Abstract
Cofilin plays an important role in actin turnover in cells by severing actin filaments and accelerating their depolymerization. The role of pH in the severing by cofilin was examined using fluorescence microscopy. To facilitate the imaging of actin filaments and to avoid the use of rhodamine phalloidin, which competes with cofilin, α-actin was labeled with tetramethylrhodamine cadaverine (TRC) at Gln41. The TRC-labeling inhibited actin treadmilling strongly, as measured by εATP release. Cofilin binding, detected via an increase in light scattering, and the subsequent conformational change in filament structure, as detected by TRC fluorescence decay, occurred 2–3 times faster at pH 6.8 than at pH 8.0. In contrast, actin filaments severing by cofilin was pH-independent. The pH-independent severing by cofilin was confirmed using actin labeled at Cys374 with Oregon Green® 488 maleimide. The depolymerization of actin by cofilin was faster at high pH.
Keywords: actin, severing, pH, cofilin
INTRODUCTION
Rapid actin polymerization/depolymerization, and filament branching and bundling are critical to cell motility and other cellular processes [Pollard and Earnshaw, 2004]. The rates of filaments assembly and disassembly in vivo are far greater than those based on in vitro measurements for purified actin [Pollard, 1986]. Cofilin/ADF (actin depolymerizing factor) family of proteins is one of the most important cellular factors affecting both actin polymerization and depolymerization.
Actin-severing activity of cofilin was recently shown to be necessary for normal cell growth [Moriyama and Yahara, 2002]. The severing by cofilin increases the number of free barbed ends of actin filaments [Maciver et al., 1998; Ichetovkin et al., 2000], accelerating actin polymerization at the leading edge of a moving cell. At the pointed ends of filaments, cofilin accelerates the rate of actin depolymerization, possibly, through increasing the actin off-rate [Carlier et al., 1997]. These effects of cofilin on actin filaments result most likely from conformational changes induced in F-actin. Striking changes in F-actin structure by cofilin have been documented in several electron microscopy and solution studies. Cofilin destabilizes F-actin via a shift in the mean twist of actin filaments [McGough et al., 1997; Galkin et al., 2001] and cooperative changes in longitudinal and lateral interprotomer contacts [McGough and Chiu, 1999; Galkin et al., 2001; McGough et al., 2001; Bobkov et al., 2002; Galkin et al., 2003; Bobkov et al., 2004]. Cooperative effects of cofilin on F-actin are evident also from the presence of “tilted” filament structure in segments free of cofilin [Galkin et al., 2002], differential scanning calorimetry (DSC) measurements [Dedova et al., 2004; Bobkov et al., 2006], and phosphorescence decay (anisotropy) studies with Cys374-labeled actin [Prochniewicz et al., 2005].
It has been reported that cofilin binding to F-actin is sensitive to pH [Vandekerckhove, 1990; Moon et al., 1993]. Other reports show that cofilin inhibits nucleotide exchange on G-actin and depolymerizes F-actin in a pH-dependent manner [Nishida, 1985; Yonezawa et al., 1985], relatively fast at pH 8.0, and much slower at pH 6.8 [Pope et al., 2004]. The mechanism of this pH effect on F-actin depolymerization by cofilin has been the subject of considerable interest [Carlier et al., 1997; Theriot, 1997; Didry et al., 1998]. Despite its physiological significance [Bernstein et al., 2000], the question of possible effect of pH on F-actin severing by cofilin has not been resolved yet. Although Du and Frieden [1998], and later Yeoh et al., [2002], concluded that F-actin severing by cofilin is similar at pH 6.5 and 8.0, the methods employed by these authors did not monitor the severing process in real time and did not observe the same filaments before and after addition of cofilin. [Chen et al., 2004] also reported that actin severing by cofilin was pH-independent, but did not measure severing directly and concluded that their results combined contributions from both severing and depolymerization events. On the other hand, observations of filaments severing by light microscopy were questioned because of their multipoint attachment to glass surface by myosin. This may either generate force and break filaments, or prevent the relaxation of cofilin-induced instability and, therefore, induce F-actin severing [Carlier et al., 1997; Maciver et al., 1998].
In this work, we investigated the effect of pH on three stages of cofilin interaction with F-actin: binding, severing, and depolymerization. Fluorescence microscopy was used for direct observation of filament severing, taking advantage of actin covalently labeled at Gln41 with tetramethylrhodamine cadaverine (TRC). This allowed us to use F-actin at low concentrations without phalloidin, which inhibits cofilin binding to actin [Bamburg et al., 1999]. We confirmed the results of these experiments by measuring at selected time points the severing of actin labeled at Cys374 with Oregon Green® 488 maleimide (OG). We found that while F-actin severing by cofilin is pH-independent, other aspects of cofilin–actin interactions are pH-dependent.
MATERIALS AND METHODS
Reagents
ATP, ADP, DTT, Sephadex were purchased from Sigma Chemical (St. Louis, MO). TRC, Oregon Green® 488 maleimide, rhodamine phalloidin and ε-ATP were purchased from Invitrogen (Carlsbad, CA).
Proteins
Skeletal myosin and actin were prepared from rabbit back muscle according to Godfrey and Harrington [1970] and Spudich and Watt [1971], respectively. Heavy meromyosin (HMM) was prepared from myosin using the protocol of Kron et al. [1991]. α-actinin was purchased from Cytoskeleton (Denver, CO). Gelsolin was purchased from Sigma Chemical Co. (St. Louis, MO).
The labeling of Gln41 on skeletal actin with TRC was performed using Ca2+-independent bacterial transglutaminase (TGase) (a generous gift of Dr. K. Seguro (Ajinomoto, Kawasaki, Japan)). TGase (1 unit/ml) was added to 50–100 μM G-actin free of DTT or β-mercaptoethanol. TRC was added immediately after that at 2.5-molar excess over actin. The reaction was performed in a dark tube, for 2 h, at room temperature. Excess reagent was removed on a Sephadex G-50 spin column pre-equilibrated with G-buffer (2.0 mM Tris, 0.2 mM CaCl2, 0.5 mM ATP, 0.005% (w/v) NaN3, pH 8.0). DTT was added then at 10 mM, and the actin concentration was measured using the Bradford assay [Bradford, 1976]. The extent of labeling (~100%) was determined using TRC extinction coefficient of E544 nm = 78 mM−1 cm−1. Actin was then polymerized with 2.0 mM MgCl2 and used within 1 week. Gelsolin-capped filaments were obtained by polymerizing actin in the presence of gelsolin (1:3000–1:200 mole ratios of gelsolin to actin) with 1.0 mM CaCl2, 50 mM KCl and 2.0 mM MgCl2.
