Expression quantitative trait loci (eQTL) mapping studies have become a widely used tool for identifying genetic variants that affect gene regulation. In these studies, expression levels are viewed as quantitative traits, and gene expression phenotypes are mapped to particular genomic loci by combining studies of variation in gene expression patterns with genome-wide genotyping. Results from recent eQTL mapping studies have revealed substantial heritable variation in gene expression within and between populations. In many cases, genetic factors that influence gene expression levels can be mapped to proximal (putatively cis) eQTLs and, less often, to distal (putatively trans) eQTLs. Beyond providing great insight into the biology of gene regulation, a combination of eQTL studies with results from traditional linkage or association studies of human disease may help predict a specific regulatory role for polymorphic sites previously associated with disease.
The study of gene expression phenotypes
Variation in gene expression is abundant in all organisms studied to date [1–3]. It has been suggested repeatedly that modifications in gene regulation are responsible for much of the observed phenotypic variation in natural populations. Indeed, like substitutions at the protein level, changes in gene regulation have been found to underlie numerous adaptive phenotypes in a variety of organisms, from beak morphology in Darwin finches [4], bristle number, wing pigmentation and trichome patterns in fruit flies [5–7], branching structure in maize [8], skeletal patterning and pelvic reduction in sticklebacks [9,10] to parental care in rodents [11]. Moreover, mutations in putative regulatory regions have been associated with > 100 human phenotypes including diverse aspects of behavior, physiology and disease (for a review, see Refs. [12,13]). Despite accumulating evidence that regulatory changes contribute to many important phenotypes, we still know little about the architecture of gene regulation (see Glossary) or about the genetic basis for variation in gene expression levels.
In particular, although we understand how mutations in coding regions affect the amino acid composition of proteins and, sometimes, how these mutations lead to differences in phenotypes, the effect of variation at the DNA level on transcript abundance remains elusive. In fact, it is difficult to identify regulatory regions in the genome, let alone to predict how polymorphisms in regulatory regions affect gene expression levels temporally or spatially [13]. This task becomes particularly important in humans because many of the loci identified in recent genome-wide association studies of complex human diseases are located outside of coding regions and hence are expected to have a function in gene regulation (e.g. Refs. [14–17]).
Expression quantitative trait loci (eQTL) mapping is one approach to determine which genomic regions help to regulate transcription and to study the impact of polymorphisms within these regions. In such studies, gene expression levels are treated as quantitative traits, and their genetic basis can be studied using well-established linkage and association mapping tools (Figure 1; Box 1). However, unlike traditional QTL mapping, which is typically limited to a few quantitative traits (e.g. Refs. [18,19]), DNA microarrays make it possible to measure the expression phenotypes of most genes in a genome simultaneously and map these phenotypes to specific genomic regions using genome-wide genetic markers.
Figure 1.
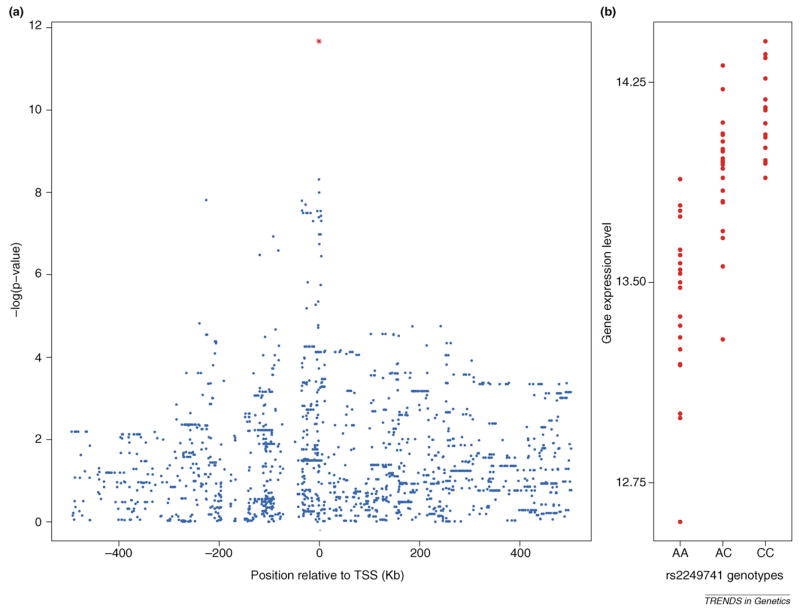
Example of an expression quantitative trait loci (eQTL) for the HLA-C gene in the HapMap European samples (data from Ref. [55]). (a) Plot of •log(P values) for the association between individual single nucleotide polymorphisms (SNPs) and expression of HLA-C. The location of the gene is indicated by the small red bar at the bottom of the figure, and the x-axis measures location relative to the transcription start site (TSS). Each data point is for a single SNP. (b) Individual expression levels of HLA-C, grouped according to the genotype of the most significant SNP in the region (rs2249741; indicated by the red data point in panel (a). Interestingly, one of the SNPs in the signal peak in panel (a) (rs92644942) has also been associated with HIV set point, suggesting that higher expression of HLA-C can help reduce HIV viral load [62].
