory epithelium and doral of WT and HET Fgf8 hypomorphs (Fig. 3, A and B). Although this invagination is greatly reduced in HOMO Fgf8 hypomorphs, Olf-1-IR ORNs remained present in the corresponding medial-ventral olfactory epithelium (Fig. 3C).
Peripherin-IR fibers were detected in the nasal region of WT (n = 3) and HOMO (n = 2) E14.5 Fgf8 hypomorphs. Peripherin-IR fibers from the nasal region target and infiltrate the olfactory bulb in WT Fgf8 hypomorphic embryos (Fig. 3, D, E, F, and H). However, peripherin-IR fibers in HOMO Fgf8 hypomorphic embryos did not enter the brain or extend past the cribriform plate (Fig. 3, G and I).
Apoptosis in Fgf8 hypomorphs
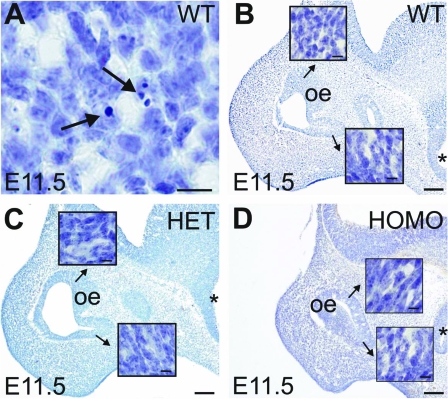
Consistent with previous studies (19,20,21), we detected apoptotic cell bodies (i.e. pyknotic cell bodies) in the E11.5 olfactory epithelium (Fig. 4A) of the nasal region. However, the number of apoptotic cell bodies in the Olf-1 positive region of the medial-ventral olfactory epithelium, as visualized by cresyl violet, was very low (less than three per section). There were no gross differences in the incidence of apoptosis among WT (n = 3), HET (n = 3), and HOMO (n = 3) Fgf8 hypomorphs (Fig. 4, B–D).
Figure 4.
Representative photomicrographs of E11.5 olfactory epithelium (oe) in cresyl violet-stained parasagittal sections of Fgf8 hypomorphs. A, Apoptotic cells in WT olfactory epithelium (arrows). Scale bar, 20 μm. B–D, Olfactory epithelium in WT, HET, and HOMO embryos. Note the absence of apoptotic cells in the medial-ventral and medial-dorsal olfactory epithelium at higher magnification (insets). The base of the arrows indicate the regions magnified in the insets. Scale bar, 200 μm or 20 μm (insets). Asterisk indicates preoptic recess.
GnRH neurons in Fgfr1 hypomorphs
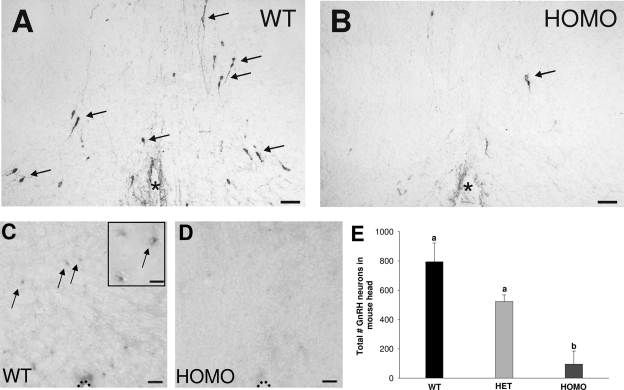
On PN 0, HOMO, Fgfr1 hypomorphs (n = 4; 95 ± 90) showed significantly decreased total number of GnRH-IR neurons, compared with WT (n = 4; 795 ± 128) and HET (n = 4; 524 ± 44) Fgfr1 hypomorphs (Fig. 5, A, B, and E; P < 0.01). Interestingly, three of the four HOMO animals had only four to eight GnRH-IR neurons per head, whereas one HOMO animal had 365 GnRH neurons, indicating incomplete penetrance of this phenotype. There was no significant difference between the total number of GnRH-IR neurons in WT and HET Fgfr1 hypomorphs (Fig. 5E). GnRH ISH showed the presence of GnRH mRNA containing preoptic area neurons in WT but not HOMO Fgfr1 hypomorphs (Fig. 5, C and D).
Figure 5.
GnRH neurons in Fgfr1 hypomorphs on PN 0. A and B, Representative photomicrographs of GnRH-IR neurons (arrows). C and D, GnRH mRNA positive (arrows) in coronal sections of the preoptic area in WT and HOMO Fgfr1 hypomorphs. Asterisk indicates third ventricle. Dotted line indicates ventral border of preoptic area. Scale bar, 50 μm or 20 μm (inset). E, Total number of GnRH-IR neurons in the whole head of WT (n = 4), HET (n = 4), and HOMO (n = 4) Fgfr1 hypomorphs. Different letters indicate significant differences (P < 0.01). Data are presented as mean ± sem.