The labeling of Cys374 on skeletal actin with Oregon Green® 488 maleimide was performed in dark, for 2 h, at room temperature. Oregon Green® 488 maleimide (80 μM) was added (from 5 mM stock in dimethyl formamide) to 20 μM G-actin free of DTT or β-mercaptoethanol. MgCl2 (2 mM) was added immediately after the addition of the reagent to polymerize actin. Labeled actin was pelleted; the pellet was rinsed with and homogenized in G-buffer (2.0 mM Tris, 0.2 mM CaCl2, 0.5 mM ATP, 0.005% (w/v) NaN3, 10 mM β-mercaptoethanol, pH 8.0), and dialyzed against it for 3 days in cold room (in dark) to remove the reagent excess. Dialyzed, labeled actin was clarified from nondepolymerized filaments using ultracentrifuge, and the protein concentration was measured prior to polymerization using the Bradford assay [Bradford, 1976]. The extent of labeling (~70–90 %) was determined using Oregon Green® 488 maleimide extinction coefficient of E515 nm = 81 mM−1 cm−1. The labeled actin was polymerized with 2.0 mM MgCl2 and used within 1 week.
Skeletal actin was labeled with pyrene by Frieden’s method [Frieden et al., 1980] as modified by Northrop et al. [1986]. The extent of labeling was determined with the pyrene extinction coefficient of E344 nm = 22 mM−1 cm−1.
WT yeast cofilin was expressed in Escherichia coli BL21 (DE3) cells under the T7 promoter (pBAT4 plasmid (a generous gift of Dr. Steven Almo, Department of Biochemistry, Albert Einstein College of Medicine, New York, NY)) [Ojala et al., 2001]. Cells were grown to a density of 0.6 absorbance units (600 nm), induced with 0.4 mM IPTG, harvested by centrifugation, resuspended in 20 mM Tris–HCl (pH 7.5), and lysed by sonication. The cell lysate was clarified by centrifugation, the supernatant applied to a QAE-52 column (Pharmacia Biotech) equilibrated with 20 mM Tris–HCl (pH 7.5), and the column was developed with a linear 0–0.5 M NaCl gradient. Peak fractions containing cofilin were pooled, concentrated, and applied to a Sephacryl S300 gel-filtration column (Pharmacia Biotech), equilibrated with 10 mM Tris–HCl (pH 7.5), 50 mM NaCl. The peak fractions were pooled and polished by MonoQ ion-exchange chromatography using a buffer system similar to that described for the QAE-52 chromatography. The concentrations of cofilin and unlabeled skeletal muscle α-actin were determined spectrophotometrically, using extinction coefficients and , respectively.
Tropomodulin was a generous gift from Dr. Velia M. Fowler (The Scripps Research Institute). Mouse capping protein, CapZ (α1 and β2 subunits), was expressed and purified as described by Palmgren et al. [2001].
Stopped-Flow Experiments
Stopped-flow measurements were carried out at 20°C in a buffer containing 20 mM KCl, 25 mM MOPS, pH 6.8 and 8.0, 2.0 mM MgCl2, 2.0 mM K+-EGTA or Ca2+-K+-EGTA, and 2.0 mM DTT. Final concentrations of F-actin (intact and TRC-or OG-labeled) and cofilin were between 0.5 and 5.0 μM. These experiments were not extended below pH 6.8 because of some TRC-F-actin bundling at the lower pH (see below). The formation of actin–cofilin complexes was followed via an increase in light scattered at 90° to the exciting beam with the excitation and emission wavelengths set at 345 nm. Conformational changes occurring in TRC-labeled actin upon cofilin binding were detected via a decrease in TRC fluorescence, with excitation and emission wavelengths set at 544 and 580 nm, respectively.
In Vitro Severing Assays
In vitro severing assays were performed similarly to a protocol previously described for in vitro motility experiments [Miller et al., 1996]. A glass slide with spacers from double side scotch tape and nitrocellulose-coated coverslip formed a ~70 μl chamber opened on two sides. HMM (5–10 μg/ml), α-actinin (1.0 μM), CapZ (0.6 μM), or tropomodulin (0.6 μM) were applied to the chamber; then BSA (2.0 mg/ml) was added to block the uncovered area on glass surface. TRC-labeled F-actin (TRC-FA) was then applied in an assay buffer (2.0 mM K+-EGTA, 20 mM KCl, 2.0 mM MgCl2, 10 mM DTT, 25 mM MOPS, pH 7.4; total ionic strength 50 mM) containing oxygen scavenging system [Umemoto and Sellers, 1990] and allowed to bind to one of the above proteins at 25°C. The unbound filaments were washed off with the assay buffer containing no cofilin. The slide was mounted on the microscope and fixed with a scotch tape. Severing was initiated by adding cofilin in an assay buffer at the desired pH (6.0–8.0), at concentrations ranging from 20 to 50 nM. Using this procedure, we avoided the occasional TRC-F-actin bundling encountered when actin was adsorbed to the coverslip directly at pH 6.0. Assay buffer containing cofilin was carefully applied to an opened side of the working chamber, and the excess liquid was removed from the other side, while the field of view remained unchanged and the filaments on the surface remained in focus. The filaments were exposed to light for short periods of time (10–15 s, every 30–60 s) during image recording by VCR. Several snapshots were taken before cofilin addition, and over the first 2–5 min after its addition.
Alternatively, 2 μM TRC-FA or OG-FA was quickly, but gently mixed at different mole ratios with cofilin. At selected time points, aliquots of such actin–cofilin mixtures were diluted 100-fold in assay buffer containing 1–5 μM phalloidin and placed on a slide for visualization.