Genome-wide mapping of eQTLs can provide great insight into the genetic architecture of gene expression variation because it simultaneously captures many regulatory interactions. The long-term goal of eQTL mapping studies is to elucidate how genotypic variation underlies morphological or physiological consequences by using gene expression levels as intermediate molecular phenotypes. For example, by combining eQTL mapping with results from traditional linkage or association studies of human disease, one can assign a specific regulatory role to polymorphic sites in a genomic region known to be associated with disease (e.g. Ref. [20]).
Here we discuss the general principles of regulatory mechanisms that are emerging from recent QTL mapping efforts in humans and other organisms and explore the challenges of mapping regulatory variation in different species. Perhaps the most salient findings of eQTL studies thus far are that (i) variation in gene expression levels is both widespread and highly heritable; (ii) gene expression levels are highly amenable to genetic mapping and (iii) most strong eQTLs are found near the target gene, suggesting that variation in cis regulatory elements underlies much of the observed variation in gene expression levels.
Variation in gene expression is widespread in human populations
Gene expression levels measured by microarrays can be affected by many nongenetic factors, including environmental variation, epigenetic modifications and random fluctuations in expression, as well as by experimental issues including measurement error, staging and (for many of the studies) variation that arises in transformed cell lines [21]. For that reason, it was not clear initially how heritable these measured expression levels would be, and early eQTL studies spent considerable effort addressing this basic question [22–25]. Indeed, measured gene expression levels of most genes were found to have statistically significant heritability.
For example, Göring et al. [24] analyzed expression data for lymphocytes isolated from 1240 individuals representing 30 large families. After removing genes whose expression levels were below a baseline threshold established by negative control samples, they found that 86% of probes mapping to RefSeq genes showed significantly heritable expression levels [at a false discovery rate (FDR) of 1%]. However, it is important to note that the actual level of heritability is moderate for most of these genes. For example, 41% of the RefSeq probes had estimated heritability >0.3, but just 5% had heritability >0.5. These results suggest that, although measured mRNA levels of most expressed genes are indeed correlated across family members, nongenetic factors are also likely to be important in determining expression levels. Moreover, because heritability estimates cannot distinguish between familial correlations due to shared genetic factors as opposed to correlations caused by shared environment, the environmental contribution could be somewhat larger than the heritability estimates would imply.
Box 1. Expression quantitative trait loci mapping methods
Mapping approaches for expression quantitative trait loci (eQTLs) in humans and other natural populations can be classified into linkage methods and association methods (reviewed in Ref. [63]). Briefly, linkage mapping uses a study design that is based on tracking the transmission of chromosomes through families. This approach aims to identify markers, or chromosomal segments, whose transmission patterns are correlated with the phenotype – implying that they are linked to QTLs. By contrast, association mapping, in its simplest form, uses samples of unrelated individuals. Here the goal is to identify markers whose genotype is correlated with the phenotype of interest, again implying that those markers are linked to QTLs.
The major advantage of linkage mapping is that a genome-wide scan can be performed using small numbers of markers [e.g. <1000 microsatellites, or slightly larger numbers of single nucleotide polymorphisms (SNPs), are usually sufficient for linkage mapping in humans]. However, for detecting common variants that affect gene expression, association mapping is a far more powerful approach [64], provided that the causal variants are in strong linkage disequilibrium (LD) with genotyped SNPs. Hence, with sufficiently dense genotyping, association mapping is much more likely to identify eQTLs with small or medium effect sizes. One plausible concern for association mapping is the possibility of false positives owing to population structure [65,66]. However, this issue can be overcome for most eQTL studies by applying recently developed methods for using genome-wide SNP data to correct for population structure [67].
Association mapping can also provide fine-scale resolution on the locations of functional variants (usually within a few tens of kb in humans, depending on the local extent of linkage disequilibrium). By contrast, linkage mapping provides much more coarse-grained localization because one relies on the occurrence of recombination events within the pedigrees to help fine map the relevant variants. Now that high-density genome-wide genotyping is readily available, association mapping will likely be the method of choice for future eQTL studies.
