Abstract
Intrinsically disordered proteins play critical but often poorly understood roles in mediating protein interactions. The interactions of disordered proteins studied to date typically entail structural stabilization, whether as a global disorder-to-order transition or minimal ordering of short linear motifs. The disordered cyclin-dependent kinase (CDK) inhibitor Sic1 interacts with a single site on its receptor Cdc4 only upon phosphorylation of its multiple dispersed CDK sites. The molecular basis for this multisite-dependent interaction with a single receptor site is not known. By NMR analysis, we show that multiple phosphorylated sites on Sic1 interact with Cdc4 in dynamic equilibrium with only local ordering around each site. Regardless of phosphorylation status, Sic1 exists in an intrinsically disordered state but is surprisingly compact with transient structure. The observation of this unusual binding mode between Sic1 and Cdc4 extends the understanding of protein interactions from predominantly static complexes to include dynamic ensembles of intrinsically disordered states.
Keywords: disorder, dynamic complex, multisite phosphorylation, NMR, protein interaction
Intrinsically disordered proteins often participate in regulatory interactions with structured receptor proteins (1–3). Such interactions are frequently modulated by phosphorylation, which requires disorder in the target protein both for optimal kinase accessibility and for subsequent accessibility of the binding motif (4). Disorder-to-order transitions of varying extents occur upon binding of disordered proteins to their receptors. Coupled binding and folding have been observed in a number of systems (5–8). In contrast, short linear binding motifs may exhibit only local ordering upon interaction with modular binding domains (9–11). In an intermediate situation, disordered protein regions with a measured propensity for helical secondary structure have been found to act as preformed molecular recognition elements (12–16). Disorder in protein complexes may thus be a common feature and may give rise to such important properties as plasticity and reversibility (17). The spectrum of different binding modes associated with intrinsically disordered proteins may even include highly dynamic complexes (16), such as proposed for interaction between the yeast cyclin-dependent kinase (CDK) inhibitor Sic1 and its cognate receptor, the F-box protein Cdc4 (18). However, the existence of a truly dynamic complex has not yet been biophysically demonstrated.
Phosphorylation by G1 CDK activity (Cln1/2–Cdc28) targets Sic1 to the SCFCdc4 ubiquitin ligase, resulting in Sic1 ubiquitination and degradation by the proteasome (19–22). The WD40 domain of Cdc4 recognizes short threonine- or serine-phosphorylated sequences in Sic1 and other substrates, termed Cdc4 phosphodegrons (CPDs) (23). A crystal structure of Cdc4 with a singly phosphorylated peptide (GLLpTPPQSG, from cyclin E) that closely matches the optimal CPD reveals a single deep pSer/Thr–Pro-binding pocket (24). Mutation of residues in this pocket results in loss of binding to Sic1 in vitro and loss of function in vivo (24). Sic1 itself contains 9 suboptimal CPD sites [supporting information (SI) Table S1], all of which contain mismatches to the optimal CPD consensus (18). Individual CPD peptides derived from Sic1 interact with Cdc4 with affinities in the high micromolar range (25, 26) (Table S2). In contrast, the deep pSer/Thr—Pro-binding pocket in Cdc4 binds with high affinity (Kd ≈1 μM) to a singly phosphorylated peptide that closely matches the CPD consensus (24). An adjacent shallow binding pocket can also interact with a downstream phosphate at the P+3/P+4 position (relative to the phosphorylated residue at the P0 position) in diphosphorylated CPD peptides, which further stabilizes local interactions (26). High-affinity binding of Sic1 to Cdc4, however, appears to require multiple weak CPD sites to be phosphorylated, which in principle renders Sic1 recognition and degradation ultrasensitive with respect to G1 CDK activity (18). These observations suggest a binding mode in which the multiple phosphoepitopes in Sic1 engage the single receptor site of Cdc4 in dynamic equilibrium (18, 24). Here, we use high-resolution NMR methods to shed light on the biophysical basis of the interaction of the polyvalent, intrinsically disordered Sic1 with Cdc4.
Results
Sic1 Is Intrinsically Disordered but Compact.
