Abstract
The small fourth chromosome of Drosophila melanogaster (3.5% of the genome) presents a puzzle. Cytological analysis suggests that the bulk of the fourth, including the portion that appears banded in the polytene chromosomes, is heterochromatic; the banded region includes blocks of middle repetitious DNA associated with heterochromatin protein 1 (HP1). However, genetic screens indicate 50–75 genes in this region, a density similar to that in other euchromatic portions of the genome. Using a P element containing an hsp70-white gene and a copy of hsp26 (marked with a fragment of plant DNA designated pt), we have identified domains that allow for full expression of the white marker (R domains), and others that induce a variegating phenotype (V domains). In the former case, the hsp26-pt gene shows an accessibility and heat-shock-inducible activity similar to that seen in euchromatin, whereas in the latter case, accessibility and inducible expression are reduced to levels typical of heterochromatin. Mapping by in situ hybridization and by hybridization of flanking DNA sequences to a collection of cosmid and bacterial artificial chromosome clones shows that the R domains (euchromatin-like) and V domains (heterochromatin-like) are interspersed. Examination of the effect of genetic modifiers on the variegating transgenes shows some differences among these domains. The results suggest that heterochromatic and euchromatic domains are interspersed and closely associated within this 1.2-megabase region of the genome.
Heterochromatin, defined as that portion of the eukaryotic genome that remains condensed as the cell progresses from metaphase to interphase, has a high proportion of repetitious DNA, contains relatively few genes, and characteristically is replicated late during S phase (1, 2). In Drosophila melanogaster, ≈30% of the genome is heterochromatic, including large blocks surrounding the centromeres and, in somatic cells, all of the Y chromosome (3–5). The juxtaposition of genes normally found in euchromatin with heterochromatic material can result in gene silencing. In D. melanogaster, an inversion in the X chromosome that places the white+ gene (required for a red-eye phenotype) adjacent to a break point in the pericentric heterochromatin results in a mosaic pattern of expression referred to as position effect variegation (PEV); the gene has been silenced in some of the cells in which it is normally expressed (2, 6). Silencing within heterochromatic domains has been associated with local alterations in chromatin structure and/or shifts in chromatin packaging that render the DNA less accessible to cleavage reagents (7–10). Specific multimeric protein complexes seem to be involved. Once established, the assembly of such complexes may propagate to include more extended sections of the DNA (as in PEV); however, such spreading is not a normal occurrence in the intact genome, suggesting the presence of boundaries to heterochromatin assembly (11).
Using D. melanogaster as the model organism, we have screened for domains that induce a variegating phenotype by mobilizing a P element vector P[hsp26-pt, hsp70-w] containing an hsp70-driven white gene and a tagged copy of the hsp26 gene (fused to a fragment of barley cDNA, designated pt, at +490 bp; ref. 10). The endogenous hsp26 gene can be readily induced to high levels of expression in most tissues at most developmental stages with heat-shock treatment. The chromatin structure of hsp26 has been well characterized (12, 13), providing a base line for examination of the chromatin structure of the transgene in different domains. The hsp70-driven white gene provides a visual marker to monitor transgene expression. A uniform red-eye phenotype (at 25°C) is observed after insertion of this P element into a euchromatic domain (sites within the banded regions of chromosomes X, 2, and 3), whereas insertion into a heterochromatic domain results in a variegating eye phenotype (10). Over 50 variegating lines have been recovered, and the position of the transgene has been mapped on the polytene chromosomes by in situ hybridization. Of these, 12 are associated with the pericentric heterochromatin (14); 17 are associated with the telomeres (15); and 24 have been observed within the banded region of the fourth chromosome.
Chromosome 4 in D. melanogaster has an overall length of 4.5–5.2 megabases (16). Most of the chromosome, 3–4 megabases, is comprised of simple satellite repeats and does not contain any known genes. The remaining portion (estimated at 1.2 megabases) has a banded pattern in polytene chromosomes (cytological region 101E–102F; ≈1% of the total bands); it appears to be a mosaic of middle and low copy repetitive DNA (often transposons) interspersed with unique DNA (17–20). Immunofluorescent staining with antibodies against HP1 gives a banded pattern across this region (21). The chromosome as a whole has been reported to be late replicating (22). However, genetic analysis suggests that the gene density within the banded region of the fourth chromosome is not very different from that in the other chromosome arms. In an attempt to saturate the region, Hochman (23) identified 37 separate vital loci (lethal, semilethal, or sterile when homozygous for the mutation) and 6 other recessive mutations with visible phenotypes; he estimated the total number of genes on chromosome 4 to be between 50 and 75. Taken together, the data suggest that the fourth chromosome of D. melanogaster may consist of interspersed euchromatic and heterochromatic domains. To test this idea, we have used the P element described above to screen for transgenic lines with insertion sites on the fourth chromosome that show full expression and properties of euchromatin-like function. Six such lines have been recovered and characterized. Mapping studies clearly show that the heterochromatin-like domains, inducing a variegating phenotype, and the euchromatin-like domains, allowing full expression of the transgenes, are interspersed along the chromosome.