The recorded images were averaged, enhanced, and analyzed using the SigmaScan Pro 5 program (SPSS).
Fluorescence Measurements
All fluorescence measurements were done on a PTI spectrofluorometer (Photon Technology Industries, South Brunswick, NJ). Measurements of ε-ATP release were performed with the excitation and emission wavelengths set at 350 and 412 nm, respectively. Measurements of TRC fluorescence were done with the excitation and emission wavelengths set at 544 and 580 nm, respectively. Measurements of OG fluorescence were made with the excitation and emission wavelengths set at 491 and 515 nm, respectively.
Nucleotide Release—Depolymerization Measurements
Nucleotide exchange due to depolymerization/treadmilling of F-actin, labeled or unlabeled with TRC, was observed at 20°C by monitoring the decay in fluorescence upon release of etheno-nucleotide (ε-ATP) from actin. Excess ATP was removed from monomeric actin on G-50 Sephadex spin columns, after which the protein was supplemented with a 10-to 20-fold molar excess of etheno-nucleotide. After incubation for 1 h on ice, actin was applied to a G-50 Sephadex spin column for transfer into G-buffer containing 10 μM etheno-nucleotide (instead of ATP) and was then polymerized by addition of KCl (50 mM) and MgCl2 (2.0 mM). The release of ε-ATP from the nucleotide-binding cleft on actin was monitored after addition of 20-fold molar excess of ATP over ε-ATP in actin (with or without cofilin).
RESULTS
The main goal of this study was to measure directly the effect of pH on severing of actin filaments by cofilin. To this end, we used TRC-F-actin, which allows for fluorescence imaging of actin filaments without the commonly employed rhodamine phalloidin. Since phalloidin competes with cofilin for binding to F-actin, we avoided using this drug in most of our experiments. Another important advantage of the TRC label at Gln41 (and other probes at this site; dansyl cadaverine [Takashi, 1988], dansyl ethylenediamine [Kim et al., 1995]) is that it lowers the critical concentration for actin polymerization and stabilizes filaments. Thus, TRC-actin filaments were stable and could be easily viewed in a fluorescence microscope at nanomolar concentration (5.0–20 nM) without significant depolymerization. To confirm that TRC-F-actin is a suitable model for probing filament severing by cofilin at different pH values, we examined several aspects of cofilin interaction with this actin and also measured the severing of OG-F-actin.
Measurements of Cofilin Binding
Stopped-flow measurements of cofilin binding to TRC-F-actin, as monitored via light scattering increase at 20°C, show ~2–3 faster binding rate at pH 6.8 than at pH 8.0 (Fig. 1A). This result is independent of TRC actin labeling; similar binding rate differences between pH 6.8 and 8.0 were observed also with unlabeled actin and OG-F-actin.
Fig. 1.
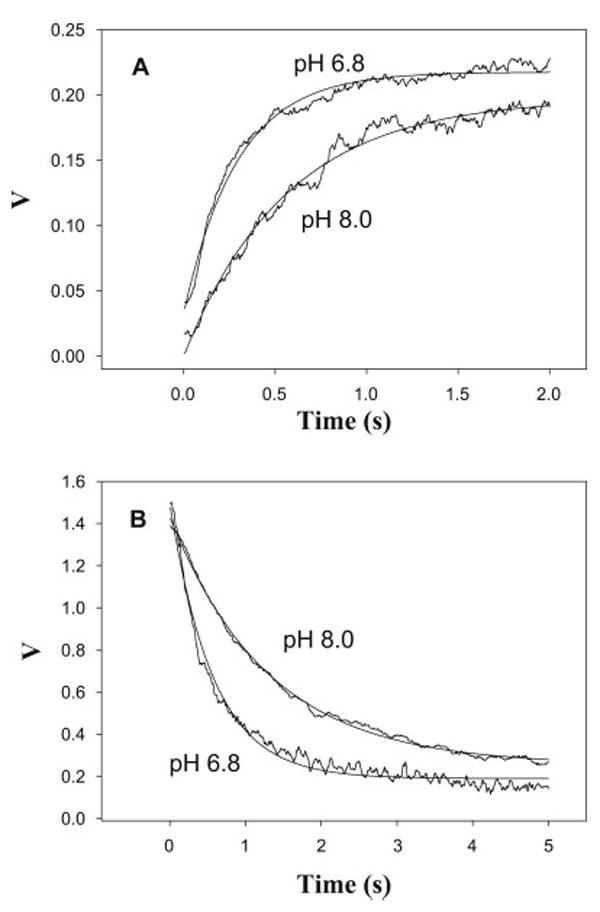
Cofilin binding to TRC-F-actin at pH 6.8 and pH 8.0. (A) Time course of increase in light scattering from 5.0 μM TRC-F-actin mixed with 5.0 μM cofilin. The light scattering traces were best fitted with a single exponential expression, yielding association rate constants kon= 3.40 ± 0.05 s−1 for pH 6.8 and 1.77 ± 0.03 s−1 for PH 8.0. (B) Decrease in the fluorescence of 5.0 μM TRC-F-actin upon mixing with 5.0 μM cofilin. The fluorescence trace was fitted to a single exponential expression. The rate constants of fluorescence change were 1.75 ±0.04 s−1 for pH 6.8 and 0.77 ± 0.01 s−1 for pH 8.0. In each case r2 > 0.95. Each reaction trace is the average of 3–5 separate recordings. The solid smooth curves are the exponential fits to data, yielding the indicated rates. Prior to experiments all proteins were dialyzed against 25 mM MOPS, 20 mM KCl, 2.0 mM MgCl2, 2.0 mM K+-EGTA, 2.0 mM DTT, pH 6.8 or pH 8.0.
TRC-actin, which binds cofilin stronger than unlabeled actin, was also used to estimate the pH dependence of cofilin-actin “off” rates. In a mixture of cofilin with a 10-fold excess of unlabeled F-actin most of the cofilin is bound. Addition of TRC-labeled F-actin in the amount equimolar to that of cofilin to such a mixture causes cofilin dissociation from unlabeled actin and it’s binding to TRC-F-actin. TRC quenching in this case reflects the rate limiting release of cofilin from unlabeled actin since the dissociation step is several hundred times slower than the binding step. The rate of cofilin release was ~40% faster at pH 8.0 than at pH 6.0 (data not shown), confirming that the overall affinity of cofilin for actin is higher at pH 6.8 than 8.0.