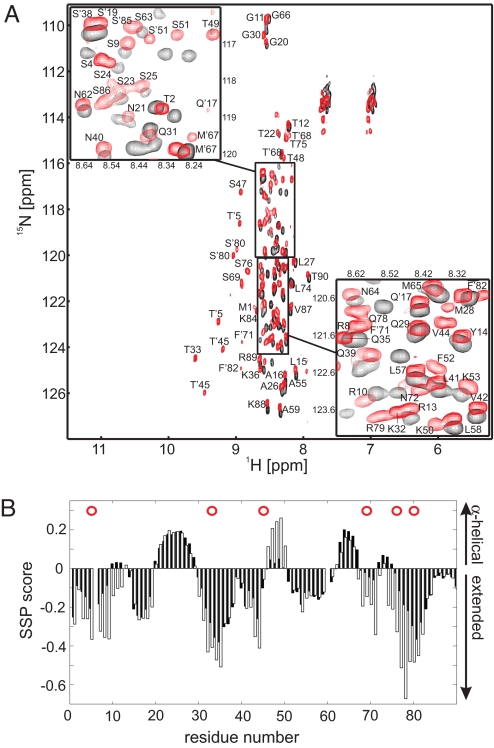
To probe the nature of the interaction between phospho-Sic1 (pSic1) and Cdc4, we used NMR spectroscopy to monitor the N-terminal 90 residues of Sic1, which do not exhibit appreciable stable secondary structure in either the unphosphorylated or phosphorylated state as determined by CD spectroscopy (18). This region contains 7 consensus CDK sites (Thr-2, Thr-5, Thr-33, Thr-45, Ser-69, Ser-76, and Ser-80) and is sufficient for high-affinity binding when fully phosphorylated (27). 1HN-15N correlation spectra (Fig. 1A) of Sic1 (black) and pSic1 (red) display limited amide proton chemical shift dispersion and sharp lines; such spectra are diagnostic of disordered proteins because the rapid interconversion of different conformers, many of which lack tertiary contacts, results in an averaged magnetic environment of the nuclei. Phosphorylation of Sic1 causes a downfield shift of amide protons of the phosphorylated residues Thr-5, Thr-33, Thr-45, Ser-69, Ser-76, and Ser-80. The lack of a downfield shift for Thr-2 indicates that this residue was not phosphorylated; the NMR spectra also demonstrate that no other serine or threonine residues were phosphorylated in these experiments. The chemical shift changes of phosphorylated residues and small changes in adjacent residues argue strongly against an overall disorder-to-order transition upon phosphorylation. Rather, both Sic1 and pSic1 are intrinsically disordered proteins, best described as ensembles of rapidly interconverting, diverse conformers. Notably, the phosphorylated state exhibits multiple resonances for residues adjacent to prolines, including those immediately after the phosphorylated residues within the suboptimal CPDs. These peaks are indicative of sampling of both trans- and cis-prolyl peptide bonds in the unbound state of pSic1 (Fig. 1A; for the Sic1 sequence, see Table S3).
Fig. 1.
Sic1 is an intrinsically disordered protein. (A) 1HN-15N correlation spectra of Sic1 (black) and pSic1 (red). In the pSic1 sample, resonances of residues Thr-5, Thr-33, Thr-45, Ser-69, Ser-76, and Ser-80 exhibit proton downfield shifts accompanied by small chemical shift changes in adjacent residues. Residues with multiple resonances, primarily from cis- and trans-prolyl isomers, are denoted with a prime. (B) Experimental SSP values (28) for Sic1 (black bars) and pSic1 (open bars). Red circles indicate phosphorylation sites.
We determined experimental secondary structural propensity (SSP) values to provide a measure of the weighted estimate of the fraction of conformers in the disordered ensemble that sample backbone torsion angles indicative of either α-helical or extended (β) structure; values of unity over a stretch of residues reflect fully formed secondary structure (28). The SSP values (Fig. 1B) for Sic1 (black bars) and pSic1 (open bars) as a function of residue position are similar, with a significant tendency toward extended conformations. However, the presence of some regions with up to a 20% weighted likelihood to adopt helical conformations suggests that Sic1 could exist as a relatively compact ensemble of interconverting conformers. We investigated this by pulsed-field gradient (PFG) diffusion NMR experiments, which measure translational diffusion coefficients and, indirectly, the hydrodynamic radius (Rh) of a protein. Although both Sic1 and pSic1 are intrinsically disordered, each appears to be surprisingly compact (Table S4), with calculated values of Rh for Sic1 and pSic1 of 21.5 ± 1.1 and 19.3 ± 1.4 Å, respectively. pSic1, in particular, is much more compact than expected for a fully denatured 90-residue polypeptide (28.7 Å) and only slightly larger than expected for a 90-residue globular protein (17.5 Å) (29, 30).