GnRH neurons in Fgfr3 KO mice
Comparison of the total number of GnRH neurons in the whole head of PN 0 WT (501 ± 33; n = 7) and HOMO Fgfr3 KO (465 ± 79; n = 5) mice revealed no significant difference between the genotypes. The lower of number GnRH-IR neurons detected in WT Fgfr3 KO compared with WT Fgf8 hypomorphs and WT Fgfr1 hypomorphs may be due to a difference in background strains.
Discussion
Our studies show that deficiencies in FGF8 or FGFR1, but not in FGFR3, obliterated all (FGF8) or a large portion (FGFR1) of developing GnRH-IR neurons. In HOMO Fgf8 hypomorphs, GnRH neurons never emerge from the olfactory epithelium and are absent in every subsequent developmental stage examined, strongly suggesting that GnRH neurons were missing from the very beginning during embryonic development. Together, our data suggest FGF8 signals predominantly through FGFR1 to promote the genesis or maintain the survival of GnRH neurons/progenitors in the olfactory placode.
Fgf8 hypomorphy completely eliminated the presence of GnRH neurons, whereas Fgfr1 hypomorphy severely reduced the total number of GnRH neurons. This discrepancy may be due to the variability in the knockdown of Fgfr1 mRNA in Fgfr1 hypomorphs, which ranges between 66 and 80%. Alternatively, other FGFRs may partially compensate for the decreased levels of FGFR1. Indeed, FGF8 signaling can also be facilitated by FGFR3. Nonetheless, our data indicate that FGF8 signaling is mainly facilitated through FGFR1. This does not rule out a role for other FGF signaling components.
Neuronal migration is an important aspect of GnRH system development. In humans, defective GnRH neuronal migration has commonly been accepted as a cause of hypogonadotropic hypogonadism in KS (22). Previous data showed that migrating GnRH neurons closely follow the scaffold of peripherin-IR olfactovomeronasal fibers to reach the forebrain (23). The abnormal organization of peripherin-IR fibers in the HOMO Fgf8 hypomorphs is consistent with an earlier report (24). Peripherin-IR fibers do not enter the brain of HOMO Fgf8 hypomorphs, whereas an extensive plexus of peripherin-IR fibers was detected in the olfactory bulbs and anterior forebrain of WT Fgf8 hypomorphs. Based on these observations, one might conclude that migratory defects underlie the absence of GnRH neurons in HOMO Fgf8 hypomorphs. However, GnRH neurons were already absent on E11.5. In other words, no GnRH neurons were present to enter migration. Therefore, we hypothesize that defects in early GnRH development, not migration, caused the absence of GnRH neurons in HOMO Fgf8 hypomorphs.
Previous studies showed that HOMO Fgf8 hypomorphs exhibited malformation of the nasal region (15). Therefore, the absence of GnRH-IR neurons from E11.5 onward may be the result of a general FGF-dependent dysgenesis of the nasal region. Although the homozygous state of Fgf8 hypomorphy eliminated all GnRH neurons, the presence of the olfactory epithelium, as indicated by the presence of Olf-1 positive neurons, persisted. Furthermore, the heterozygous state of Fgf8 hypomorphy, which obliterated more than 50% of all GnRH neurons, continued to have normal morphogenesis of the nasal region and olfactory bulbs (19). Together these data strongly suggest that deficits in FGF8 function affect some olfactory epithelium-derived cell populations (i.e. GnRH neurons) more than others. Of interest, we showed that a ventral recess of the olfactory epithelium from which GnRH neurons emerge is greatly reduced in HOMO Fgf8 hypomorphs on E11.5. This recess is highly FGF8 sensitive, and its malformation is likely the cause for the failure of GnRH neurons to emerge. This recess has also been shown to give rise to the vomeronasal organ (VNO) (25). Our preliminary data showing that the VNO is missing in the HOMO Fgf8 hypomorphs (data not shown) are consistent with the notion that this recess fails to develop and give rise to differentiated derivatives. Our data also suggest the presence of discrete developmental zones in the olfactory placode (26), with FGF8 signaling deficiency more likely to impact the ventral zone that gives rise to the VNO and GnRH neurons.