Study designs in model organisms, such as yeast, flies and mice, often share characteristics of both linkage and association mapping. For example, Brem and Kruglyak [45] created 40 haploid yeast strains that were segregants from a cross between two parental strains. They wanted to see if there was any correlation between expression levels and marker genotypes (the latter effectively identify the parental origin of the chromosomal segment). Their approach can be viewed as either linkage (correlation between transmitted chromosomes and phenotype) or association (correlation between genotype and phenotype). Because this type of design allows one to test directly for association between genotype and phenotype, it achieves the statistical power of an association study; however, it usually provides poor localization of the associated variants because the numbers of recombination events are very limited [45].
Finally, allele-specific expression assays (e.g. Refs. [27,68]) offer a fundamentally different approach to discovering factors that might affect gene expression levels. In these studies, one tests whether the two copies of a gene from a single individual are expressed in equal amounts. Assuming that both chromosomes are exposed to the same soup of trans-acting factors, a difference between the expression levels of the two copies implies that there is a functional difference in cis between the two chromosomes. This type of approach can, in principle, identify effects that would not be detected by standard eQTL methods: for example, variation that is present in just a single member of a sample or noninherited epigenetic factors. However, the method provides no guidance as to the location of the causal variation, beyond the implication that it acts in cis.
Recently, several groups have also found that, for a large fraction of loci, mean expression levels vary among populations [24,26,27]. For instance, Stranger et al. [28] estimated that 17–29% of loci have significant differences in mean expression levels between pairs of HapMap populations. One possible explanation for this observation is that these expression differences are caused by polymorphic sites with divergent allele frequencies between the HapMap populations. However, given that few single nucleotide polymorphisms (SNPs) in the genome have large frequency differences between populations [29], it seems likely that much of the expression variation across populations is caused by environmental factors. Indeed, it has been shown that even very closely related populations living in different environments can have substantially different expression profiles. For example, Idaghdour et al. [30] studied gene expression in leukocyte samples from Moroccans living in three different environmental conditions: in a city, in a mountain village and in a desert. Although these three groups are similar at the genetic level, the authors estimated that 37% of expressed genes show significant differences in mean expression levels among the three groups (at an FDR of 1%). Additionally, technical differences in the preparation or propagation of samples might also create apparent differences in gene expression between populations (as has been suggested for the much older CEU cell lines compared with the other HapMap samples [28]).
Cis and trans regulation of gene expression
A common observation, from a variety of both linkage and association eQTL studies, is that numerous genes have proximal eQTLs, likely in cis regulatory elements (see Box 2 for a discussion of the definitions of cis and trans). For example, Stranger et al. [28] measured gene expression in the transformed lymphoblastoid cell lines that were prepared by the International HapMap Project [29]. Using 2.2 million common SNPs genotyped by the HapMap to test for association in 210 of these cell lines (derived from unrelated individuals), they identified 831 genes with a significant proximal eQTL (at an FDR of 5%; proximal regions were defined as a 2-MB window containing the gene). More recently, Emilsson et al. [25] used microsatellites to map eQTLs and identified proximal eQTLs for 9% of genes in blood and 6% of genes in adipose tissue after applying linkage analysis to samples of 938 and 570 individuals, respectively (proximal signals in this case were defined as signals that were significant at the nearest microsatellite to the gene).
The precision of localization of eQTLs in association-based studies is limited by the extent of strong linkage disequilibrium (LD) and often provides resolution to within 10–20 kb in humans. Two studies, using different samples of transformed lymphoblastoid cell lines, concluded that most proximal eQTLs lie close to the actual genes [23,28]. For example, data from Stranger et al. [28] indicate that most of the proximal eQTLs map within •100 kb of the transcription start site of the regulated gene. Similarly, Dixon et al. [23] found only a few eQTLs that were located >100 kb from the relevant gene on the same chromosome. In addition, they observed that more distal eQTLs were usually much weaker than were signals close to the gene.
Box 2. Transcriptional regulation: cis and trans elements
The terms cis and trans were introduced to genetics by Haldane [69] to describe differences in the configurations of mutant alleles in heterozygotes, in analogy with cis and trans isomers in chemistry. The terms cis and trans were essentially replacements for Bateson’s terms ‘coupling’ and ‘repulsion’. In the cis (coupling) configuration, two mutations were inherited together, whereas in the trans (repulsion) configuration, the two mutations were found on different members of a pair of homologous chromosomes. Morgan et al. [70] proposed that linkage between the mutations was responsible for unequal numbers of the two types of heterozygotes in crosses, and Lewis [71] operationalized the definition in his cis-trans test for position effects, which has been widely used as a method for detecting whether two mutants lie in the same gene.
