Abstract
Objectives
To investigate how the SOS response, an error-prone DNA repair pathway, is expressed following subinhibitory quinolone treatment of Mycobacterium tuberculosis.
Methods
Genome-wide expression profiling followed by quantitative RT (qRT)–PCR was used to study the effect of ciprofloxacin on M. tuberculosis gene expression.
Results
Microarray analysis showed that 16/110 genes involved in DNA protection, repair and recombination were up-regulated. There appeared to be a lack of downstream genes involved in the SOS response. qRT–PCR detected an induction of lexA and recA after 4 h and of dnaE2 after 24 h of subinhibitory treatment.
Conclusions
The pattern of gene expression observed following subinhibitory quinolone treatment differed from that induced after other DNA-damaging agents (e.g. mitomycin C). The expression of the DnaE2 polymerase response was significantly delayed following subinhibitory quinolone exposure.
Keywords: SOS repair, mutation, gene expression
Introduction
Quinolones are commonly used for the treatment of a wide range of bacterial infections. Resistance has emerged rapidly in important bacterial pathogens including methicillin-resistant Staphylococcus aureus (MRSA)1 largely through transmission, Pseudomonas aeruginosa1 and Streptococcus pneumoniae.2 Resistance emerging in Mycobacterium tuberculosis could threaten the development of these agents for the treatment of susceptible and multidrug-resistant disease that is currently underway.3
Fluoroquinolones act on DNA gyrase resulting in lethal double-stranded DNA breaks.4 In response to DNA damage, there is potent induction of the SOS regulon: a set of genes involved in DNA repair, recombination and mutagenesis.5 The first step in the pathway depends on activation by recA and repression by lexA. Quinolones are less mutagenic in cells that cannot mount an SOS response (for example, stationary-phase cells, RecA− mutants6 and gyrA mutants5). The low fidelity polymerases induced during the SOS response enhance survival through the emergence of quinolone resistance mutations.7
Members of the quinolone family with C-8 position substituents, for example, moxifloxacin, have increased efficacy at killing resistant gyrase and topoisomerase IV (a quinolone target in bacteria other than mycobacteria) mutants.8 This is important as the bactericidal action of the quinolones with C-8 substituents will not only kill mutants but will prevent further mutation. While ciprofloxacin has been part of the regimen for treatment of multidrug-resistant tuberculosis (TB), the superior activity of moxifloxacin (and the probability that it is less likely to select resistance) against pulmonary TB has led to its further evaluation in clinical studies.9
We have shown that exposure of Mycobacterium fortuitum to subinhibitory concentrations of quinolone increases the mutation rate.10 When cultures were exposed to 1/2 MIC ciprofloxacin, there was a 73–120-fold increase in mutation rate. This effect was found to be dose-dependent with a smaller increase associated with 1/4 and 1/8 MIC. The increase in mutation rate was found for all selecting agents including rifampicin, erythromycin, gentamicin and moxifloxacin, suggesting that the mutagenic effect of fluoroquinolones is across the whole genome. These data are in accord with studies that showed that subinhibitory doses of fluoroquinolones result in an increased mutation rate in Escherichia coli, MRSA and P. aeruginosa.1,4,5
The implications for anti-TB therapy are clear. The prolonged use of fluoroquinolones in an anti-TB regimen may risk treatment with sub-optimal doses, increasing the selection pressure to resistance.11 To characterize the mechanism behind the induction of mutation by subinhibitory fluoroquinolone exposure in mycobacteria, the induction of the error-prone SOS system was investigated by whole genome expression profiling and quantitative RT (qRT)–PCR in M. tuberculosis.
Materials and methods
Quinolone treatment of cultures
Exponentially growing M. tuberculosis H37Rv (NCTC 7416) (∼106 cfu/mL) cultured in Middlebrook 7H9 broth, containing 0.2% Tween 80 (BDH) and 10% albumin dextrose catalase enrichment (BD), was exposed to 0.25 mg/L (1/2 MIC) or 0.125 mg/L (1/4 MIC) of ciprofloxacin (Cellgro Herndom) over a period of 24 h. Untreated culture controls were included at 0 h where sterile distilled water was added instead of ciprofloxacin. As a positive control of the SOS response, a sample was also treated with 0.2 mg/L mitomycin C (Sigma Aldrich) for 24 h. This experiment was repeated three times.