Conformational changes occurring in TRC-F-actin upon cofilin binding were detected via TRC fluorescence quenching. In a previous study [Bobkov et al., 2004], using actin labeled at the same position (Gln41) with a dansyl probe, we observed fast cofilin binding (via light scattering), and a slower conformational change in the subdomain 2 region of actin (via fluorescence change). In analogy to that result, the quenching of TRC-F-actin fluorescence occurred slower than the increase in light scattering (Fig. 1B). As in the case of binding, the rate constant for this conformational change was faster at pH 6.8 (1.75 ± 0.04 s−1) than at pH 8.0 (0.77 ± 0.01 s−1). Thus, the early stages of actin–cofilin interaction occur faster at the lower pH.
Actin Depolymerization and Treadmilling Induced by Cofilin
The acceleration of F-actin depolymerization and treadmilling is frequently monitored via nucleotide (εATP) exchange on actin [Nishida, 1985], which is accompanied by the quenching of εATP fluorescence. This exchange with external ATP occurs fast in G-actin and extremely slowly in filamentous actin. Addition of depolymerizing agents, such as cofilin, to F-actin accelerates the release of εATP due to quickly forming actin monomers, thereby reporting on actin depolymerization. In the absence of cofilin, the release of εATP from F-actin was slow under both pH conditions (Fig. 2A). The rate of εATP release increased with addition of cofilin and was several fold greater at pH 8.0 (1.16 ± 0.01) × 10−3 s−1 than at pH 6.8 (0.22 ± 0.01) × 10−3 s−1 (Fig. 2A). These results are in agreement with previous observations [Bobkov et al., 2002].
Fig. 2.
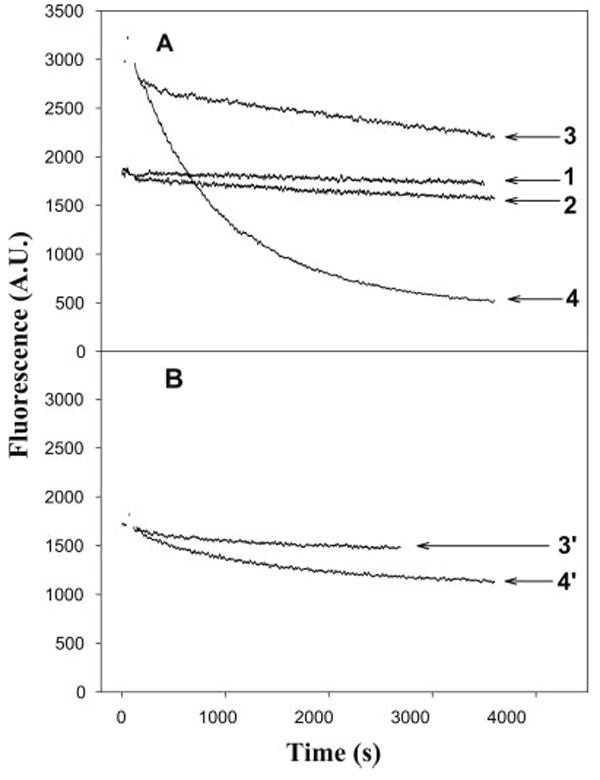
Release of εATP from skeletal F-actin in the presence and absence of cofilin. The release of εATP from F-actin was monitored via a decrease in εATP fluorescence. The reaction was initiated by the addition of 0.2 mM ATP to 5.0 μM F-actin (A) or TRC-F-actin (B) [free of ATP and polymerized in the presence of εATP] in the presence of 5.0 μM cofilin (curves 3 and 4 in A; curves 3′ and 4′ in B) or in the absence of cofilin (curves 1 and 2 in A). The reaction buffer contained 2.0 mM K+-EGTA, 20 mM KCl, 2.0 mM MgCl2, 10 mM DTT, 25 mM MOPS (total ionic strength- 50 mM) at pH 6.8 (curves 1 and 3 in A; curve 3′ in B) or 8.0 (curves 2 and 4 in A; curve 4′ in B). The depolymerization rates for F-actin with cofilin at pH 6.8 and 8.0 were (0.22 ± 0.01) × 10−3 and (1.16 ± 0.01) × 10−3, respectively.
TRC labeling stabilizes F-actin, slowing its depolymerization and treadmilling. Although the nucleotide exchange rate in TRC-F-actin was greatly reduced and difficult to measure, the overall cofilin effect was similar to that observed for unlabeled actin i.e., faster εATP release was induced at pH 8.0 than 6.8 (Fig. 2B).
To prevent the rebinding of εATP-G-actin to the barbed ends of filaments, we polymerized G-actin in the presence of gelsolin (at a mole ratio of 200:1 actin to gelsolin). Moreover, filaments with a length of 250 monomers or less are presumably not easily broken further down by cofilin [Pope et al., 2000], hampering the formation of free ends. The depolymerization of these gelsolin-capped short filaments by cofilin was still faster at pH 8.0 than 6.0 (data not shown). Thus, the depolymerization of actin filaments detected via nucleotide exchange occurs faster at pH 8.0, unlike the first stages of actin–cofilin interaction (Fig. 1). The slow depolymerization of TRC-F-actin helps to separate this process from filament severing in the fluorescence microscopy experiments.
F-actin Severing by Cofilin: pH 6.0 and 8.0
For the observation of severing, actin filaments were attached to a glass coverslip surface through binding to HMM adsorbed to that surface. The density of HMM was such as to provide a sufficient number of immobilizing contacts for actin filaments and yet permit local diffusion of severed filament ends, thereby facilitating the detection of severing. The optimal concentration of HMM used for adsorption to the glass surface was 5.0 μg/ml, which on the basis of prior calibrations [Wakabayashi et al., 1975; Uyeda et al., 1990] should result in ~6 ± 2 myosin heads bound per 1.0 μm of actin filaments. Filaments could be attached more tightly to the surface with a greater number of myosin heads, but this interfered with easy detection of the severed filament fragments.