Molecular studies of gene regulation have classified regulatory interactions based on their effects in cis or trans, and the terms have been co-opted to describe particular types of regulatory elements. Consistent with the original definitions, cis regulatory elements have an allele-specific effect on gene expression, whereas trans elements affect the regulation of both alleles. Examples of trans elements may be transcription factors or insulators that regulate transcription initiation or small interfering RNA that regulates RNA stability. Examples of cis elements include promoter regions, enhancers and boundary elements, which regulate transcription initiation, or poly-A signals and siRNA binding sites, which regulate RNA stability [72].
In the expression quantitative trait loci (eQTL) mapping literature, regulatory polymorphisms are often said to be in cis or in trans on the basis of their physical distance from the regulated gene. Regulatory variation that is mapped near the target gene is classified as being cis. Although such a distance-based definition probably provides a reasonable broad classification of cis and trans regulatory elements (e.g. the ENCODE project found that •60% of transcription factor–binding sites – one type of cis regulatory elements, reside within 3 kb of transcription start sites [57]), this definition becomes problematic if trans regulatory elements exist near their gene target (e.g. a transcription factor that regulates adjacent target genes). Alternatively, the distance-based definition of regulatory elements might lead to the misclassification of long distance cis elements as ‘trans’ owing to conservative definitions of the proximal ‘cis windows’ (e.g. defining as cis only eQTLs that reside within 100 kb of the transcription start site). In any case, distance-based classification of eQTLs as cis or trans requires empirical validation.
Given that LD tends to spread signals out, these observations argue that most determinants of gene regulation tend to be close to the target gene, putatively in cis, and that long-range regulators are either much less frequent or exert much smaller effects. Consistent with this conjecture, distal-acting eQTLs were found to have much smaller effect sizes compared with proximal eQTLs (e.g. Ref. [23]).
The enrichment of proximal eQTLs that are near the genes they regulate is consistent with the view that changes in cis regulatory elements are less likely to have deleterious effects than changes in trans, because mutations in cis elements are more likely to affect the regulation of only one gene [13,26]. Thus, these observations suggest that genetic variation in cis regulatory elements might have a disproportional effect on gene expression variation and perhaps on gene expression evolution [27].
However, the enrichment of proximal compared with distal eQTLs might be overestimated because of statistical and technical reasons. For example, it should be noted that it is more difficult to detect a distal eQTL than a proximal eQTL of the same effect-size because the tests for distal effects are subject to a much greater burden of multiple testing (because, whereas the window size for proximal eQTLs is small, distal eQTLs can be found anywhere in the genome). Consistent with the view that many distal eQTLs are missed, at the time of writing, the human data do not seem to indicate strong clustering of distal-acting eQTLs into ‘master regulators’ as reported in other organisms inclduing mice and Drosophila [23,25] (although possible examples in humans have been reported [28,31]).
In addition, sequence polymorphisms at the microarray probes that are used to measure gene expression might be responsible for some of the observed proximal eQTLs. Because sequence mismatches between the target and the probe affect microarray hybridization intensity [32], one might estimate different expression levels for samples with different alleles at the probe sequence. These apparent differences in gene expression will be associated with any marker that is in LD with polymorphisms in the probe and thus will be mapped as spurious proximal eQTLs. The number of proximal eQTLs that can be explained by this technical artifact is unknown. Alberts et al. [33] estimated that 24% of probe sets on Affymetrix human gene expression arrays contain a SNP in at least one probe (using the HapMap SNPs) and that 4% of probe sets contain a SNP in three or more probes. These numbers are likely to be underestimates of the true proportion of probes that contain SNPs, because current databases of human variation such as HapMap and dbSNP still do not contain many of the common variants in the genome [34]. For that reason, excluding probes with known SNPs can is only a partial solution to this problem. To date, however, most eQTL studies do not even exclude probes with known SNPs.
Mapping in model organisms: evidence for eQTL hotspots
Genome-scale eQTL mapping studies in nonhuman organisms have predominantly focused on three objectives: (i) to identify QTLs associated with variation in transcript abundances in defined mapping populations and categorize them as proximal or distal to the locus of the transcript they affect, (ii) to determine the numbers, genomic distributions and magnitudes of eQTL effects on transcript levels and (iii) to evaluate whether eQTLs interact additively to control transcript levels. Despite differences in experimental designs and analysis methods, the relatively young eQTL mapping literature suggests some basic answers, at least with respect to the first two objectives.