RNA extraction
RNA was extracted from each culture at 0, 4, 12 and 24 h using the method of Mangan et al.12 and the Fast RNA Pro-Blue Kit (Q-Biogene Inc.). Samples were purified using the RNeasy MiniElute Cleanup Kit for bacteria (Qiagen Ltd). The RNA was quantified using the Agilent 2100 Bioanalyser System (Agilent Technologies Inc.).
Microarray protocol
M. tuberculosis whole genome arrays, consisting of PCR products representing 3924 genes from M. tuberculosis H37Rv, were supplied by the Bacterial Microarray Group at St George's Hospital, University of London (ArrayExpress accession number: A-BUGS-2; http://bugs.sgul.ac.uk/A-BUGS-2). Aliquots of 5 µg (equivalent to ∼2–5 × 108 bacteria) of total RNA were used as a template for cDNA synthesis and fluorescent dye analogues incorporated during reverse transcription using Cy3-dCTP. This was hybridized against Cy5-labelled M. tuberculosis H37Rv genomic DNA (provided by Colorado State University). For each sample, two technical replicates were performed. Arrays were scanned using an Affymetrix™ 428 array scanner (MWG). All images were overlaid for feature extraction in ImaGene 5.5 (BioDiscovery Inc.) and further processed with the MAVI Pro 2.6.0 software package (MWG Biotech). Data were entered into GeneSpring GX™ 7.2 (Agilent Technologies) software to perform normalization and statistical analysis.
Statistical analysis
The ratios of Cy3/Cy5 intensities were median normalized using only the data flagged as present or marginal by ImaGene. These ratios were further normalized to the control samples to provide a more meaningful expression ratio that reflected up- or down-regulation compared with the control. Differentially expressed genes were identified by the ANOVA-based approach using a parametric test in which variances were not assumed equal (Welch t-test) with a false discovery rate of <0.05.
Fully annotated microarray data has been deposited in BµG@Sbase (accession number: E-BUGS-63; http://bugs.sgul.ac.uk/E-BUGS-63) and also ArrayExpress (http://www.ebi.ac.uk/microarray-as/aer/entry; accession number: E-BUGS-63).
qRT–PCR
qRT–PCR was performed using primers and dual-labelled probes to amplify dnaE2 from Boshoff et al.,13 recA (forward primer, aatgaccggcgcgctga; reverse primer, cgttgaagttctacgcgtc; probe, ttcgggcaccacggcgatcttcat), lexA (primers from Brooks et al.;14 probe, tcttcccgctgccgcgtgagc) and sigA (forward primer and probe from Hampshire et al.;15 reverse primer, ttctcgacctgatccaggaag).
RNA (5 µL of ∼1 pg/reaction) was added to 1× QuantiTect Probe RT–PCR Master Mix (Qiagen) and made up to a final volume of 25 µL with RNase-free water (Qiagen). The reaction was performed on a Rotorgene™ 3000 thermal cycler (Corbett Research). The reaction components and the cycling conditions were all as outlined in the manufacturer's instructions in the QuantiTect™ Probe RT–PCR Handbook. Each sample was processed in triplicate, including a control without RT for each sample also performed in triplicate. All qRT–PCR experiments included a positive control of RNA from M. tuberculosis H37Rv. The threshold level was set to 0.1, which was above the background fluorescence of the ‘no template’ control in the exponential phase of the curve. The efficiency of each probe and primer pair was >99%. Relative gene expression was calculated using the 2−ΔΔCt method,16 normalizing all genes to sigA, which is considered to be constant under stress conditions.17
Results and discussion
Gene expression analysis by microarrays following exposure to subinhibitory concentrations of ciprofloxacin revealed differential expression across the whole genome. The 4 h treatment with 1/2 MIC ciprofloxacin resulted in the greatest number of genes differentially expressed. The expression of genes involved in DNA repair, mutagenesis and recombination in M. tuberculosis was further analysed [Table S1, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)]. One hundred and ten genes were identified using the gene list presented by Mizrahi and Andersen,18 and a search of the Entrez Genome, NCBI database of the COG (clusters of orthologous groups of proteins) classification DNA replication, recombination and repair. The microarray data indicated that, following subinhibitory concentrations of ciprofloxacin, 16/110 genes involved in DNA repair, mutagenesis and recombination were up-regulated (Table 1) and 11/110 genes were down-regulated (data not shown). Following this microarray analysis, we investigated further the expression of recA and lexA, representing the primary genes involved in the SOS response, and dnaE2, the polymerase thought to be associated with mutagenesis.13 For these experiments, ciprofloxacin exposure was extended to 24 h and mitomycin C, a DNA-damaging agent known to induce the SOS response, was included as a control.