Addition of cofilin in assay buffer at any pH (6.0–8.0) led to a fast degradation of actin filaments. Break points were observed along the entire filament; the fragments degraded then further until almost complete disappearance (Fig. 3). Severing activity was measured only on those filaments, which could be identified in fluorescence images taken before and after cofilin addition. Full statistical analysis was carried out for filaments exposed to cofilin for 2 min, at which point their fragmentation could be easily measured. Such an analysis revealed similar increase in the number of actin filaments and a decrease in their mean length due to cofilin action at pH 6.0–8.0 (Fig. 4; Table I). The excellent agreement (at both pH values) between the increase in filaments numbers and the decrease in their mean length shows the absence of depolymerization effects (filament shortening) under our experimental conditions. Thus, no pH effect on filament severing was detected in these experiments. Using TRC-F-actin polymerized in the presence of gelsolin did not change this result.
Fig. 3.
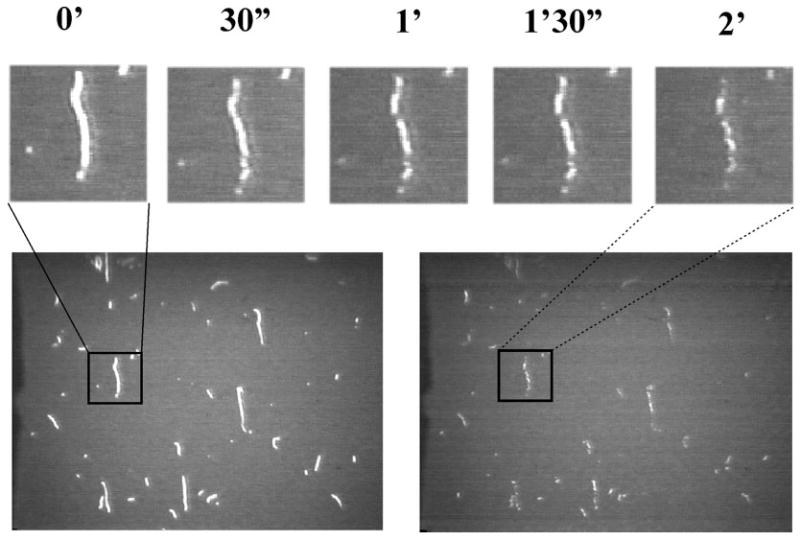
TRC-F-actin severing by cofilin. Snapshot images of TRC-F-actin filaments bound to the HMM-covered surface were taken before addition of cofilin and during the incubation for 2 minutes with cofilin. The top panel shows magnified images of a single filament degradation taken every 30 s after addition of cofilin. The assay buffer contained 2.0 mM K+-EGTA, 20 mM KCl, 2.0 mM MgCl2, 10 mM DTT, 25 mM MOPS (total ionic strength- 50 mM), 14 mM glucose, 9 × 10003 units of catalase/mL, and 240 units of glucose oxidase/mL at pH 6.0 or 8.0.
Fig. 4.
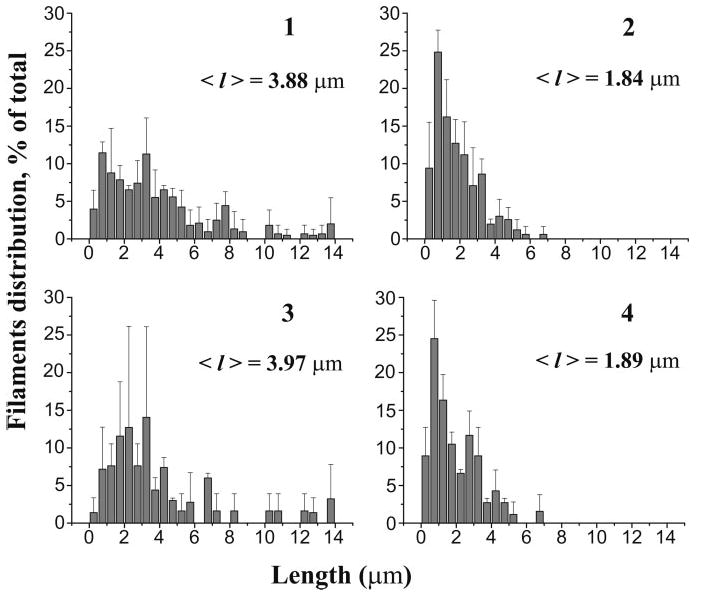
Distribution of actin filaments length upon severing by cofilin at pH 6.0 and 8.0. Snapshot images of TRC-F-actin filaments attached to HMM were enhanced and processed using Sigma Scan Pro 5 image analysis program. The length of at least 200 filaments was measured in each case after the screen was calibrated using a grid-containing slide. Length distributions at pH 6.0 (1, 2) and pH 8.0 (3, 4) are shown for filaments before the addition of 20 nM cofilin (1, 3) and after 2 minutes of incubation at 25°C (2, 4). The data obtained at pH 6.8 are given in Table 1.
TABLE I.
Changes in TRC-actin Filaments Length and Number Upon Severing by Cofilin
| pH
|
||
|---|---|---|
| 6.0 | 8.0 | |
| Increase in number of filaments after 2 min of severing (%) | 102 ± 17 | 110 ± 27 |
| Decrease in filaments length after 2 min of severing (%) | 49 ± 19 | 50 ± 20 |
A mean increase in the number of filaments (%) on the screen was obtained by comparing the number of short filament fragments resulting from severing of longer filaments by cofilin with the number of filaments counted in each slide area prior to cofilin addition. The decrease in the mean length of filaments (%) shows the change in their mean length after 2 min incubation with cofilin.
Although actin labeling by TRC allowed for easy filament visualization and real-time severing study, this modification stabilizes actin filaments and thus, may desensitize them to a pH effect on severing. To decrease the risk of such a putative effect of TRC on filaments, we copolymerized at different mole ratios fully labeled TRC-G-actin and unlabeled G-actin (the minimum content of TRC-actin for imaging the severing was ~25%). We did not detect any significant difference in severing at pH 6.0 or 8.0 using the 25% TRC-labeled F-actin.