Materials and Methods
Drosophila Culture and P Element Mobilization.
Flies were cultured on standard cornmeal-sucrose-yeast-agar medium at 25°C. The screen to recover flies showing a PEV phenotype has been described (10). Seven of these lines having the P element within the banded region of the fourth and one line (118E-10) with the P element within the pericentric region of the fourth were studied. Flies showing a full-red-eye phenotype, carrying the P element on the fourth chromosome, were recovered by crossing females from line 39C-12 (transgene at position 102B on the fourth chromosome, variegating phenotype) to males carrying the Δ2-3 transposase; male progeny that were Sb, Δ2-3; P[hsp26-pt, hsp70-w] were independently crossed to y w67c23; net; sbd; spapol females, and the resulting male progeny with full red eyes were screened (by using the same female stock) for segregation of the P element with the fourth chromosome. From 1,200 males examined, 5 were recovered carrying a single P element on the fourth. One line (1-M707) was recovered from a similar screen starting with females from line 39C-X (transgene on the X at 2D; red-eye phenotype); this screen was ≈10-fold less efficient.
Determination of P Element Insertion Sites.
The cytological positions of the transgene inserts were determined with polytene chromosomes isolated from late third instar larvae (24) hybridized with the P element construct as a probe (10). Map positions were established at higher resolution by hybridization of cloned fragments of adjacent genomic DNA to cosmid and bacterial artificial chromosome (BAC) clones from the fourth chromosome (20, 45). The DNA flanking the P element inserts was cloned in vector pCR3.1 (Invitrogen) by using inverse PCR (14). The primers used were 5′-GCTTCGGCTATCGCGGGACCACCACCTTATGTTA-3′ and 5′-GACGAAATGAACCACTCGGACCATTTTGAGCG-3′. DNAs isolated from various BAC/cosmid clones were dotted on to a GeneScreen Plus membrane (Schleicher & Schuell) and then treated for 10 min in 0.5 M NaOH, followed by 10 min in 1.0 M Tris⋅HCl (pH 7.5); the filter then was dried. Membranes were hybridized with denatured, 32P probe in 6× SSC, 1% SDS, with 50 mM phosphate buffer (pH 7.5) at 65°C overnight. Membranes were washed at 65°C once in 2× SSC with 0.1% SDS and twice in 0.2× SSC with 0.1% SDS before autoradiographic exposure.
Northern Analysis.
Total RNA was isolated from hemizygous female flies (4–5 days after emergence) that had been heat-shocked at 37°C for 1 h. The RNA was fractionated on a 1.2% agarose gel with formaldehyde and transferred to a nylon membrane (25). Expression of the transgene was detected by using the plant DNA fragment (pt) isolated from plasmid pGH19 (26) as a probe. RNA loading was monitored with the rp49 probe (27). Probes were labeled with [α-32P]dATP and [α-32P]dCTP by using random primers. Expression levels of the transgenes in R and V lines were assessed with a PhosphorImager and normalized to control line 39C-X (set at 100%).
Chromatin Structure Analysis.
Chromatin structure was assessed by using XbaI digestion of nuclei from third-instar larvae as described (28, 29). Purified DNA from XbaI-treated nuclei was digested to completion with SalI, run in a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with the plant DNA fragment pt labeled with [α-32P]dATP and [α-32P]dCTP. Hybridization products were detected by autoradiography. Proximal XbaI site accessibility was calculated as the ratio of the amount of signal from the gel band resulting from that cleavage to the total from all three bands within the lane (signal assessed with a PhosphorImager). Values were normalized to the value obtained from line 39C-X.
Testing the Effects of Known Modifiers of PEV.