Table 1.
Up-regulated genes from DNA repair, recombination and mutagenesis gene list (110 genes) with statistically significant differences among time and MIC for 4 and 12 h treatments with 1/2 and 1/4 MIC ciprofloxacin including the t-test P value
| Gene name | Rv no. | Normalized expression (P value) | Treatment | Predicted function |
|---|---|---|---|---|
| tagA | Rv1210 | 1.44 (3.10E–02) | 12 h 1/2 MIC | DNA-3-methyladenine glycosidase I |
| recF | Rv0003 | 1.50 (1.22E–03) | 4 h 1/4 MIC | DNA replication and SOS induction |
| Rv3202c | 1.51 (6.95E–03) | 12 h 1/2 MIC | possible ATP-dependent DNA helicase | |
| uvrD2 | Rv3198c | 1.51 (3.40E–03) | 4 h 1/4 MIC | putative UvrD |
| 1.65 (1.16E–02) | 12 h 1/2 MIC | |||
| nei | Rv3297 | 1.60 (1.66E–02) | 12 h 1/2 MIC | probable endonuclease VIII |
| dnaB | Rv0058 | 1.66 (2.79E–02) | 12 h 1/4 MIC | DNA helicase (contains intein) |
| Rv2896c | 1.68 (1.89E–05) | 4 h 1/2 MIC | predicted Rossmann fold nucleotide-binding protein involved in DNA uptake | |
| 2.11 (3.92E–03) | 4 h 1/4 MIC | |||
| mutT4 | Rv3908 | 1.76 (8.24E–04) | 12 h 1/2 MIC | mutator protein MutT |
| recA | Rv2737c | 1.78 (5.16E–04) | 4 h 1/4 MIC | recombinase (contains intein) |
| 4.13 (8.31E–06) | 12 h 1/2 MIC | |||
| gyrA | Rv0006 | 1.87 (4.59E–03) | 12 h 1/4 MIC | DNA gyrase subunit A |
| dut | Rv2697c | 1.89 (5.34E–03) | 4 h 1/2 MIC | deoxyuridine triphosphatase |
| 2.00 (1.99E–05) | 4 h 1/4 MIC | |||
| xthA | Rv0427c | 2.11 (9.16E–06) | 4 h 1/4 MIC | exodeoxyribonuclease III |
| 4.30 (1.33E–02) | 12 h 1/2 MIC | |||
| 3.13 (1.96E–03) | 12 h 1/4 MIC | |||
| radA | Rv3585 | 2.14 (1.24E–02) | 12 h 1/2 MIC | probable DNA repair RadA homologue |
| dnaE1 | Rv1547 | 2.72 (2.43E–03) | 12 h 1/2 MIC | DNA polymerase III, [alpha] subunit |
| Rv2821c | 2.84 (6.83E–05) | 4 h 1/2 MIC | uncharacterized protein predicted to be involved in DNA repair (RAMP superfamily) | |
| 2.27 (7.26E–06) | 4 h 1/4 MIC | |||
| xthA | Rv0427c | 3.13 (1.96E–03) | 12 h 1/4 MIC | exodeoxyribonuclease III |
Genes are ranked by ascending levels of expression when compared with the untreated control at time 0 h.