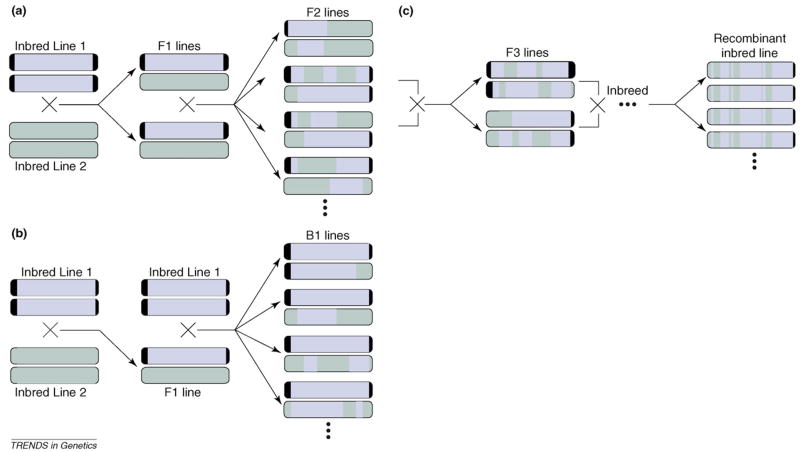
Unlike in humans, populations with defined genetic relationships can be constructed in model organisms. Such study designs increase the statistical power of QTL studies [35] but can decrease their generality because a restricted subset of alleles ultimately segregate in the mapping population, leaving many potentially consequential loci monomorphic. There are three commonly used designs in the eQTL literature (Figure 2). In the ‘F2 design’, two inbred lines are mated to form a heterozygous F1 generation. These F1 organisms are then interbred to form an F2 generation. Independent assortment and recombination will scramble the genomes in these F2 organisms generating genotypic variation (e.g. Refs. [36–39]). In the ‘back-cross design’, the F1 organisms are mated to one of the two parents to form a B1 generation [40]. For nonclonal organisms, F2 and B1 mapping populations are ephemeral. To create a permanent mapping population, F2 individuals can be selfed or brother-sister mated for several generations to create a set of ‘recombinant inbred lines’. Such lines are largely homozygous but have distinct combinations of parental alleles [3,39,41,42].
Figure 2.
Breeding designs commonly used in model organism expression quantitative trait loci (eQTL) studies. The original inbred lines are shaded in different colors to track transmission of segments from one pair of homologous chromosomes. However, as inbred lines 1 and 2 undoubtedly are not polymorphic for many alleles, the actual genotypes at many marker loci on the grey and black chromosomes will be the same. (a) F2 design. Two inbred lines are crossed to form a heterozygous but identical F1 generation. These F1 individuals are then crossed to form an F2 generation. (b) Backcross design. Two inbred lines are crossed to form an F1 generation. F1 individuals are then backcrossed to one of the inbred parents to form a B1 generation. (c) Recombinant inbred lines. Inbreeding from the F2 generation on eventually results in near homozygous individuals with a mixture of markers from the original inbred lines. One such line is depicted starting from a brother-sister pair of F2 individuals. Different recombinant lines can be created by crossing different brother-sister pairs.
The power to identify loci associated with transcriptional variation depends in part on the size of the mapping population and the size of the effect of allelic variation on transcript abundances. As with most statistical procedures, larger sample sizes yield better estimates, but, because of the expense of genome-wide measurements, most eQTL mapping studies in model organisms analyze relatively few lines•almost always <100. This severely limits the ability to detect eQTLs with small effects, particularly when significance levels are adjusted for multiple testing. Because the number of tests performed for distal eQTLs is much greater than for proximal eQTLs (as explained earlier), and because eQTL mapping studies routinely find that the magnitudes of the effects of proximal (putatively cis) eQTLs are larger than those that affect distant (putatively trans) loci [21,43,44], the true ratio of putative trans to putative cis eQTLs is difficult to estimate.