Homozygous females from V lines were crossed to males carrying modifiers of PEV: (line 1) y w67c23 (control), (line 2) y w67c23; Su(var)2-101/Cy, (line 3) y w67c23; Su(var)2-502/Cy, (line 4) y w1118; Su(var)3-7/T(2, 3)apXa, (line 5) y w67c23; E(var)3-93D/TM3, y+ Ser Sb, and (line 6) y w67c23, E(var)3-93E/TM3]. Female progeny (4–5 days after emergence) carrying both the transgene and the modifier mutation were photographed. The level of eye pigmentation of the hemizygous female V line flies with the transgene in a y w background was used as the standard control; siblings containing the balancer chromosome were not used for comparison, because the balancers frequently have accumulated mutations, including some that modify PEV. To measure eye pigmentation, five hemizygous adult female flies (4–5 days after emergence) were homogenized in 0.5 ml of 0.01 M HCl in ethanol (30). The homogenate was placed at 4°C overnight and then warmed at 50°C for 5 min. After centrifugation in a microfuge for 10 min, the supernatant was recovered, and the OD at 480 nm was recorded. The average value from female y w67c23 flies was used to establish background values. Four to eight independent samples were analyzed for each stock; means and standard deviations are reported.
Results
Recovery of Transgenic Lines on the Fourth Chromosome.
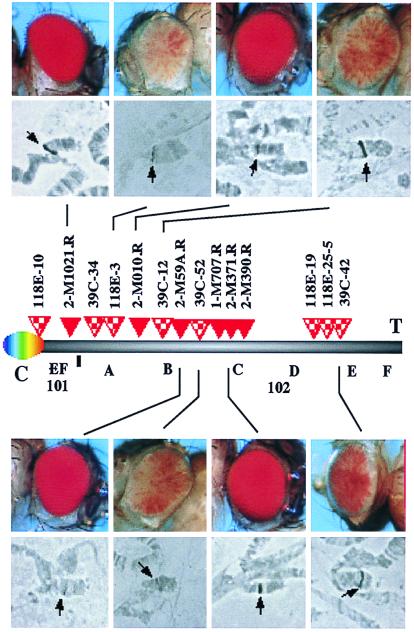
A screen was designed to recover lines with P element inserts on the fourth chromosome showing the desired phenotype; six such “R” lines, showing a full red eye and containing a single P element (determined by Southern analysis), were obtained (see Materials and Methods). These six lines have been studied in a comparison with eight lines that have a single P element within the fourth chromosome that show a variegating phenotype (V lines), recovered in the earlier screen (10). The V lines are all homozygous viable; four of the R lines are homozygous lethal, whereas lines 2-M390R and 2-M1021R are weakly viable. In situ hybridization with the P element as a probe places the insert in line 118E-10 in the pericentric heterochromatin of chromosome 4; all other lines studied carry the transgene within the banded region of the fourth chromosome (Fig. 1).
Figure 1.
Transgenes within the banded region of chromosome 4 can have a variegating or a nonvariegating phenotype. Six R lines and eight V lines have been characterized. Photographs of the eye phenotypes (representative samples) were taken from hemizygous y w67c23; P[hsp26-pt, hsp70-w]/+ adult female flies. Map positions of the transgene inserts were established as described (Materials and Methods). The order of insertion sites is unambiguous, except that the relative positions of 118D-19 and 118E-25-5 cannot be distinguished. Abbreviations: C, centromere; T, telomere.
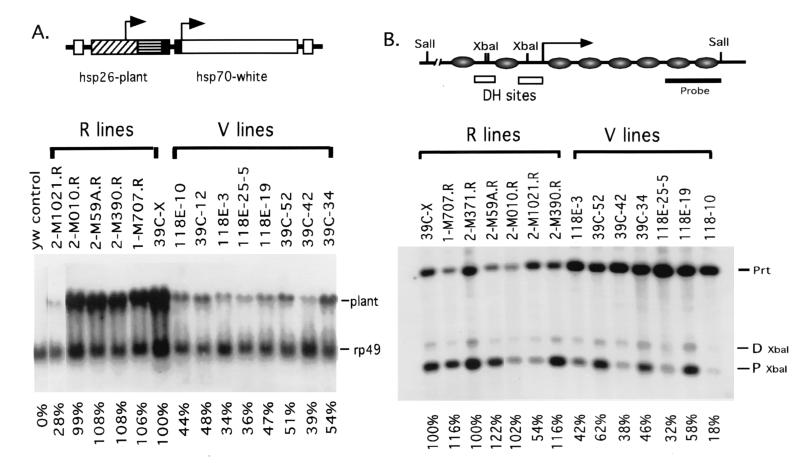
To determine whether the transcriptional activity of the transgenes in the R lines is similar to that of the transgenes in euchromatic domains on other chromosomes, we have characterized the expression level of hsp70-white by measuring eye pigments and the expression level of hsp26-pt after heat-shock induction by using Northern analysis. Line 39C-X, which contains the P element at position 2D on the X chromosome, was used as the control. Five of the six R lines showed mean eye pigment levels of 94% to 126% and mean hsp26-pt expression of between 108% and 112% relative to 39C-X (see Table 1 and Fig. 2A). Line 2-M1021R, which has a P insert close to a negative regulatory element of the ci gene (see below), shows 87% eye pigment levels and 48% hsp26-pt induction. Aside from this exceptional case, the results indicate the presence of euchromatic environments on the fourth chromosome that mimic the expression properties of those found elsewhere in the genome. The mean eye pigment levels for the eight V lines vary from 8% to 16% of 39C-X control values, whereas mean heat-shock-inducible expression of hsp26-pt varies from 30% to 43% of 39C-X control values. These eye pigment values and values of hsp26-pt expression are similar to those observed for the transgenes inserted into pericentric heterochromatin (10, 14).