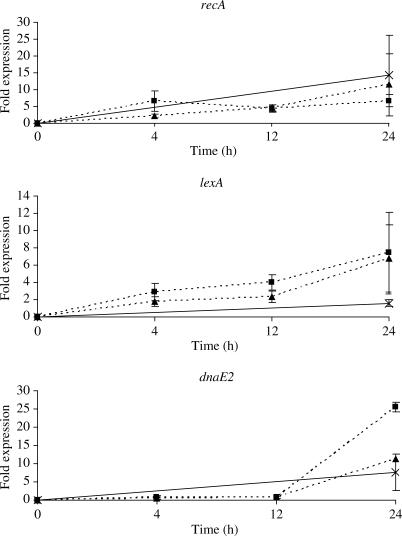
A 2-fold up-regulation of recA was observed by microarray analysis after 4 h of treatment with 1/4 MIC ciprofloxacin and 4-fold after 12 h of treatment with 1/2 MIC ciprofloxacin, indicating induction of the SOS response (Table 1). This up-regulation was confirmed by qRT–PCR. Expression of recA increased after 4 h by 2-fold and continued to increase to 4-fold after 12 h of treatment with 1/4 MIC ciprofloxacin (Figure 1). Differences in lexA expression during treatment were not identified by microarray analysis but a gradual increase in fold expression over 24 h was observed by qRT–PCR, from 1.8 to 2.3 to 6.8 and 2.9 to 4 to 7.4 with 1/4 and 1/2 MICs, respectively (Figure 1). Subtle shifts of gene expression may not be detected by microarray-based transcriptome analysis because of a smaller dynamic range than qRT–PCR so it is possible that more genes involved in DNA repair may be up-regulated.19 Small changes in LexA expression have been observed in M. tuberculosis after various DNA-damaging treatments but only after extended periods of exposure.20
Figure 1.
Relative fold change in expression of recA, lexA and dnaE2 normalized to sigA calculated by the equation 2−ΔΔCt, after treatment with 1/4 MIC ciprofloxacin (triangles), 1/2 MIC ciprofloxacin (squares) or 0.2 mg/L mitomycin C (crosses) over 24 h. The values are the means of triplicate samples on at least two independent inductions; the error bars indicate the standard error of the mean.
The expression of dnaE2 was found unchanged in the microarray analysis; however, a delayed response, only observed after 12 h, was detected by qRT–PCR with an increase from 0.8-fold to 25.5-fold and 0.9-fold to 11.2-fold between 12 and 24 h, after 1/2 and 1/4 MIC treatments, respectively (Figure 1).
Following 24 h of treatment with mitomycin C, recA, dnaE2 and lexA were overexpressed (Figure 1). The levels of recA expression (14.2-fold) were comparable to those found in other studies;14,20 however, induction of dnaE2 (7.7-fold) and lexA (1.6-fold) were lower than that reported previously,13,21 but one of these studies used a 90 min exposure to mitomycin C.
The mechanism whereby quinolones increase the mutation rates is thought to be the error-prone SOS system.22,23 Microarray-based analysis of M. tuberculosis gene expression response to sub-MIC of quinolones did not show a consistent pattern of SOS up-regulation. It was noted that both microarray and qRT–PCR analyses demonstrated a significant delay in DnaE2 response. These data suggest that the SOS response following subinhibitory quinolone exposure differs from that with other DNA-damaging agents. Further work is required to dissect these pathways in view of their critical role in the mechanism of hypermutability. The risk of selecting for resistance must be considered when introducing a quinolone to a TB treatment regimen, especially if sub-optimal dosing occurs.
Funding
The whole genome M. tuberculosis microarray was constructed and analysed at St George's, University of London, as part of the multi-collaborative microbial pathogen microarray facility (BµG@S), for which funding from The Wellcome Trust's Functional Genomics Resources Initiative is acknowledged (grant number 062511).
Transparency declarations
S. H. G. and T. D. M. have received research funding from Bayer-Schering HealthCare who manufacture quinolone antibiotics. The remaining authors have none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Supplementary Material
Acknowledgements
We thank Dr Lucy Brooks for help with depositing data in BµG@Sbase and ArrayExpress.