FGF8 is a potent cell survival factor. Indeed, the elimination of FGF8 signaling in Fgf8 conditional KOs resulted in abnormally high levels of apoptosis in the olfactory epithelium between E10.5 and E12.5 (19). On E11.5, we showed that the incidence of apoptosis in the olfactory epithelium was comparable among WT, HET, and HOMO Fgf8 hypomorphs. This discrepancy may be due to the less severe FGF8 deficiency in hypomorphs, compared with the conditional KOs (19). Alternatively, the difference in apoptotic incidence may be due to the nonlinear dosage effect of Fgf8. Previous data showed that the complete elimination or overexpression of FGF8 protein causes higher levels of apoptosis, whereas a decrease in functional FGF8 protein expression results in cell survival (27). This U-shaped effect could cause the incidence of apoptosis to differ dramatically between these two transgenic mice. Lastly, our data do not completely rule out the possibility that Fgf8 hypomorphy can cause increased levels of apoptosis in the early or presumptive olfactory placode, in which Fgf8 mRNA expression was first detected from E8.5 onward (28). Therefore, an intriguing consequence of Fgf8 hypomorphy may be the premature elimination of progenitor cells that are fated to become GnRH neurons before E11.5. Alternatively, Fgf8 hypomorphy may have hampered progenitor cells from becoming GnRH neurons. Unfortunately, the lack of a specific marker for GnRH progenitor cells prevents further studies along this line.
Although both Fgfr1 and Fgfr3 are expressed in developing GnRH neurons (11) and bind FGF8 equally well (14), the total number of GnRH neurons decreased in only HOMO Fgfr1 hypomorphs but not HOMO Fgfr3 KO mice. These data strongly suggest FGF8’s effects on the emergence of GnRH neurons from the olfactory placode are mediated through FGFR1. FGF8 signaling may be direct on GnRH neurons because GnRH neurons express FGFR1 during embryonic development (11).
The targeting of olfactovomeronasal fibers has been hypothesized to be required for the morphogenesis of the olfactory bulbs (29). In Fgf8 hypomorphs, the inability of peripherin-IR fibers to enter and contact the most anterior region of the brain may have caused the failure of olfactory bulb morphogenesis (15). Currently the mechanisms underlying the inability of peripherin-IR fibers to enter the embryonic HOMO Fgf8 hypomorphic brain are unknown. The elongation of olfactory fibers has been shown to be accompanied by migrating olfactory ensheathing cells, which are glia that originate from the olfactory epithelium (30,31). These olfactory ensheathing cells express high levels of FGFR1 and have been shown to be permissive for the elongation/growth of olfactory axons (32,33). Together with our results, we hypothesize that the abnormal organization of the peripherin-IR fibers may, in part, be due to decreased FGF8 signaling on FGFR1-expressing olfactory ensheathing cells, possibly impairing their functions, and hence disrupting the growth of peripherin-IR fibers. Alternatively, FGF8 deficiency may inhibit the emergence or number of olfactory ensheathing cells from the olfactory epithelium, resulting in the observed disorganization of peripherin-IR fibers.
Although genetic analysis showed that KS is causally linked to loss-of-function mutations in Fgfr1, and more recently in Fgf8 (13), the cellular effects of these mutations on human GnRH system development are unknown. Interestingly, these loss-of-function mutations are heterozygous in nature. Therefore, it has been recently proposed that the severity and pathogenesis of KS may depend on the compounding effects of heterozygous loss-of-function mutations on multiple causative genes (34). However, our experimental results clearly indicate that the heterozygous state of Fgf8 hypomorphy alone is sufficient to cause significant disruptions GnRH neuronal development. Furthermore, our studies suggest that Fgfr1 and Fgf8 mutations in KS may compromise human GnRH system function by abrogating the initial emergence of GnRH neurons from the olfactory epithelium. Finally, the abnormal organization of peripherin-IR olfactory fibers may explain the occurrence of anosmia in KS.
Footnotes
This work was supported by Grant RO1 HD042634 from the National Institutes of Health and the Endocrine Society Bridge Award (to P.-S.T.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online June 19, 2008
Abbreviations: E, Embryonic day; FGF, fibroblast growth factor; FGFR, FGF receptor; HET, heterozygous; HOMO, homozygous; ICC, immunocytochemistry; IR, immunoreactive; ISH, in situ hybridization; KO, knockout; KS, Kallmann syndrome; ORN, olfactory receptor neuron; PN, postnatal day; VNO, vomeronasal organ; WT, wild type.