Table 1.
Characteristics of R and V lines
| Lines | Insertion site of the P element | Eye pigmentation level, % | Expression of the hsp26 transgene, % | XbaI accessibility, % | Identified cosmid or BAC |
|---|---|---|---|---|---|
| 39C-X | 2D | 100 (n = 4) | 100 (n = 4) | 100 (n = 3) | ND |
| 2-M1021R | 101F | 87 ± 9 (n = 4) | 48 ± 13 (n = 3) | 57 ± 11 (n = 4) | L7L |
| 2-M010R | 102B | 100 ± 5 (n = 4) | 111 ± 8 (n = 2) | 103 ± 6 (n = 2) | BAC R1E21 |
| 2-M59AR | 102B | 126 ± 12 (n = 4) | 111 ± 7 (n = 3) | 101 ± 8 (n = 2) | 14b14 |
| 1-M707R | 102C | 98 ± 7 (n = 4) | 108 ± 4 (n = 3) | 120 ± 9 (n = 3) | 37107 |
| 2-M371R | 102C | 122 ± 6 (n = 4) | 112 ± 10 (n = 2) | 104 ± 6 (n = 3) | 37107 |
| 2-M390R | 102C | 94 ± 11 (n = 4) | 108 ± 9 (n = 3) | 127 ± 5 (n = 3) | 37107 |
| 118E-10 | Centromere | 8 ± 1 (n = 8) | 31 ± 9 (n = 3) | 18* | ND |
| 39C-34 | 102A | 11 ± 1 (n = 8) | 41 ± 12 (n = 3) | 38 ± 8 (n = 3) | BAC 17A1, 55e5 |
| 118E-3 | 102A | 11 ± 1 (n = 4) | 34 ± 9 (n = 3) | 44 ± 6 (n = 3) | BAC N42J09 |
| 39C-12 | 102B | 10 ± 2 (n = 5) | 42 ± 5 (n = 2) | 47* | 19o03 |
| 39C-52 | 102B-C | 12 ± 1 (n = 6) | 43 ± 6 (n = 3) | 48 ± 14 (n = 3) | ND |
| 118E-19 | 102D-E | 15 ± 1 (n = 12) | 37 ± 11 (n = 3) | 47 ± 8 (n = 3) | 19h01 |
| 118E-25-5 | 102D-E | 11 ± 2 (n = 8) | 39 ± 7 (n = 2) | 37 ± 5 (n = 2) | 19h01 |
| 39C-42 | 102D-E | 16 ± 1 (n = 8) | 30 ± 8 (n = 3) | 35 ± 7 (n = 2) | 19h01, 12o5 |
ND, not determined.
See also ref. 10.
Figure 2.
(A) The hsp26-pt transgene in an R line shows full heat-shock-inducible expression, whereas that in a V line shows low heat-shock-inducible expression. A map of the P element used is shown (Upper; ref. 10). Expression of the transgene after heat shock was detected by using the plant DNA fragment to hybridize a Northern blot (values normalized to control line 39C-X). (B) hsp26-pt transgenes in V lines are packaged in a less accessible chromatin structure than are those in R lines. A schematic representation of the hsp26 promoter region of the euchromatic gene is shown (Upper); circles represent the approximate locations of nucleosomes, and the two nucleosome-free regions (DNase I hypersensitive sites containing the heat shock regulatory elements) upstream of the transcription start site (arrow) are indicated. Chromatin structure was assessed by using XbaI digestion of nuclei from third-instar larvae (see Materials and Methods); proximal XbaI site accessibility is given normalized to that of line 39C-X.