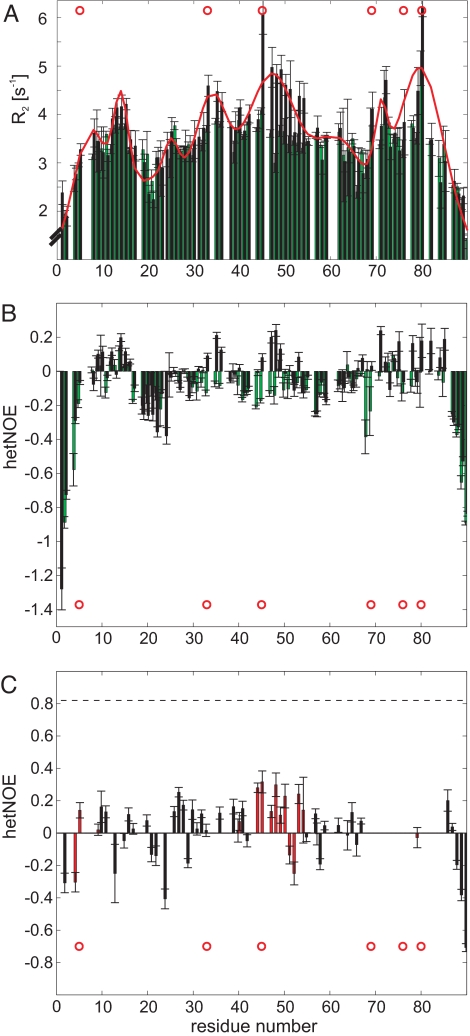
The conformational flexibilities of Sic1 and pSic1 were probed by backbone 15N relaxation experiments that report on picosecond to nanosecond time scale motion (Fig. 2 A and B; Fig. S1). Nonuniform values for R2 rates reflect structural contacts, with a fit of the data demonstrating maxima near residues with restricted motion (31). Maxima in the fit to the R2 rates are observed at a number of positions, including near Tyr-14 and Thr-45, residues hypothesized to be involved in transient tertiary contacts based on broadening observed for Tyr-14 upon interaction with Cdc4 (see below; Fig. S2). To test this hypothesis, we measured the hydrodynamic radii by using PFG diffusion experiments for phosphorylated Sic1 having mutations in these residues. Substitution of Tyr-14 or Thr-45 for Ala in pSic1 increases the Rh values to 23.2 ± 0.5 Å or 23.1 ± 0.3 Å at 25 °C, respectively (Table S4), an expansion of 20% compared with wild-type, suggesting that specific tertiary contacts contribute to the compactness. Paramagnetic relaxation enhancement (PRE) data obtained for isolated pSic1 (Fig. S3) provide additional evidence for multiple transient tertiary contacts. The predominantly negative heteronuclear NOEs (hetNOEs; Fig. 2B) confirm that Sic1 is intrinsically disordered. The same regions with enhanced R2 rates show slightly positive hetNOEs in pSic1. Both R2 and hetNOE report on the same motional restriction of fast dynamics in disordered proteins (H. Schwalbe, personal communication). The overall low values of the hetNOEs rule out folding of pSic1 and confirm the intrinsically disordered nature of the protein. The compactness and significant amount of transient secondary and tertiary structure may influence the binding properties of pSic1.
Fig. 2.
Sic1 contains transient structure, yet is highly dynamic in both free and complexed states. (A) Transverse 15N relaxation rate constants of Sic1 (green bars) and pSic1 (black bars). Clusters of residues exhibiting restricted motion described by Gaussian distributions were used to fit (red line) the transverse relaxation rates of pSic1 (31) (see also SI Materials and Methods). (B) HetNOEs of Sic1 (green bars) and pSic1 (black bars). (C) HetNOEs of pSic1 in complex with Skp1–Cdc4 at a molar ratio (Skp1–Cdc4:pSic1) of 1.2, at which 99% of pSic1 is bound. Negative and small positive values demonstrate disorder of pSic1 even in the complex. Trans- and cis-prolyl isomers are reported as black and red bars, respectively. The hetNOE expected for a rigid large complex at this field strength is depicted as a dashed line at 0.82 (46). Red circles indicate phosphorylation sites.
Multiple Suboptimal CPD Sites of Sic1 Engage Cdc4 Without Global Ordering.
We next evaluated the interaction of 6-fold phosphorylated Sic1 with a monomeric Skp1–Cdc4 complex; the Skp1 subunit of SCFCdc4 is required for soluble expression of Cdc4 but does not interact with substrates (24). Using an intrinsic Trp fluorescence-binding assay, we measured an apparent overall Kd value of ≈0.6 μM between the pSic1 preparation and Skp1–Cdc4, whereas no binding was detected for Sic1. Titration of unlabeled Skp1–Cdc4 into a solution of 15N-labeled pSic1 allowed us to monitor the binding interaction by broadening of signals in NMR 1HN-15N correlation spectra. Disordered proteins give rise to sharp NMR resonances because of fast internal motions and short effective correlation times. Interactions of disordered regions with large folded proteins lead to resonance line broadening of interacting residues because of a larger effective correlation time, restricted local motion, and possible exchange between free and bound states on millisecond to microsecond time scales. In addition, we suspect effects from conformational exchange within Skp1–Cdc4, which is not amenable to high-resolution NMR because of its very broad NMR resonances. At a Skp1–Cdc4 to pSic1 molar ratio of 1.2, for which 99% of Sic1 is bound at the concentrations used, many observable NMR signals are evident (Fig. S4). This result is in striking contrast to the overall severe line broadening that would be expected in a stable protonated 80-kDa complex at 5 °C. We measured hetNOEs for the complex and found overall low-negative or low-positive hetNOE values for the residues that are not broadened beyond detection in the complex (Fig. 2C), confirming the disordered nature of large parts of pSic1 in complex with Skp1–Cdc4. The unaltered chemical shifts of the observable signals of pSic1 in the presence of Skp1–Cdc4 further demonstrate that pSic1 is disordered when in complex with Skp1–Cdc4.