To further confirm that our severing results are not biased by the TRC label on Gln41, we employed actin labeled with a different probe and at different residue i.e., with Oregon Green® 488 maleimide covalently attached to Cys374. Although OG labeling may also induce some conformational changes in actin, the different structures of TRC and OG, and their attachments to different parts of actin molecule, provide an appropriate test for labeling-induced artifacts in filament behavior. We mixed 2 μM OG-F-actin with 0.5 μM cofilin in a working buffer with a desired pH, and took an aliquot of the mixture after 30 s of incubation. The aliquot was immediately diluted 100-fold in the working buffer containing 5 μM phalloidin and then placed on the HMM-coated slide for visualization. Image analysis showed no difference in actin severing at pH 6.8 and 8.0 (Fig. 5). Taken together, our observations with OG-F-actin and 25% labeled TRC-F-actin show that the pH-independent severing of actin filaments by yeast cofilin is not related to a particular labeling of actin, but is an intrinsic property of the actin–yeast cofilin system.
Fig. 5.
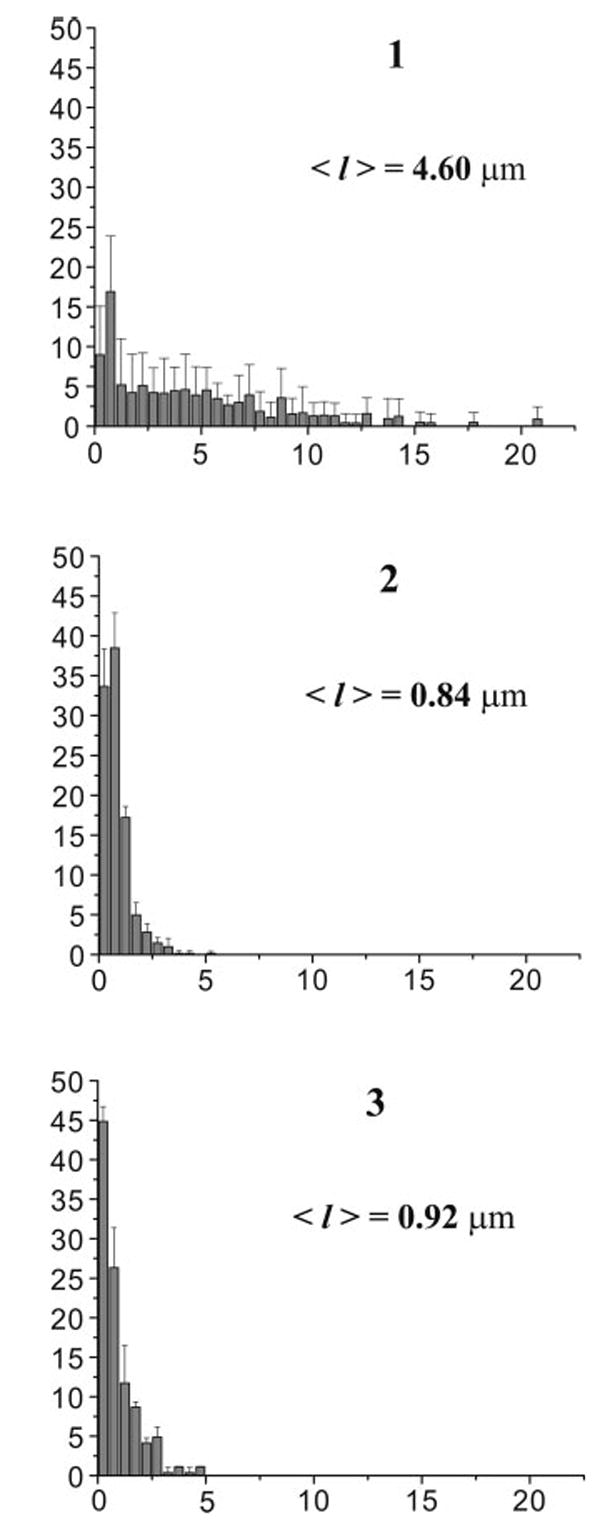
Distribution of actin filaments length upon severing by cofilin at pH 6.8 and 8.0. Snapshot images of OG-F-actin filaments were enhanced and processed using Sigma Scan Pro 5 image analysis program. The length of at least 200 filaments was measured in each case after the screen was calibrated using a grid-containing slide. Length distributions at pH 6.8 (2) and pH 8.0 (3) are shown for the control filaments (1) and after 30 seconds of incubation of 2 μM OG-F-actin with 0.5 μM cofilin at 25°C (2,3).
The results shown in Fig. 4 were obtained at the low concentrations of proteins used in our fluorescence microscopy experiments; the final concentration of TRC-F-actin before its application to HMM-coated surface was 10 nM and cofilin was used at 20–50 nM. These conditions were optimal for accurate quantitative analysis of severing activity on the glass surface, i.e. higher amounts of added cofilin caused fast filaments degradation, complicating the quantitative analysis of this process. To observe F-actin severing at protein concentrations closer to physiological, we added 1–2 μM of unlabeled F-actin to the assay buffer. This solution, along with 0.4–2.0 μM cofilin was introduced into HMM-coated surface containing TRC-F-actin. In analogy to the results described above, we observed no effect of pH on cofilin performance, also under these conditions. To distinguish between severing and depolymerization effects of cofilin on filament length, we used CapZ/ tropomodulin-capped TRC-F-actin (where both ends of filaments are capped, and the cofilin-induced depolymerization is inhibited), and stabilized the resulting filament fragments against depolymerization with phalloidin. Thus, we preincubated TRC-F-actin with tropomodulin and CapZ (at 2 μM each), and then added cofilin. After 1 min incubation with cofilin, actin was diluted to ~20 nM in an assay buffer (pH 6.8 and 8.0) containing phalloidin, to stop depolymerization and severing, and applied to the HMM-coated surface. We observed only very short fragments of actin in the solution and on the surface at pH 6.8 and 8.0. Taken together, both by directly monitoring the severing process under the microscope or taking time point aliquots of the actin severing occurring in solution for microscopy observations, we did not detect a pH dependence of F-actin severing by cofilin.