We next examined the chromatin structure of the hsp26-pt transgenes, looking at accessibility of the XbaI site located in the proximal regulatory region (Fig. 2B). This site is part of a DNase-I-hypersensitive region in the endogenous gene and is highly accessible to nucleases in transgenes within euchromatic domains. The regulatory region of the hsp26-pt transgene in five of six R lines is highly accessible to the restriction enzyme XbaI, with mean values ranging from 101% to 127% (average 111%) relative to the value for 39C-X; again, line 2-M1021R gives a relatively low value, averaging 57%. (Genomic sequencing showed the hsp26 regulatory sequence in the 2-M1021R transgene to be intact and unaltered.) In contrast, the mean accessibility of the XbaI site in the eight V lines is between 18% and 48% of that of the control 39C-X, similar to results obtained earlier for the transgene in pericentric heterochromatin (10). The results support the hypothesis that both euchromatic and heterochromatic domains are present within the banded portion of chromosome 4 and provide further data indicating that the local chromatin structure has an important role in regulating gene expression.
Euchromatic and Heterochromatic Domains Are Interspersed.
As shown in Fig. 1, in situ hybridization of the polytene chromosomes indicates that the transgenes analyzed herein are located in the banded portion of the fourth chromosome. (Control line 118E-10 is in the pericentric heterochromatin.) The assignments made in this study (Table 1) are based on our analysis of multiple experiments. In some cases, in situ hybridization was also carried out with cloned DNA from toy or ey as a reference probe; similar results were obtained (39C-12,102B; 39C-52,102C; 118E-19,102D-E; 118E-25-5,102C; 39C-42,102D-E) (P. Callaerts, U. Kloter, S. Flister, and W. Gehring, personal communication). However, the resolution that can be obtained with this approach is limited. To generate a higher resolution map, providing an unambiguous order for the insertion sites, the flanking DNA for each transgene was cloned by inverse PCR (14); these “tag clones” were used to screen an ordered collection of cosmid and BAC clones from the fourth chromosome (20, 45). By assigning each tag clone to one or more cosmid and/or BAC clones (Table 1), we have been able to derive the relative positions of the transgene insertion sites for the R and V lines (Fig. 1). The results show that R and V insertion sites are indeed interspersed, indicating that domains that allow full expression (euchromatin) and those inducing a variegating phenotype (heterochromatin) are interspersed along the fourth chromosome.
This higher resolution analysis of the data indicates some clustering of P element insertion events. Three of the V lines show insertion sites in cosmid clone 19h01, suggesting a “hot spot” for insertion in the relatively large block of heterochromatin in band 102E. Two of these tag clones (from lines 39C-42 and 118E-25-5) apparently include highly repetitious DNA, as shown by using these clones to probe a Southern blot of genomic DNA (data not shown). Three of the R lines show insertion sites in cosmid clones 37107 and 16j20, which are known to contain the zfh-2 gene (20). Lines 2-M371R, 1-M707R, and 2-M663R (the latter was not studied in detail, because it contains two copies of the P element) form a complementation group (O. Peter and K. Basler, personal communication). Sequencing of the tag clones and comparison to the sequence of cosmid 16j20 place the insertion site for 2 M-371R ≈1 kilobase upstream of the 5′ end of the zfh-2 transcript and that for 2 M-390R within the first intron.
The tag clone from line 2-M1021R hybridizes to cosmid L7L, which is known to contain three genes, including cubitus interruptus (ci; ref. 20). Genetic analysis indicated that line 2-M1021R is an allele of ci; homozygotes have a fusion of wing veins 4 and 5 because of inappropriate expression of ci (R. Holmgren, personal communication). Sequencing of the tag clone places the insertion site within a cluster of repeats upstream of the ci transcript (20). This 3-kilobase region has a similar, nonidentical, AT-rich sequence present approximately every 200 bp. The P element insertion site is between the promoter and the 5′ sequences required to prevent ectopic expression of ci in the posterior compartment (31). The proximity to this negative regulatory element may explain the low (but uniform, nonvariegating) level of expression of the transgenes in line 2-M1021R.
Although it has not yet been possible to define the sites of insertion for the other R lines, the results obtained to date are consistent with the hypothesis that the transgenes in the R lines are located close to genes that are normally expressed. In the one case where we are aware of a direct experimental test, a genomic clone including the ci gene provided normal gene function when placed in other euchromatic domains by P element transformation (32, 33). Thus, both our results and those of others argue that the euchromatic domains on chromosome 4 are functionally similar to those elsewhere in the genome.