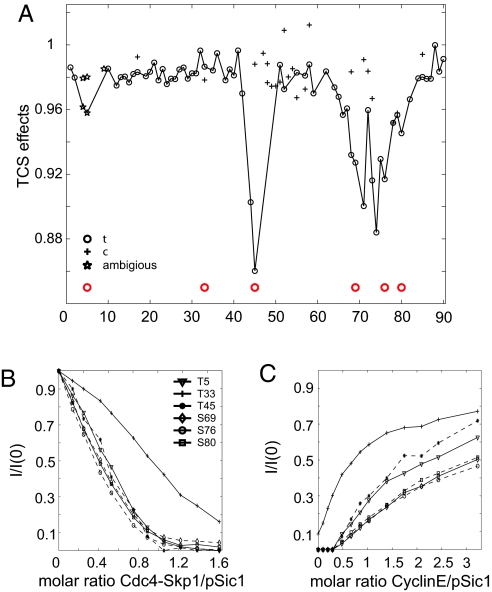
The interaction between pSic1 and Cdc4 was probed in a site-specific manner by using alternative NMR techniques. The binding interface of a protein complex can be mapped by transferred cross-saturation (TCS), which measures decreases in the resonance intensities caused by the transfer of magnetization saturation (32, 33). TCS experiments were used to monitor the interaction of deuterated pSic1 and protonated Skp1–Cdc4 complex via the amide resonances of the free pSic1 (Fig. 3A and Fig. S5). At a pSic1:Cdc4 molar ratio of ≈26:1, we detected significant TCS effects that mapped exactly to the phosphorylation sites pThr-5, pThr-45, pSer-69, pSer-76, and pSer-80, reflecting direct transfer of saturated magnetization over a short distance from protonated Skp1–Cdc4 to Sic1. The strongest TCS effects are observed at pThr-45 and pSer-76 with no significant TCS effect seen for pThr-33 (note that Thr-2 was again not phosphorylated). These data provide strong evidence that at least 5 of the 6 phosphorylated suboptimal CPD sites interact directly with Cdc4.
Fig. 3.
Multiple phosphorylation sites interact with Cdc4 in a dynamic equilibrium. (A) TCS effects on 2H,15N pSic1 (610 μM) in the presence of unlabeled Skp1–Cdc4 for trans- (circles) and cis-prolyl isomers (crosses) at a molar ratio (pSic1:Skp1–Cdc4) of ≈26:1. Ratios of resonance intensities from backbone amide groups with and without irradiation are normalized by a control experiment without Skp1–Cdc4 present [(Iirr/I0)+Cdc4/(Iirr/I0)−Cdc4] to correct for effects from residual protonation in pSic1. In cases of >2 isomers, only effects for cis- and trans-isomers with respect to the P+1 proline of the CPD are reported (while data are solely from trans-isomers of other prolines). Where only 1 symbol is plotted, cis- and trans-prolyl isomers were not resolved. Because isomers could not be assigned unambiguously in the region around pThr-5, effects for all isomers in this region are depicted as stars. Lines are plotted through the most broadened isomers as a guide. Phosphorylation sites are indicated by red circles. (B) Intensity ratios of HN resonances of Sic1 phosphorylation sites (trans-isomers) as a function of Cdc4:pSic1 molar ratio. (C) Relative intensities of HN resonances of Sic1 phosphorylation sites upon back-titration with a high-affinity cyclin E-derived phosphopeptide (CycE9pT380, Kd 1μM).