We also tested the hypothesis that specific actomyosin interaction may affect the severing action of cofilin by replacing HMM with α-actinin. As in the case of HMM-attached filaments, severing by cofilin of TRC-F-actin attached to α-actinin was pH-independent.
DISCUSSION
With the exception of Acanthamoeba actophorin [Maciver et al., 1998] and depactin [Bamburg et al., 1999] all ADF/cofilin proteins disrupt actin filaments more efficiently at higher pH. This aspect of ADF/ cofilin function attracted considerable attention because of the potential impact of local pH changes that occur in cells on cofilin–actin interactions. Despite this interest, the mechanism and the nature of the pH-sensitivity of filament disruption by cofilin remains unknown.
The difficulty in analyzing the pH-sensitivity of the cofilin–actin system has been related in part to the complex effects of cofilin on filament severing, depolymerization, and nucleation. Two of these effects are clearly pH-dependent, with the nucleating activity of cofilin being stronger at low pH (6.8), while the depolymerizing activity is much stronger at high pH (7.8). In most cases, the information on filament severing at pH 6.8 and 7.8 has been obtained from indirect spectroscopic measurements of F-actin depolymerization in the presence of capping proteins [Maciver et al., 1998], or microscopy imaging of populations of filaments before and after their incubation with cofilin. The overall slower F-actin shortening by cofilin at pH 6.8 than 7.8 has been attributed to either a combination of nucleation and depolymerization rates [Bonet et al., 2000; Blondin et al., 2002] or the reduced severing and depolymerization of filaments [Chen et al., 2004]. However, indirect assays of filament severing by ADF showed little, if any, pH-dependence of this process [Yeoh et al., 2002]. Concerns were raised also about the use of surface-attached filaments in microscopy studies of their severing by cofilin, speculating that the restricted ability of these filaments to twist and flex could have caused their breakdown [Carlier et al., 1999].
Although recent studies suggest a possible role of conformational changes in actin [Blondin et al., 2002] or cofilin [Pope et al., 2004] in the pH-sensitivity of F-actin depolymerization by cofilin, they have not examined the severing reaction itself. In this study, fluorescence microscopy observations of Gln41-TRC-labeled actin filaments enabled the direct monitoring of their severing. To ensure proper counting of such events, only those filaments that could be tracked both before and after cofilin infusion were used in severing measurements. The main result of these observations is that actin filaments are severed by yeast cofilin at similar rates over the pH range from 6.0 to 8.0. This is documented by a similar increase in the number of filaments and the change in the mean filament length over time independent of the site of actin labeling or the label itself (Figs. 4 and 5; Table I).
While actin filament severing by yeast cofilin is pH-independent, other aspects of cofilin–actin interaction show the expected pH-dependence. The rates with which cofilin binds and induces conformational changes in F-actin are faster at pH 6.8 than 8.0, and actin depolymerization is faster at pH 8.0 than 6.8, consistent with prior observations [Hawkins et al., 1993; Maciver et al., 1998]. The fact that the depolymerization of F-actin by cofilin is faster at the high pH, despite somewhat weaker cofilin binding and the lack of severing sensitivity to pH, focuses our attention on the pointed ends of filaments, the main site of their depolymerization. Recent electron microscopy image analysis [Galkin et al., 2003] has suggested that the structure of pointed end segments of actin filaments resembles that of cofilin-bound filaments in terms of the disrupted interprotomer subdomain 1/2 contacts. If the intrinsically faster actin depolymerization at high pH is linked to a greater propensity of such contact disruption then cofilin, which favors this filament state, may be binding better to pointed ends at pH 8.0 than 6.8, in contrast to it’s binding to other filament parts. Cofilin could accelerate the depolymerization at pointed ends by facilitating the dissociation of small blocks of actin protomers rather than individual actin units [Fujiwara et al., 2002]. Although this hypothesis needs to be tested, it could explain the apparent contradiction in pH effects on cofilin binding and depolymerization of F-actin.
Acknowledgments
Contract grant sponsor: USPHS; Contract grant numbers: 077190, 38542; Contract grant sponsor: NSF; Contract grant number: MCB 0316269.
Abbreviations used
- ADF
actin depolymerizing factor
- HMM
heavy meromyosin
- ε-ATP
1,N6-ethenoadenosine 5′-triphosphate
- TRC
tetramethylrhodamine cadaverine
- OG
Oregon Green® 488 maleimide
Footnotes
Published online 14 July 2006 in Wiley InterScience (www.interscience.wiley.com).