Genetic Characterization of the Variegating Lines.
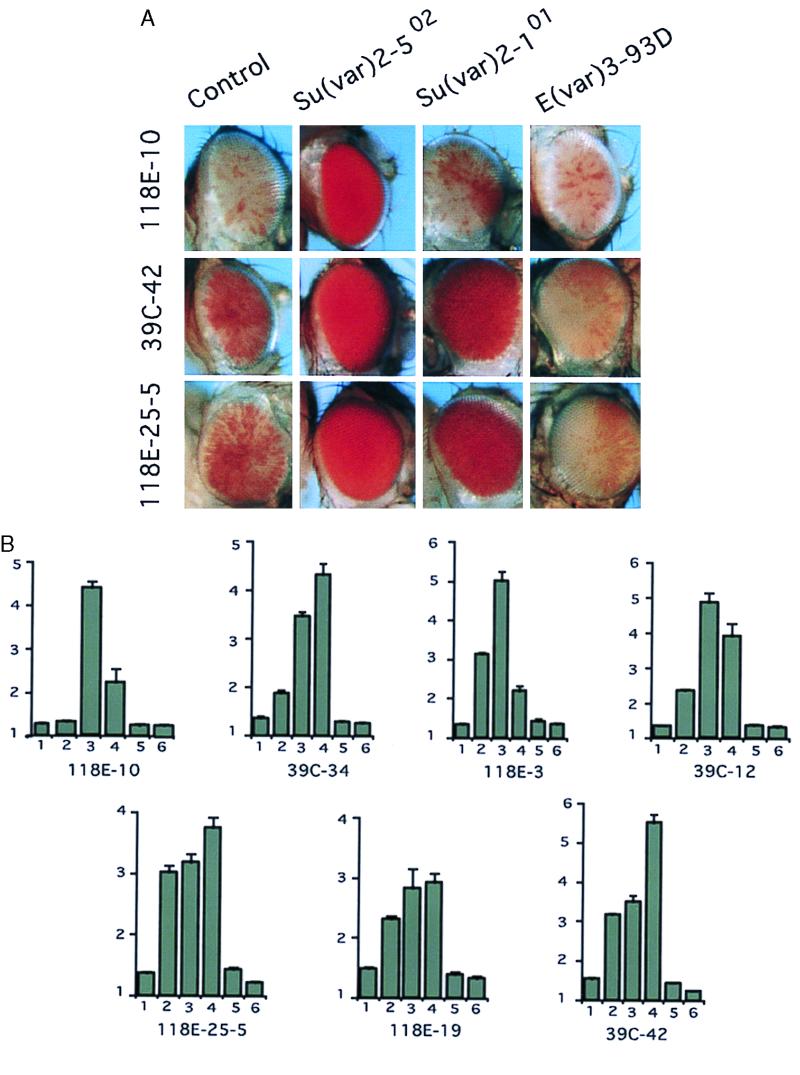
Several of the genetic loci known to suppress or enhance a PEV phenotype [Su(var) or E(var) mutations] have been characterized in detail and shown to encode chromosomal proteins or to affect the modification of chromosomal proteins (2, 6). Having obtained a set of variegating lines carrying the same P element insert at different positions on chromosome 4, one can ask whether all of these fourth chromosome heterochromatic domains seem to have a similar functional composition; if so, we would expect a similar response to modifiers of PEV. To examine this question, we set up crosses between several Su(var) and E(var) mutations and seven V lines, assessing the change in eye pigmentation levels. Modifiers examined included Su(var)2-101, which is associated with increased levels of histone acetylation (34); Su(var)2-502, a missense mutation in HP1 (35); Su(var)3-7, a null mutation (deficiency) for a zinc-finger protein known to interact with HP1 (36); E(var)3-93E, a hypomorph of the E2F transcription factor gene (37); and E(var)3-93D, a mutation in the gene encoding Mod(mdg4) (38, 39). The results are shown in Fig. 3. Silencing of the transgene was suppressed in all of the V lines by the Su(var)2-502 and Su(var)3-7 mutations, suggesting that these proteins, thought to play a structural role in pericentric heterochromatin, contribute generally to the heterochromatic structures on the fourth chromosome. However, the impact of Su(var)2-101, which is associated with increased histone acetylation, varied; 118E-10 (centromeric insertion site) showed no significant response, whereas all other V lines showed a response. None of the V lines showed any response to the E(var)3-93E mutation. Lines 39C-42 and 118E-25-5 showed a significant enhancement of silencing in the presence of the E(var)3-93D mutation, whereas the other six V lines were not affected significantly. Quantitative differences in the responses were noted consistently; however, a similar pattern of response is observed for lines 118E-25-5, 118E-19, and 39C-42, which are close to each other on chromosome 4 (tags identify the same cosmid) and thus may fall within a single heterochromatic domain. The results suggest that the different blocks of heterochromatin on the fourth chromosome, although they have a common structural basis, may vary in the specific protein complexes associated with a given site. Such a pattern is reminiscent of the one observed for silencing complexes based on the Pc-G proteins (40). The overall pattern of responses indicates that the variegating transgenes on the fourth chromosome are packaged in a heterochromatin that resembles that of the pericentric region.