References
- Livne I, Gibson MJ, Silverman AJ 1993 Biochemical differentiation and intercellular interactions of migratory gonadotropin-releasing hormone (GnRH) cells in the mouse. Dev Biol 159:643–656 [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW 1989 Origin of luteinizing hormone-releasing hormone neurons. Nature 338:161–164 [DOI] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H 1989 Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA 86:8132–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Nieburgs A, Elkabes S 1989 Spatiotemporal cell expression of luteinizing hormone-releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Brain Res Dev Brain Res 46:309–318 [DOI] [PubMed] [Google Scholar]
- Wray S, Key S, Qualls R, Fueshko SM 1994 A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev Biol 166:349–354 [DOI] [PubMed] [Google Scholar]
- Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP 2006 Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet 2:e175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick D, Franco B, Sherins RJ, Heye B, Pike L, Crawford J, Maddalena A, Incerti B, Pragliola A, Meitinger T, et al 1992 Brief report: intragenic deletion of the KALIG-1 gene in Kallmann’s syndrome. N Engl J Med 326:1752–1755 [DOI] [PubMed] [Google Scholar]
- Hardelin JP, Julliard AK, Moniot B, Soussi-Yanicostas N, Verney C, Schwanzel- Fukuda M, Ayer-Le Lievre C, Petit C 1999 Anosmin-1 is a regionally restricted component of basement membranes and interstitial matrices during organogenesis: implications for the developmental anomalies of X chromosome-linked Kallmann syndrome. Dev Dyn 215:26–44 [DOI] [PubMed] [Google Scholar]
- Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pecheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP 2003 Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet 33:463–465 [DOI] [PubMed] [Google Scholar]
- Tsai PS, Moenter SM, Postigo HR, El Majdoubi M, Pak TR, Gill JC, Paruthiyil S, Werner S, Weiner RI 2005 Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol Endocrinol 19:225–236 [DOI] [PubMed] [Google Scholar]
- Gill JC, Moenter SM, Tsai PS 2004 Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology 145:3830–3839 [DOI] [PubMed] [Google Scholar]
- Mason I 2007 Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev Neurosci 8:583–596 [DOI] [PubMed] [Google Scholar]
- Falardeau J, Chung WCJ, Beekeen A, Plummer L, Sidis Y, Raivio T, Dwyer A, Na S, Hall J, Huot C, Alois N, Quinton R, Cole LW, Hughes V, Mohammadi M, Tsai PS, Pitteloud N 2008 Decreased FGF8 signaling causes GnRH deficiency in human and mice. J Clin Invest 118:2822–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M 2006 Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev 20:185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR 1998 An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet 18:136–141 [DOI] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM 1996 Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet 12:390–397 [DOI] [PubMed] [Google Scholar]
- Partanen J, Schwartz L, Rossant J 1998 Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev 12:2332–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WCJ, Swaab DF, De Vries GJ 2000 Apoptosis during sexual differentiation of the bed nucleus of the stria terminalis in the rat brain. J Neurobiol 43:234–243 [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL 2005 Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development 132:5211–5223 [DOI] [PubMed] [Google Scholar]
- Pellier V, Astic L 1994 Cell death in the developing olfactory epithelium of rat embryos. Brain Res Dev Brain Res 79:307–315 [DOI] [PubMed] [Google Scholar]
- Voyron S, Giacobini P, Tarozzo G, Cappello P, Perroteau I, Fasolo A 1999 Apoptosis in the development of the mouse olfactory epithelium. Brain Res Dev Brain Res 115:49–55 [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Bick D, Pfaff DW 1989 Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Mol Brain Res 6:311–326 [DOI] [PubMed] [Google Scholar]
- Fueshko S, Wray S 1994 LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev Biol 166:331–348 [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Young C, Calof A Requirement for Fgf8 in generation of GnRH-expressing hypothalamic neurons during mouse development. Proc Annual Meeting of the Society for Neuroscience 37, San Diego, CA, 2007, (Abstract 562.7) [Google Scholar]
- Wray S 2002 Development of gonadotropin-releasing hormone-1 neurons. Front Neuroendocrinol 23:292–316 [DOI] [PubMed] [Google Scholar]
- Wray S 2001 Development of luteinizing hormone releasing hormone neurones. J Neuroendocrinol 13:3–11 [DOI] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL 2006 Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133:1831–1844 [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR 1995 The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121:439–451 [DOI] [PubMed] [Google Scholar]
- Gong Q, Shipley MT 1995 Evidence that pioneer olfactory axons regulate telencephalon cell cycle kinetics to induce the formation of the olfactory bulb. Neuron 14:91–101 [DOI] [PubMed] [Google Scholar]
- Tisay KT, Key B 1999 The extracellular matrix modulates olfactory neurite outgrowth on ensheathing cells. J Neurosci 19:9890–9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennent R, Chuah MI 1996 Ultrastructural study of ensheathing cells in early development of olfactory axons. Brain Res Dev Brain Res 95:135–139 [DOI] [PubMed] [Google Scholar]
- Chuah MI, Au C 1994 Olfactory cell cultures on ensheathing cell monolayers. Chem Senses 19:25–34 [DOI] [PubMed] [Google Scholar]
- Hsu P, Yu F, Feron F, Pickles JO, Sneesby K, Mackay-Sim A 2001 Basic fibroblast growth factor and fibroblast growth factor receptors in adult olfactory epithelium. Brain Res 896:188–197 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W 2007 Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest 117:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]