References
- Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9(9):364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Painter WB, Chen H, Minamide LS, Abe H, Bamburg JR. Intracellular pH modulation of ADF/cofilin proteins. Cell Motil Cytoskeleton. 2000;47(4):319–336. doi: 10.1002/1097-0169(200012)47:4<319::AID-CM6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Blondin L, Sapountzi V, Maciver SK, Lagarrigue E, Benyamin Y, Roustan C. A structural basis for the pH-dependence of cofilin. F-actin interactions. Eur J Biochem. 2002;269(17):4194–4201. doi: 10.1046/j.1432-1033.2002.03101.x. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. J Mol Biol. 2002;323(4):739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Shvetsov A, Benchaar S, Scoville D, Almo SC, Reisler E. Cofilin (ADF) affects lateral contacts in F-actin. J Mol Biol. 2004;337(1):93–104. doi: 10.1016/j.jmb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356(2):325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- Bonet C, Ternent D, Maciver SK, Mozo-Villarias A. Rapid formation and high diffusibility of actin-cofilin cofilaments at low pH. FEBS J. 2000;267(11):3378–3384. doi: 10.1046/j.1432-1327.2000.01372.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J Cell Biol. 1997;136(6):1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Ressad F, Pantaloni D. Control of actin dynamics in cell motility. Role of ADF/cofilin. J Biol Chem. 1999;274(48):33827–33830. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Sneider JM, Boyle JA, Minamide LS, Bamburg JR. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 2004;43(22):7127–7142. doi: 10.1021/bi049797n. [DOI] [PubMed] [Google Scholar]
- Dedova IV, Nikolaeva OP, Mikhailova VV, dos Remedios CG, Levitsky DI. Two opposite effects of cofilin on the thermal unfolding of F-actin: A differential scanning calorimetric study. Biophys Chem. 2004;110(12):119–128. doi: 10.1016/j.bpc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Didry D, Carlier MF, Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J Biol Chem. 1998;273(40):25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Du J, Frieden C. Kinetic studies on the effect of yeast cofilin on yeast actin polymerization. Biochemistry. 1998;37(38):13276–13284. doi: 10.1021/bi981117r. [DOI] [PubMed] [Google Scholar]
- Frieden C, Lieberman D, Gilbert HR. A fluorescent probe for conformational changes in skeletal muscle G-actin. J Biol Chem. 1980;255(19):8991–8993. [PubMed] [Google Scholar]
- Fujiwara I, Takahashi S, Tadakuma H, Funatsu T, Ishiwata S. Microscopic analysis of polymerization dynamics with individual actin filaments. Nat Cell Biol. 2002;4(9):666–673. doi: 10.1038/ncb841. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153(1):75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure. Curr Biol. 2002;12(7):570–575. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J Cell Biol. 2003;163(5):1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey JE, Harrington WF. Self-association in the myosin system at high ionic strength. I. Sensitivity of the interaction to pH and ionic environment. Biochemistry. 1970;9(4):886–893. doi: 10.1021/bi00806a025. [DOI] [PubMed] [Google Scholar]
- Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin-filaments. Biochemistry. 1993;32(38):9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- Ichetovkin I, Han J, Pang KM, Knecht DA, Condeelis JS. Actin filaments are severed by both native and recombinant dictyostelium cofilin but to different extents. Cell Motil Cytoskeleton. 2000;45(4):293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Kim E, Motoki M, Seguro K, Muhlrad A, Reisler E. Conformational changes in subdomain 2 of G-actin: Fluorescence probing by dansyl ethylenediamine attached to Gln-41. Biophys J. 1995;69(5):2024–2032. doi: 10.1016/S0006-3495(95)80072-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Toyoshima YY, Uyeda TQ, Spudich JA. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 1991;196:399–416. doi: 10.1016/0076-6879(91)96035-p. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur J Biochem. 1998;256(2):388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. J Mol Biol. 1999;291(3):513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138(4):771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Pope B, Weeds A. The ADF/cofilin family: Accelerators of actin reorganization. Results Probl Cell Differ. 2001;32:135–154. doi: 10.1007/978-3-540-46560-7_10. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Wong WW, Bobkova E, Rubenstein PA, Reisler E. Mutational analysis of the role of the N-terminus of actin in actomyosin interactions. Comparison with other mutant actins and implications for the cross-bridge cycle. Biochemistry. 1996;35(51):16557–16565. doi: 10.1021/bi962388+. [DOI] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120(2):421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci. 2002;115(Pt 8):1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- Nishida E. Opposite effects of cofilin and profilin from porcine brain on rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1985;24(5):1160–1164. doi: 10.1021/bi00326a015. [DOI] [PubMed] [Google Scholar]
- Northrop J, Weber A, Mooseker MS, Franzini A, Bishop MF, Dubyak GR, Tucker M, Walsh TP. Different calcium dependence of the capping and cutting activities of villin. J Biol Chem. 1986;261(20):9274–9281. [PubMed] [Google Scholar]
- Ojala PJ, Paavilainen V, Lappalainen P. Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry. 2001;40(51):15562–15569. doi: 10.1021/bi0117697. [DOI] [PubMed] [Google Scholar]
- Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155(2):251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Assembly and dynamics of the actin filament system in nonmuscle cells. J Cell Biochem. 1986;31(2):87–95. doi: 10.1002/jcb.240310202. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Earnshaw WC. Cell Biology. Philadelphia, PA: Saunders; 2004. [Google Scholar]
- Pope BJ, Gonsior SM, Yeoh S, McGough A, Weeds AG. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J Mol Biol. 2000;298(4):649–661. doi: 10.1006/jmbi.2000.3688. [DOI] [PubMed] [Google Scholar]
- Pope BJ, Zierler G, Kuhne R, Weeds AG, Ball LJ. Solution structure of human cofilin: Actin binding, pH sensitivity, and relationship to actin-depolymerizing factor. J Biol Chem. 2004;279(6):4840–4848. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- Prochniewicz E, Janson N, Thomas DD, De La Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. J Mol Biol. 2005;353(5):990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction, Part I: Biochemical studies of the interaction of the tropomyosin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246(15):4866–4871. [PubMed] [Google Scholar]
- Takashi R. A novel actin label: A fluorescent probe at glutamine-41 and its consequences. Biochemistry. 1988;27(3):938–943. doi: 10.1021/bi00403a015. [DOI] [PubMed] [Google Scholar]
- Theriot JA. Accelerating on a treadmill: ADF/cofilin promotes rapid actin filament turnover in the dynamic cytoskeleton. J Cell Biol. 1997;136(6):1165–1168. doi: 10.1083/jcb.136.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto S, Sellers JR. Characterization of in vitro motility assays using smooth muscle and cytoplasmic myosins. J Biol Chem. 1990;265(25):14864–14869. [PubMed] [Google Scholar]
- Uyeda TQP, Kron SJ, Spudich JA. Myosin Step Size: Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J Mol Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J. Actin-binding proteins. Curr Opin Cell Biol. 1990;2(1):41–50. doi: 10.1016/s0955-0674(05)80029-8. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T, Huxley HE, Amos LA, Klug A. Three-dimensional image reconstruction of actin-tropomyosin complex and actin-tropomyosin-troponin T-troponin I complex. J Mol Biol. 1975;93:477–497. doi: 10.1016/0022-2836(75)90241-7. [DOI] [PubMed] [Google Scholar]
- Yeoh S, Pope B, Mannherz HG, Weeds A. Determining the differences in actin binding by human ADF and cofilin. J Mol Biol. 2002;315(4):911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Sakai H. pH control of actin polymerization by cofilin. J Biol Chem. 1985;260(27):14410–14412. [PubMed] [Google Scholar]