Despite low power to identify eQTLs with small effects, transcript abundances are generally found to be polygenic traits [43,45,46]. Moreover, in contrast to current results in humans, expression QTLs in several studies in model organisms were found to be unevenly distributed across chromosomes, with several examples of regions of high distal eQTL concentration (i.e. eQTL hotspots). Although some of these might be artifacts of microarray normalization [47], regions with a high number of distal eQTLs might harbor ‘master regulators’ that affect the expression levels of many genes. For example, Mehrabian et al. [48] dissected an eQTL hotspot in mice and identified a locus that affects the regulation of several metabolic traits associated with obesity and bone density. Similarly, West et al. [43] identified several genomic hotspots in Arabidopsis in which different alleles were associated with the regulation of a large number of genes. Interestingly, transcription factors were not overrepresented in the hotspots, reinforcing the idea that changes in any part of a network could percolate through to affect many other genes.
Beyond eQTL screens in model organisms?
One of the long-term goals of eQTL studies in model organisms is to delineate which eQTLs regulate metabolic and developmental pathways [49] – not an easy task, given that gene expression is often a polygenic trait with genes of both major and minor effects. Furthermore, transcriptional networks can be plastic [50,51] and, as a consequence, eQTLs can be extremely context dependent, differing because of a large number of factors, such as temperature [41], sex [38], developmental stage and tissues [44]. The task of mapping entire regulatory networks given these difficulties seems daunting [52].
One other complication of eQTL mapping studies in model organisms is that they predominantly use populations derived from two inbred lines. Quantitative genetic estimates are only applicable to the population from which they are derived and depend fundamentally on causative genetic variation being present in the population. The ranges of expression levels in mapping populations often fall outside the parental range, suggesting rampant genetic interactions including potential epistatic and/or compensatory effects. The use of constructed populations is typically a great advantage of working with model organisms, but in this case, it limits the applicability of eQTL mapping results. Distal eQTLs are difficult to replicate even from independent crosses derived from the same parents [39], and to date, no studies have attempted to replicate eQTLs using crosses of different genotypes. Transcriptional networks in natural populations, including humans, operate in much richer genotypic and environmental contexts.
In contrast to studies in model organisms, eQTLs are being mapped in humans using natural populations. Thus, although eQTL studies in humans face the same challenges regarding the plasticity of the polygenic expression phenotypes, results obtained in human studies are not expected to be restricted only the samples used (when population structure is taken into account).
Concluding remarks and future perspectives
The considerable advances in expression quantitative trait loci (eQTL) studies notwithstanding, there are still open questions about the biology and applications of eQTL mapping. First, there are important technical questions about the extent to which eQTLs are replicated across independent samples and independent platforms for measuring gene expression (see Box 3 for a discussion on different approaches to compare results across studies). Second, most of the human eQTL studies to date have analyzed transformed lymphoblast cell lines or lymphocyte samples, because these are the most readily available tissues [22–24,28,31,53–55]. However, because expression patterns differ dramatically across tissues, there is now great interest in collecting similar data for a much wider variety of tissues. Indeed, three recent studies have started moving beyond cell types in blood by characterizing eQTLs in cortical [56], adipose [25] and liver [20] tissues.
Expression QTL mapping can provide great insight into the biology of gene regulation. One can view eQTL mapping as a sort of large-scale mutagenesis experiment, in which •10 million common single nucleotide polymorphisms (SNPs) have been sprinkled down on the human genome, and each individual receives a random collection of these. Measurements of gene expression provide us with a tool for learning which types of SNPs are most likely to affect gene regulation, in a way that complements other experimental approaches such as allele-specific expression studies [57] or reporter gene assays [58].
Box 3. How to compare results across expression quantitative trait loci studies?
Although several robust patterns emerge when results from multiple expression quantitative trait loci (eQTL) studies are considered, one discouraging observation is that specific eQTLs are not generally replicated across studies [24,39]. For example, Gö ring et al., [24] reported that, although they replicated 11 of the top 13 proximal linkage signals from Morley et al., [31], they failed to replicate even the top distal signals from Morley et al. The discrepancy can partly be explained by the overall low power to detect eQTLs, particularly distal ones. However, an alternative (yet related) explanation for the lack of replication might be the method used to compare results across studies. In most studies, eQTLs are identified using an arbitrary statistical cut-off to ensure a minimum number of false positives. This approach leads to numerous false negatives, which might increase the discrepancies between the lists. For example, consider a true eQTL with small effect that has been detected as significant in one study (e.g. with P < 0.05) but did not reach the arbitrary statistical cut-off in another study (e.g. P = 0.06). An effective comparison of such two studies would not consider these results to be a discrepancy.