We also analyzed the ratios of resonance intensities of labeled pSic1 in the free form and in the presence of Skp1–Cdc4 as a function of residue position (Fig. S2a) from our initial titration of unlabeled Skp1–Cdc4 into 15N-labeled pSic1. We observed local minima of up to 9 residues in length around the phosphorylation sites pThr-5, pThr-45, pSer-69, pSer-76, and pSer-80, consistent with the engagement of these sites with Cdc4 and primarily local ordering of the binding motif. The relative signal intensities at each particular position approximately reflect the proportion of unbound sites, with the resonance at pThr-33 being less broadened during the titration than other CPD sites (Fig. 3B), consistent with its low local affinity. Although Thr-45 is genetically the most important site, our broadening data point to it being engaged to a lesser degree than the closely spaced phosphoserine CPDs. The strong TCS effect for pThr-45 is determined by rates of binding and release in addition to the affinity. The secondary and tertiary structure involving this residue may explain its in vivo significance, likely for overall affinity, and could also compete with local binding. Importantly, the reduced signal intensity at each CPD site in the Cdc4 complex is reversed by addition of a high-affinity phosphopeptide derived from cyclin E (CycE9pT380, GLLpT380PPQSG) that has a Kd of 1 μM for Skp1–Cdc4 (Fig. 3C) (24). Together, the line broadening and TCS data clearly indicate an interaction between pSic1 and Cdc4 in which most, if not all, of the CPD sites engage in proportion to their individual local affinities (Table S2).
Specific Interaction of Trans-Prolyl Isomers.
In the crystal structure of Cdc4 in complex with the CycE9pT380 phosphopeptide (24), the proline residue occupying the P+1 binding pocket (relative to the phosphorylated residue at the P0 position) adopts a strict trans configuration. In contrast, engagement of a downstream phosphorylated residue (P+4) at a secondary binding surface in diphosphorylated peptide–Cdc4 structures does not constrain residues at the P+5 position (26), even if the residue is a proline (X. Tang, et al., unpublished data). Thus, the trans-prolyl isomer is specific for interactions with the primary binding pocket on Cdc4. For both the TCS and broadening data, where the two prolyl isomers of individual CPDs can be resolved, interaction effects are significantly more pronounced for resonances of the trans-prolyl isomers (Fig. 3A, circles as opposed to crosses; Fig. S2a). The prevalence of trans-prolyl isomers after the phosphorylation sites interacting with Cdc4 is firm evidence that the complex between pSic1 and Cdc4 is mediated largely by the interaction of multiple suboptimal CPD sites with the core pSer/pThr–Pro binding pocket.
Discussion
Sic1 and Cdc4 Interact in a Dynamic Complex.
NMR resonance broadening and TCS effects demonstrate that the high-affinity pSic1–Cdc4 interaction is mediated by multiple suboptimal CPDs. The preferential engagement of trans-prolyl isomers further demonstrates that multiple CPD motifs bind in the core pocket of Cdc4 and not at secondary sites. The transfer of saturation in the TCS experiments is observed because of exchange between free and Cdc4-interacting Sic1, ruling out a collection of static complexes. These findings can only be explained by a model in which multiple CPDs rapidly exchange on and off of the Cdc4-binding surface (Fig. 4). The individual weak affinities of isolated short CPD phosphopeptides derived from Sic1 (ref. 25; Table S2) translate to a difference in free energy of binding that is on the order of thermal energy. For example, the ΔΔGbinding of 4.3 kJ/mol between the pSer-69/pSer-76/pSer-80 and the pThr-2/pThr-5 phosphopeptides would correspond to an equilibrium binding ratio of 85:15 for these 2 peptides at 25 °C. Importantly, the proportional occupancies of the corresponding regions of pSic1 in complex with Cdc4 (Fig. 3B and Fig. S2) are even more similar than expected from the affinities of isolated phosphopeptides. These similar apparent local affinities are consistent with a dynamic equilibrium in which each individual CPD interacts transiently with the P0/P+1 binding pocket of Cdc4, thereby enabling exchange among different sites.
Fig. 4.
Dynamic complex of pSic1 and Cdc4. Multiple suboptimal CPD motifs in pSic1 engage the core Cdc4-binding site in a dynamic equilibrium. Sites not directly bound in the core binding site at any given instant can contribute to the binding energy via a second binding site or via long-range electrostatic interactions. A more detailed view of one possible interaction is shown, with the pThr-45 CPD binding to Cdc4. The linear binding motif that is ordered upon interaction is shown in stick representation [model based on the structure of the Cdc4–cyclin E peptide complex (24)]; the parts of pSic1 remaining disordered are depicted as black lines.
Elements of Dynamic Protein Interactions.