Figure 3.
Responses of the variegating transgenes on the fourth chromosome to different genetic modifiers. (A) Homozygous females from V lines were crossed to males carrying modifiers of PEV as indicated. Female progeny carrying both the transgene and the modifier mutation were photographed. (B) Eye pigmentation levels for these flies (values 10 × OD480). The number under each bar indicates that the transgene is present with y w67c23 (control) (bar 1), Su(var)2-101 (bar 2), Su(var)2-502 (bar 3), Su(var)3-7 (bar 4), E(var)3-93E (bar 5), or E(var)3-93D (bar 6).
Discussion
The P element-based reporter P[hsp26-pt, hsp70-w] provides a useful tool for assessing gene expression and chromatin structure at a given position within the Drosophila genome, allowing a characterization of the organization of the chromosomes into different functional domains. Using this tool, we have found that the small fourth chromosome of D. melanogaster seems to consist of interspersed domains of euchromatin and heterochromatin, defined in terms of the expression characteristics and chromatin structure of the transgenes as described above. The results of this functional analysis reconcile the previous cytological and genetic studies indicating that, despite the presence of blocks of heterochromatin, the genes on the fourth chromosome indeed lie in euchromatin-like domains and can function as such. Results from a study of x-ray-induced deficiencies have suggested a similar organization, with heterochromatic sequences separating groups of genes, in section 20 at the base of the X chromosome (41). HP1 staining of the polytene chromosomes indicates a small set of HP1-positive bands at the bases of chromosome arms 2R and 3L (21); thus, a similar region of interspersion may be present between the “mostly euchromatic” banded regions and “mostly heterochromatic” pericentric regions as a general feature of the larger Drosophila chromosomes (41).
One anticipates that heterochromatic domains will contain primarily repetitious sequences, whereas euchromatic domains will be largely unique. The recent sequence analysis of cosmids cosL7L and cos16j20 has identified the presence of known repetitious elements (such as Hoppel) and the short DINE repeats in and around the ci and zfh-2 genes (20); thus, the mere presence of such repetitious sequences is not sufficient to create a heterochromatic domain. Further, despite numerous findings that indicate that nuclear organization can affect gene silencing (42, 43), clearly mere proximity to heterochromatin is not sufficient. Boundary elements that flank the transcriptionally repressed HMR locus in the yeast Saccharomyces cerevisiae have been identified and characterized recently (44); these elements prevent the spread of silencing to adjacent genes. The present analysis indicates that functionally similar elements can be inferred to be at work on the D. melanogaster fourth chromosome.
Acknowledgments
We thank Jo Wuller and Lynn Podenski for technical assistance and U. Kloter, P. Caellarts, S. Flister, and W. Gehring (University of Basel), O. Peter and K. Basler (University of Zurich), and R. Holmgren (Northwestern University) for permission to cite their unpublished findings. S.C.R.E. thanks M. Ashburner, J. Roote, A. Carpenter, S. Russell, and other members of the Ashburner lab (University of Cambridge) for assistance and hospitality during a sabbatical leave. We thank the members of our research groups for reading and discussing the manuscript. This work was supported by National Institutes of Health Grant HD23844 (to S.C.R.E.) and grants from the Medical Research Council of Canada and the Natural Sciences and Engineering Research Council of Canada (to J.L.). F.-L.S. is a W. M. Keck Research Fellow.