A second problem is that significant eQTLs are classified as cis or trans based on an arbitrary distance cut-off relative to the regulated gene (see Box 2). Typically, the results are analyzed by comparing the lists of significant cis and trans eQTLs [24,73]. However, as pointed out by Williams et al., [73], a comparison of such lists is confounded by the different arbitrary decisions made in each study. Thus, a meta-analysis of the data from multiple studies, using a single, consistent method, is necessary for a meaningful comparison.
One approach for an effective comparison between studies is to estimate the power to detect any eQTL that is seen in one study using the parameters of an alternative study. Instead of comparing the entire lists of significant eQTLs in both studies, one would focus only on the eQTLs for which the lack of replication is inconsistent with the estimated statistical power [39].
Alternatively, one could use a statistical model to simultaneously analyze the data from multiple studies. Such a statistical model would be designed to estimate the proportion of true eQTLs across studies, taking into account that eQTLs that are identified as significant in at least one dataset are more likely to be eQTLs in other datasets. Such an approach is similar to methods for controlling the false discovery rate in microarray experiments, by estimating from the data the proportion of genes that are differentially expressed.
We believe that this type of data can yield much more fine-grained information about the impact of individual SNPs on expression levels. For example, the extent to which eQTLs will be shared across diverse tissues is still unknown. Similarly, eQTL data have yet to be used to infer regulatory networks. This could be done, for example, by identifying proximal eQTLs for transcription factors that also mapped as distal eQTLs for other genes (implying a regulatory interaction between the transcription factor and its target). To achieve the potential of eQTL mapping as a tool for understanding gene regulation, it will be necessary to fine map the functional eQTL sites. For some purposes, we might simply want to estimate an approximate physical location of the causal variant(s). However, it will often be of interest to use the data to shortlist those variants that might be the actual functional sites [52] and to proceed to functional validation. It is usually difficult to identify functional variants in humans with any confidence in silico, because the HapMap – the primary genome-wide resource on genotype data in humans – currently contains only approximately one third of common SNPs [34] and is therefore expected to contain only a minority of the relevant functional variants. By contrast, the forthcoming 1000 Genomes Project (http://www.1000genomes.org/) will soon provide resequencing data on large numbers of individuals, thus providing reasonably complete information on common variation throughout the genome (outside highly repetitive regions). These new data, along with new imputation methods (e.g. Refs. [52,59]), should accelerate the process of identifying the functional alleles that affect gene expression levels.
Finally, eQTL mapping can provide important information for dissecting the genetics of complex disease (Figure 1). In its simplest form, the identification of eQTLs can provide a tool for connecting SNPs that are significant in genome-wide association studies of disease to a molecular mechanism [14,20]. Expression QTLs may also be useful for linking genes and individual variants to cellular phenotypes, such as cell line sensitivity to chemotherapeutic agents [60]. More ambitiously, one might be able to use patterns of gene expression and eQTL mapping in people with and without disease to identify networks of genes that are differentially regulated in the two groups [61]. Any eQTL that up- or downregulates such a network is a natural candidate for affecting the disease phenotype itself and would be of particular interest in association studies of the disease.
Acknowledgments
The authors thank J. Borevitz for helpful discussions and Sridhar Kudaravalli for helping to generate Figure 1. Y.G. is supported by NIH Grant GM077959, J.K.P. is supported by Grant HG002772 and S.A.R. is supported by NIH fellowship 5F32GM080966.