The mechanism of Sic1 recognition by Cdc4 and its dependence on multiple phosphorylation events have been debated in the literature. One model requires Sic1 phosphorylation on multiple distinct sites to create sufficient affinity for a biological response (18), whereas a second model assumes that a single specific diphosphorylated sequence is necessary and sufficient for targeting of Sic1 (26). The results we present here are consistent with the first model, in which multiple phosphorylation sites dynamically engage the core Cdc4-binding site. Although diphosphorylated CPD motifs indeed bind Cdc4 with higher affinity (25, 26), none approaches the affinity of fully phosphorylated Sic1 (Table S2). Rather, the contribution of +3/+4 phosphorylation sites most likely affords a means to fine tune the affinity of local CPD sites, as observed for both N-terminal hydrophobic and C-terminal basic residues in the CPD consensus sequence (18). Moreover, given the flexible but compact nature of Sic1, conformational constraints and electrostatic repulsion may substantially weaken individual site contributions to overall affinity. Indeed, whereas the pSer-69/pSer-76/pSer-80 triphosphopeptide has a Kd value of 2.4 μM, which in principle might be sufficient to target pSic1 to Cdc4, this cluster alone cannot mediate the pSic1–Cdc4 interaction (20). Notably, the isolated triphosphopeptide has a net charge of −4, whereas the corresponding triphosphorylated full-length targeting region has a net charge of +5 (see Table S2). As observed for a Sic1 phosphopeptide series, a mean-field model predicts that phosphorylated states of Sic1 having less positive charge will exhibit a stronger interaction with the positively charged Cdc4-binding surface (25).
A requirement for multiple phosphorylation events in Sic1 in principle sets a high threshold for the level of active G1 CDK required to initiate transition to S phase, thereby ensuring a coordinated onset of DNA synthesis and genomic stability (18). Although our in vitro demonstration of a dynamic complex does not explain the putative ultrasensitivity of the interaction, it provides a structural foundation for understanding the models for polyvalent interactions that have been proposed to lead to ultrasensitivity (25). Long-range electrostatic interactions with averaging effects over a dynamic structural ensemble can in principle generate highly nonlinear binding in response to phosphorylation (25, 34). The transient structure and compact nature of pSic1 may also contribute to its high-affinity interaction with Cdc4. Compactness may not only facilitate exchange of the different sites, but also long-range electrostatic interactions and reduced solvent screening effects. In particular, a disordered ensemble of compact states could generate an average negative electrostatic field that would help tether pSic1 in the local vicinity of the positively charged binding surface on Cdc4 (25). Thus, the combination of intrinsic disorder and significant amounts of transient secondary and tertiary structure in Sic1 leading to compaction facilitates the observed dynamic binding mode and may lead to ultrasensitivity driven by long-range polyelectrostatic interactions.
Sic1 contains two additional phosphorylation sites remote from the targeting region (Thr-173 and Ser-191), making their participation in the described dynamic complex unlikely. It is possible, however, that Thr-173 and Ser-191 serve as decoy sites that compete with the key binding sites for phosphorylation by the targeting kinase (35), allowing different mechanisms in Sic1 to synergize to increase the ultrasensitivity of Sic1 degradation with respect to G1 CDK activity.
A recent proposal argues that multiple phosphorylation events can in principle be the basis for ultrasensitivity if (i) phosphorylation leads to reduced conformational entropy of the free disordered ligand and (ii) the number of conformations available to the bound ligand is negligible compared with its free state (36). Although we observed a small degree of motional restriction in Sic1 caused by phosphorylation (Fig. 2), transient ordering was primarily localized to the extended binding motifs (Fig. S2). The contribution of this mechanism to the ultrasensitivity of the interaction may therefore not be significant. In contrast, the multiplicity of conformations involving different sites of Sic1 in the bound complex with Cdc4, as well as the significant disorder remaining in the noninteracting regions of Sic1, increases the entropy of the bound state relative to a more typical ordered complex, contributing favorably to binding (25).
Intrinsically disordered regions frequently mediate dynamic protein interactions in signaling networks; these interactions often exhibit unusual binding characteristics, such as multisite dependence and ultrasensitivity (37, 38). We anticipate that polyvalent binding modes similar to the pSic1–Cdc4 interaction reported here will arise in other contexts, such as for the disordered regulatory region of the cystic fibrosis transmembrane conductance regulator (16), the disordered N-terminal segment of the Ets-1 transcription factor (39), and the Ste5 MAPK cascade scaffold protein (40). Based on the pSic1–Cdc4 model system, we speculate that multiple weak-binding motifs within disordered proteins may often synergize to yield high-affinity binding of the sort typically ascribed to static interactions between complementary protein interfaces. The continuum of properties of compactness, conformational heterogeneity, secondary structural propensities, and tertiary contacts within disordered protein states can in principle afford myriad variations in protein interaction modes, which may be readily shaped by evolution (41). The complex interaction dynamics of disordered proteins reshapes the conventional view of structurally rigid protein interactions, with attendant implications in protein engineering and drug discovery.
Materials and Methods
Full details are in the SI Materials and Methods.
Sic1 Sample Preparation.