Abbreviations
- BAC
bacterial artificial chromosome
- PEV
position effect variegation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF242878–AF242880).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090530797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090530797
References
- 1.Heitz E. Jahrb Wiss Bot. 1928;69:726–818. [Google Scholar]
- 2.Weiler K S, Wakimoto B T. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 3.Gatti M, Pimpinelli S. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- 4.Karpen G. Curr Opin Genet Dev. 1994;4:281–291. doi: 10.1016/s0959-437x(05)80055-3. [DOI] [PubMed] [Google Scholar]
- 5.Elgin S C R. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 6.Wallrath L L. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 7.Loo S, Rine J. Science. 1994;164:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 8.Allshire R C, Javerzat J-P, Redhead N J, Cranston G. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 9.Weiss K, Simpson R T. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallrath L L, Elgin S C R. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 11.Hennig W. Chromosoma. 1999;108:1–9. doi: 10.1007/s004120050346. [DOI] [PubMed] [Google Scholar]
- 12.Cartwright I L, Elgin S C R. Mol Cell Biol. 1986;6:779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas G H, Elgin S C R. EMBO J. 1988;7:2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cryderman D E, Cuaycong M H, Elgin S C R, Wallrath L L. Chromosoma. 1998;107:277–285. doi: 10.1007/s004120050309. [DOI] [PubMed] [Google Scholar]
- 15.Cryderman D E, Morris E J, Biessmann H, Elgin S C R, Wallrath L L. EMBO J. 1999;18:3724–3735. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locke J, McDermid H E. Chromosoma. 1993;102:718–723. doi: 10.1007/BF00650898. [DOI] [PubMed] [Google Scholar]
- 17.Miklos G L G, Yamamoto M-T, Davies J, Pirrotta V. Proc Natl Acad Sci USA. 1988;85:2051–2055. doi: 10.1073/pnas.85.7.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmena M, Gonzalez C. Chromosoma. 1995;103:676–684. doi: 10.1007/BF00344228. [DOI] [PubMed] [Google Scholar]
- 19.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R. Genome Res. 1999;9:137–149. [PMC free article] [PubMed] [Google Scholar]
- 21.James T C, Eissenberg J C, Craig C, Dietrich V, Hobson A, Elgin S C R. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 22.Barigozzi C, Dolfini S, Fraccaro M, Rezzonico-Raimondi G, Tiepolo L. Exp Cell Res. 1966;43:231–234. doi: 10.1016/0014-4827(66)90399-5. [DOI] [PubMed] [Google Scholar]
- 23.Hochman B. Cold Spring Harbor Symp Quant Biol. 1973;38:581–589. doi: 10.1101/sqb.1974.038.01.062. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M, editor. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. 37. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Heck G R, Ho T-H D. Plant Mol Biol. 1996;30:611–623. doi: 10.1007/BF00049335. [DOI] [PubMed] [Google Scholar]
- 27.Wong Y-C, O'Connell P, Rosbash M, Elgin S C R. Nucleic Acids Res. 1981;9:6749–6762. doi: 10.1093/nar/9.24.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q, Wallrath L L, Granok H, Elgin S C R. Mol Cell Biol. 1993;13:2802–2814. doi: 10.1128/mcb.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cartwright I L, Cryderman D E, Gilmour D S, Pile L A, Wallrath L L, Weber J A, Elgin S C E. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- 30.Khesin R B, Leibovitch B A. Mol Gen Genet. 1978;162:323–328. doi: 10.1007/BF00268858. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz C, Locke J, Hishida C, Kornberg T B. Development (Cambridge, UK) 1995;121:1625–1635. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- 32.Methot N, Basler K. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 33.Hepker J, Blackman R, Holmgren R. Development (Cambridge, UK) 1999;124:549–558. [Google Scholar]
- 34.Dorn R, Heymann S, Lindigkeit R, Reuter G. Chromosoma. 1986;93:398–403. [Google Scholar]
- 35.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C R. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleard F, Delattre M, Spierer P. EMBO J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seum C, Spierer A, Pauli D, Szidonya J, Reuter G, Spierer P. Development (Cambridge, UK) 1996;122:1949–1956. doi: 10.1242/dev.122.6.1949. [DOI] [PubMed] [Google Scholar]
- 38.Dorn R, Crauss V, Reuter G, Saumweber H. Proc Natl Acad Sci USA. 1993;90:11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerasimova T I, Corces V G. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 40.Pirrotta V. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 41.Lifschytz E. Genetics. 1978;88:457–467. doi: 10.1093/genetics/88.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 43.Cockell M, Gasser S M. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 44.Donze D, Adam C R, Rine J, Kamakaka R T. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Locke, J., Podemski, L., Aippersbach, N., Kemp, H. & Hodgetls, R. (2000) Genetics, in press. [DOI] [PMC free article] [PubMed]