References
- 1.Fung-Tomc J, Kolek B, Bonner DP. Ciprofloxacin-induced, low-level resistance to structurally unrelated antibiotics in Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1289–96. doi: 10.1128/aac.37.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillespie SH, Voelker LL, Ambler JE, et al. Fluoroquinolone resistance in Streptococcus pneumoniae: evidence that gyrA mutations arise at a lower rate and that mutation in gyrA or parC predisposes to further mutation. Microb Drug Resist. 2003;9:17–24. doi: 10.1089/107662903764736300. [DOI] [PubMed] [Google Scholar]
- 3.Rustomjee R, Lienhardt C, Kanyok T, et al. A Phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int J Tuberc Lung Dis. 2008;12:128–38. [PubMed] [Google Scholar]
- 4.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–92. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips I, Culebras E, Moreno F, et al. Induction of the SOS response by new 4-quinolones. J Antimicrob Chemother. 1987;20:631–8. doi: 10.1093/jac/20.5.631. [DOI] [PubMed] [Google Scholar]
- 6.Levin DE, Marnett LJ, Ames BN. Spontaneous and mutagen-induced deletions: mechanistic studies in Salmonella tester strain TA102. Proc Natl Acad Sci USA. 1984;81:4457–61. doi: 10.1073/pnas.81.14.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisognano C, Kelley WL, Estoppey T, et al. A RecA–LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J Biol Chem. 2004;279:9064–71. doi: 10.1074/jbc.M309836200. [DOI] [PubMed] [Google Scholar]
- 8.Dong Y, Xu C, Zhao X, et al. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth, survival, and resistance. Antimicrob Agents Chemother. 1998;42:2978–84. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moadebi S, Harder CK, Fitzgerald MJ, et al. Fluoroquinolones for the treatment of pulmonary tuberculosis. Drugs. 2007;67:2077–99. doi: 10.2165/00003495-200767140-00007. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie SH, Basu S, Dickens AL, et al. Effect of subinhibitory concentrations of ciprofloxacin on Mycobacterium fortuitum mutation rates. J Antimicrob Chemother. 2005;56:344–8. doi: 10.1093/jac/dki191. [DOI] [PubMed] [Google Scholar]
- 11.Ferrara AM. New fluoroquinolones in lower respiratory tract infections and emerging patterns of pneumococcal resistance. Infection. 2005;33:106–14. doi: 10.1007/s15010-005-4102-8. [DOI] [PubMed] [Google Scholar]
- 12.Mangan JA, Monahan IM, Butcher PD. Gene expression during host-pathogen interactions: approaches to bacterial mRNA extraction and labelling for microarray analysis. In: Wren B, Dorrell N, editors. Functional Microbial Genomics: Methods in Microbiology. London: Academic Press; 2002. pp. 137–51. [Google Scholar]
- 13.Boshoff HI, Reed MB, Barry CE, III, et al. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–93. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 14.Brooks PC, Movahedzadeh F, Davis EO. Identification of some DNA damage-inducible genes of Mycobacterium tuberculosis: apparent lack of correlation with LexA binding. J Bacteriol. 2001;183:4459–67. doi: 10.1128/JB.183.15.4459-4467.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampshire T, Soneji S, Bacon J, et al. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis. 2004;84:228–38. doi: 10.1016/j.tube.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Kendall SL, Movahedzadeh F, Rison SC, et al. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinb) 2004;84:247–55. doi: 10.1016/j.tube.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol. 1998;29:1331–9. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 19.Shi L, Sohaskey CD, Kana BD, et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci USA. 2005;102:15629–34. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papavinasasundaram KG, Anderson C, Brooks PC, et al. Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology. 2001;147:3271–9. doi: 10.1099/00221287-147-12-3271. [DOI] [PubMed] [Google Scholar]
- 21.Davis EO, Dullaghan EM, Rand L. Definition of the mycobacterial SOS box and use to identify LexA-regulated genes in Mycobacterium tuberculosis. J Bacteriol. 2002;184:3287–95. doi: 10.1128/JB.184.12.3287-3295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirz RT, O'Neill BM, Hammond JA, et al. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol. 2006;188:7101–10. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984;48:273–89. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.