Sic1 1–90 was expressed as a GST fusion protein at 16 °C in Escherichia coli BL21(DE3) codon plus cells grown in minimal medium with [15N]NH4Cl and/or [13C]glucose as the sole nitrogen and carbon sources, respectively, depending on the desired labeling scheme. For TCS experiments, 2H,15N-labeled Sic1 was expressed in minimal medium prepared in 99% 2H2O and with [15N]NH4Cl and [2H,12C]glucose as nitrogen and carbon sources. The GST-Sic1 protein was purified by using glutathione–Sepharose affinity chromatography. After digestion with TEV protease, GST was precipitated by lowering the pH to 5.0 and spun out at 42,000 × g. Sic1 was purified to homogeneity by using a Superdex 75 column (Amersham Biosciences), as confirmed by mass spectrometry. Phosphorylation was performed as reported (24) and monitored by mass spectrometry showing that the largest population was 6-fold phosphorylated. pSic1 was purified by using reversed-phase chromatography on a Gemini C18 column (Phenomenex). The CDK sites at Thr-5, Thr-33, Thr-45, Ser-69, Ser-76, and Ser-80 were phosphorylated, whereas Thr-2 was not phosphorylated in this construct. The Thr residues at positions 9, 48, and 49 that have been purported to generate high-affinity diphosphorylated epitopes necessary for Sic1 recognition (26) were also not phosphorylated as shown by the absence of downfield shift in NMR experiments.
Cdc4 Sample Preparation.
Skp1–Cdc4263–744 was purified as reported in ref. 24. Cdc4 expresses at low levels in E. coli BL21 codon plus cells (and in other expression systems tested), on the order of 0.5 mg/L culture. It was thus not feasible to isotope-label Cdc4 for NMR purposes. In addition, Skp1–Cdc4 is a 72-kDa complex that gives rise to very broad NMR resonances. We suspect that these broad resonances are caused by conformational exchange within Cdc4. The Sic1–Cdc4 interaction could therefore only be probed from the perspective of Sic1.
NMR Experiments.
All NMR data were collected on Varian Inova 500-MHz, 600-MHz, and 800-MHz spectrometers at 5 °C except for PFG diffusion experiments, which were carried out at 5, 10, 15, 20, and 25 °C as indicated in Table S4. The NMR samples were prepared in PBS [10 mM phosphate, 140 mM NaCl (pH 7.0)], 1 mM EDTA, 0.2% NaN3, 10% D2O.
Assignments.
Unphosphorylated and phosphorylated Sic1 1–90 wild type (WT) and pSic1pT45 samples at 0.25–0.35 mM [in PBS (pH 7.0), 1 mM EDTA, 0.2% NaN3] were used for assignment experiments according to standard procedures. Triple-resonance assignment experiments (42) were performed at 800 MHz, including CBCA(CO)NNH and HNCACB to examine Cα/Cβ chemical shifts. We used the program SSP (28) to combine chemical shifts for different nuclei into a SSP score representing the expected fraction of α-helical or extended structure at a given residue.
PFG diffusion experiments.
Experiments were carried out on 200 μM and 600 μM samples of Sic1 and pSic1, respectively, as 1D 1H pulse gradient stimulated echo longitudinal encode–decode (PG-SLED) experiments with watergate sequences (29). One percent dioxane (2.5 μL) was used as an internal standard with an effective hydrodynamic radius (Rhref) of 2.12 Å. Hydrodynamic radii were extracted as described in SI Materials and Methods.
Relaxation experiments.
15N R1, R1ρ, and hetNOE values were measured by using published pulse schemes (43) with details described in SI Materials and Methods.
Models of a polypeptide chain with unrestrained segmental motions predict R2 relaxation rates that reach a plateau in the middle of the sequence, whereas the protein termini exhibit smaller relaxation rates. The observation of regions of residues with higher relaxation rates indicates the presence of transient secondary or tertiary structure. Transverse relaxation rates of pSic1 were fitted to a model of segmental motions and deviations centered around clusters of residues as described in SI Materials and Methods (31).
TCS experiments of pSic1 WT and Skp1–Cdc4.
The TCS experiments were carried out as described (32, 44) on a 600-MHz spectrometer equipped with a cold probe. A 545 μM sample of 2H,15N-labeled pSic1 in complex with 1.3 or 3.8 mol% (7 μM or 21 μM, respectively) unlabeled Skp1–Cdc4263–744 was prepared in PBS at pH 7.0, 1 mM EDTA, 0.02% NaN3, 10% D2O. Saturation of the aliphatic protons of Skp1–Cdc4263–744 was done with a WURST-2 adiabatic pulse with a maximum radiofrequency (RF) amplitude of 1,800 Hz and a saturation frequency of 0.6 ppm at a saturation time of 2.5 s. TCS effects are presented as ratios of (Iirr/I0) with Cdc4 and (Iirr/I0) without Cdc4 [(Iirr/I0)Cdc4/(Iirr/I0)woCdc4]. A control experiment was performed that is described in SI Materials and Methods.