Glossary
- Expression quantitative trait loci (eQTL) hotspot
a locus in which genetic variation is associated with the expression variation of many genes. Because the resolution of the mapping depends on the density of markers, an eQTL hotspot may reflect the presence of a single influential regulator (such as a transcription factor) or several linked loci that affect transcript levels of different genes
- Genetic architecture of a quantitative trait
a description of the association between variation at the DNA sequence level and variation in a quantitative trait (e.g. variation in gene expression). Based or the patterns of genetic association with variation in the quantitative trait, the genetic architecture is classified as single or multilocus traits, which can interact additivity, or include epitasis, dominance, cis and/or trans effects
- Heritability
Heritability is the phenotypic variance in the population that is caused by genetic variation, divided by the total phenotypic variance. Heritability is usually estimated using the extent of similarity among relatives
- however
most studies cannot exclude the possibility that environmental factors that are shared among relatives might inflate heritability estimates
- Linkage disequilibrium (LD)
LD refers to the situation in which the alleles at one marker tend to co-segregate with particular alleles at a second marker. For example, in an extreme case, the ‘A’ allele at one single nucleotide polymorphism (SNP) might always appear together with the ‘B’ allele at a second SNP on the same chromosome, and similarly, the alternative alleles ‘a’ and ‘b’ might always co-occur. Strong LD usually occurs only between pairs of markers that are separated by less than a few tens of kilobases
References
- 1.Oleksiak MF, et al. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 2.Gilad Y, et al. Natural selection on gene expression. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Genissel A, et al. Cis and Trans Regulatory effects contribute to natural variation in transcriptome of Drosophila. Mol Biol Evol. 2008;25:101–110. doi: 10.1093/molbev/msm247. [DOI] [PubMed] [Google Scholar]
- 4.Abzhanov A, et al. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 5.McGregor AP, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 6.Stern DL. A role of Ultrabithorax in morphological differences between Drosophila species. Nature. 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gompel N, et al. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 8.Clark RM, et al. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 10.Cresko WA, et al. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci U S A. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammock EA, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- 12.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 14.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 15.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 18.Robin C, et al. hairy: A quantitative trait locus for drosophila sensory bristle number. Genetics. 2002;162:155–164. doi: 10.1093/genetics/162.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald SJ, et al. The effect of polymorphisms in the enhancer of split gene complex on bristle number variation in a large wild-caught cohort of Drosophila melanogaster. Genetics. 2005;171:1741–1756. doi: 10.1534/genetics.105.045344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 23.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 24.Göring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 25.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 26.Carroll SB, et al. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Blackwell: Science; 2004. [Google Scholar]
- 27.Wittkopp PJ, et al. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 28.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idaghdour Y, et al. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PloS Genet. 2008;4:e1000052. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilad Y, Borevitz J. Using DNA microarrays to study natural variation. Curr Opin Genet Dev. 2006;16:553–558. doi: 10.1016/j.gde.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Alberts R, et al. Sequence polymorphisms cause many false cis eQTLs. PLoS One. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer: 1998. [Google Scholar]
- 36.Damerval C, et al. Quantitative trait loci underlying gene product variation: a novel perspective for analyzing. Genetics. 1994;137:289–301. doi: 10.1093/genetics/137.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 38.Bhasin JM, et al. Sex specific gene regulation and expression QTLs in mouse macrophages from a strain intercross. PLoS One. 2008;3:e1435. doi: 10.1371/journal.pone.0001435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peirce JL, et al. How replicable are mRNA expression QTL? Mamm Genome. 2006;17:643–656. doi: 10.1007/s00335-005-0187-8. [DOI] [PubMed] [Google Scholar]
- 40.Klose J, et al. Genetic analysis of the mouse brain proteome. Nat Genet. 2002;30:385–393. doi: 10.1038/ng861. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, et al. Mapping Determinants of Gene Expression Plasticity by Genetical Genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao L, et al. Combining gene expression QTL mapping and phenotypic spectrum analysis to uncover gene regulatory relationships. Mamm Genome. 2006;17:575–583. doi: 10.1007/s00335-005-0172-2. [DOI] [PubMed] [Google Scholar]
- 43.West MA, et al. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petretto E, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brem RB, et al. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 46.Brem RB, et al. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams RW. Expression genetics and the phenotype revolution. Mamm Genome. 2006;17:496–502. doi: 10.1007/s00335-006-0006-x. [DOI] [PubMed] [Google Scholar]
- 48.Mehrabian M, et al. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- 49.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 50.van Swinderen B, Greenspan RJ. Flexibility in a gene network affecting a simple behavior in Drosophila melanogaster. Genetics. 2005;169:2151–2163. doi: 10.1534/genetics.104.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stern S, et al. Genome-wide transcriptional plasticity underlies cellular adaptation to novel challenge. Mol Syst Biol. 2007;3:106. doi: 10.1038/msb4100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PloS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 55.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 57.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chabot A, et al. Using reporter gene assays to identify cis regulatory differences between humans and chimpanzees. Genetics. 2007;176:2069–2076. doi: 10.1534/genetics.107.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 60.Huang RS, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 64.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 65.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 66.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 68.Serre D, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haldane JBS. New Paths in Genetics. Harper & Brothers; 1942. [Google Scholar]
- 70.Morgan TH, et al. The Mechanism of Mendelian Heredity. Henry Holt & Company; 1915. [Google Scholar]
- 71.Lewis EB. The relation of repeats to position effect in Drosophila melanogaster. Genetics. 1945;30:137–166. doi: 10.1093/genetics/30.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 73.Williams RBH, et al. The influence of genetic variation on gene expression. Genome Res. 2007;17:1707–1716. doi: 10.1101/gr.6981507. [DOI] [PubMed] [Google Scholar]