NMR Titrations of pSic1 WT and Mutants with Skp1–Cdc4.
HSQC spectra were recorded with 24 transients at 5 °C (as above) on 360 μM (pSic1 WT) and 230 μM (pSic1pT45) 15N/13C-labeled Sic1 samples at 800 and 500 MHz, respectively, for increasing concentrations of Skp1–Cdc4263–744. Data were analyzed by taking the ratio of bound to free signal intensities.
All data were processed with the NMRPipe/NMRDraw suite of programs (45) or “Fuda: A function and data fitting and analysis package” (S. M. Kristensen and D. F. Hansen, personal communication).
Supplementary Material
Acknowledgments.
We thank M. Borg, H. S. Chan, V. Kanelis, E. Stollar, and T. Pawson for many fruitful discussions; A. Davidson for use of his fluorometer; and R. Muhandiram for assistance with NMR experiments. T.M. is the recipient of a Terry Fox Post Ph.D. Research Fellowship. Additional support was provided by funds from the Canadian Cancer Society (to M.T. and J.D.F.-K.) and the Canadian Institutes of Health Research (CIHR) (to F.S. and J.D.F.-K.). L.E.K. is a Canada Research Chair; M.T. holds a Royal Society Wolfson Research Merit Award and is a Howard Hughes Medical Institute International Research Scholar and a Research Professor of the Scottish Universities Life Sciences Alliance.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809222105/DCSupplemental.
References
- 1.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 2.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Radivojac P, et al. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Demarest SJ, Deechongkit S, Dyson HJ, Evans RM, Wright PE. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci. 2004;13:203–210. doi: 10.1110/ps.03366504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Guzman RN, Martinez-Yamout MA, Dyson HJ, Wright PE. Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: Regulation by competition between intrinsically unstructured ligands for nonidentical binding sites. J Biol Chem. 2004;279:3042–3049. doi: 10.1074/jbc.M310348200. [DOI] [PubMed] [Google Scholar]
- 8.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 9.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 10.Neduva V, et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLOS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 12.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Oldfield CJ, et al. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 14.Sivakolundu SG, Bashford D, Kriwacki RW. Disordered p27Kip1 exhibits intrinsic structure resembling the Cdk2/cyclin A-bound conformation. J Mol Biol. 2005;353:1118–1128. doi: 10.1016/j.jmb.2005.08.074. [DOI] [PubMed] [Google Scholar]
- 15.Vacic V, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker JMR, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tompa P, Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 19.Schwob E, Boehm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 20.Verma R, et al. Phosphorylation of Sic1p by G1 CDK required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 21.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 22.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 23.Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 24.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 25.Borg M, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci USA. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7–Skp1–cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Verma R, Feldman RMR, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structural propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkins DK, et al. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 30.Kohn JE, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein-Seetharaman J, et al. Long-range interactions within a nonnative protein. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 32.Nakanishi T, et al. Determination of the interface of a large protein complex by transferred cross-saturation measurements. J Mol Biol. 2002;318:245–249. doi: 10.1016/S0022-2836(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 33.Shimada I. NMR techniques for identifying the interface of a larger protein–protein complex: Cross-saturation and transferred cross-saturation experiments. Methods Enzymol. 2005;394:483–506. doi: 10.1016/S0076-6879(05)94020-2. [DOI] [PubMed] [Google Scholar]
- 34.Klein P, Pawson T, Tyers M. Mathematical modeling suggests cooperative interactions between a disordered polyvalent ligand and a single receptor site. Curr Biol. 2003;13:1669–1678. doi: 10.1016/j.cub.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Kim SY, Ferrell JE. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 36.Lenz P, Swain PS. An entropic mechanism to generate highly cooperative and specific binding from protein phosphorylations. Curr Biol. 2006;16:2150–2155. doi: 10.1016/j.cub.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Cohen P. The regulation of protein function by multisite phosphorylation: A 25-year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 38.Serber Z, Ferrell JE. Tuning bulk electrostatics to regulate protein function. Cell. 2007;128:441–444. doi: 10.1016/j.cell.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Pufall MA, et al. Variable control of ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 40.Strickfaden SC, et al. A mechanism for cell cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerhart J, Kirschner M. The theory of facilitated variation. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8582–8589. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- 43.Farrow NA, et al. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi K, et al. Structural basis of the KcsA K+ channel and Agitoxin2 pore-blocking toxin interaction by using the transferred cross-saturation method. Structure. 2003;11:1381–1392. doi: 10.1016/j.str.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.