Abstract
Cell-free systems offer a unique platform for expanding the capabilities of natural biological systems for useful purposes, i.e. synthetic biology. They reduce complexity, remove structural barriers, and do not require the maintenance of cell viability. Cell-free systems, however, have been limited by their inability to co-activate multiple biochemical networks in a single integrated platform. Here, we report the assessment of biochemical reactions in an Escherichia coli cell-free platform designed to activate natural metabolism, the Cytomim system. We reveal that central catabolism, oxidative phosphorylation, and protein synthesis can be co-activated in a single reaction system. Never before have these complex systems been shown to be simultaneously activated without living cells. The Cytomim system therefore promises to provide the metabolic foundation for diverse ab initio cell-free synthetic biology projects. In addition, we describe an improved Cytomim system with enhanced protein synthesis yields (up to 1200 mg/l in 2 h) and lower costs to facilitate production of protein therapeutics and biochemicals that are difficult to make in vivo because of their toxicity, complexity, or unusual cofactor requirements.
Keywords: cell-free biology, in vitro translation, oxidative phosphorylation, protein synthesis, synthetic biology
Introduction
Cell-free systems provide a valuable platform for understanding, using, and expanding the capabilities of natural systems (Forster and Church, 2006, 2007; Swartz, 2006; Doktycz and Simpson, 2007; Meyer et al, 2007). As a complement to in vivo-based approaches, cell-free systems have the advantage of offering direct access to complex biological processes. New components (natural and non-natural) can be added or synthesized and can be maintained at precise ratios. The chemical environment can be controlled, actively monitored, and rapidly sampled. Moreover, systemic responses to environmental stimuli are minimized and high efficiency is afforded by directing resources toward exclusive user-defined objectives.
Cell-free protein synthesis (CFPS) systems, based on crude extracts, are one of the most prominent examples of cell-free biology. They were used in the pioneering studies of Nirenberg and Matthaei (1961) and played an essential role in deciphering the genetic code (Nirenberg, 2004). More recently, CFPS has shown remarkable utility as a protein synthesis technology (Katzen et al, 2005; Swartz, 2006), including the production of patient-specific vaccine candidates (Yang et al, 2005; Kanter et al, 2007) and pharmaceutical proteins (Yang et al, 2005; Goerke and Swartz, 2008). To test and model our understanding of how biology works, constructive cell-free approaches have also been reported. Shimizu et al (2001) reconstituted all of the factors necessary for protein synthesis from purified components. Additionally, genetic circuits have been built in crude extracts (Noireaux et al, 2003) and also from individual macromolecules (Kim et al, 2006).
Despite being used for decades as a tool in fundamental and applied research, one major disadvantage of most cell-free systems is their inability to co-activate multiple complex biochemical networks in a single integrated platform. To address this challenge, we sought to create a useful and cost-effective cell-free system that co-activates central metabolism, oxidative phosphorylation, transcription, translation, and protein folding. As catabolism coupled to respiration provides the most prolific natural energy supply, success would also address one of the principal challenges for CFPS systems; namely, activating efficient, cost-effective energy metabolism to support long-lived protein production. Historically, costly one-step phosphorylation reactions driven by phosphoenolpyruvate (PEP) and similar compounds have been used (Swartz, 2006). Unfortunately, this approach provides only brief bursts of ATP in batch reactions resulting in limited batch duration (∼60 min) and leading to inhibitory phosphate accumulation (>30 mM) (Kim and Swartz, 2001). The discovery that cofactor additions activate additional ATP supply from catabolism of the PEP by-product, pyruvate, suggested that more complicated metabolic processes were feasible (Kim and Swartz, 2001).
To pursue our objective, we chose to use an Escherichia coli-based crude extract system. An alternative approach is to construct a system of purified components (Shimizu et al, 2001). However, the time and cost associated with reconstituting every factor involved in energy metabolism, respiration, transcription, translation, and protein folding is prohibitive for most commercial applications (Swartz, 2001). Furthermore, it is not obvious that the benefits of a purified system (e.g. completely defined component parts; Shimizu et al, 2001) outweigh the challenge of reconstituting complex metabolic networks and their necessary interactions (Swartz, 2001; Xie et al, 2006), especially as membrane-associated complexes are required for respiration.
Our integrated CFPS system uses S30 extract, a DNA template, T7 RNA polymerase, non-phosphorylated energy substrates, nucleotides, and salts for effective protein production. The extract is prepared from cells harvested during mid-exponential growth at ∼3 OD (A600) (Swartz et al, 2004). Cells are completely lysed (>95%) by a single pass through a high-pressure homogenizer at ∼20 000 p.s.i.g. Then, cell lysate is centrifuged twice at 30 000 g for 30 min to remove cellular debris, remaining intact cells, and genomic DNA. The supernatant is incubated to release and degrade endogenous mRNA and a final dialysis introduces a suitable storage buffer and removes small molecule components. We reasoned that inverted inner membrane vesicles (IMVs) competent for oxidative phosphorylation would be generated by the high shear rate lysis procedure (Hertzberg and Hinkle, 1974; Nishio et al, 2002) and would also be retained in the extract. Furthermore, we believed that these vesicles could be activated to produce ATP from reducing equivalents generated by catabolism. However, our earlier studies using a CFPS system energized by PEP (the PANOx system) clearly showed that protein synthesis was unaffected by both oxygen availability and respiration inhibitors (Kim and Swartz, 2001).
We therefore adopted the hypothesis that cytoplasmic mimicry would encourage more natural metabolism. We would use natural E. coli cytoplasmic conditions to guide design and construction of the proper extract preparation procedures and physicochemical reaction conditions. Previously, we reported a new CFPS method stemming from these efforts. Called the Cytomim system, this platform more accurately mimics the E. coli intracellular environment by removing unnatural components (e.g. polyethylene glycol (PEG) and pH buffers) and reducing the concentrations of ionic components (Supplementary Table I) (Jewett and Swartz, 2004a). The results were dramatic. Yields of a model bacterial protein, chloramphenicol acetyl transferase (CAT), increased from less than 100 μg protein/ml with a previous CFPS method to more than 700 μg protein/ml when 33 mM of sodium pyruvate was used as an inexpensive energy substrate (Jewett and Swartz, 2004a).
In a follow-up study, three factors were found to be responsible, in combination, for enhanced yields: growing the source cells on a medium containing glucose and inorganic phosphate, reducing the magnesium concentration in the cell-free reaction, and replacing PEG with spermidine and putrescine (Jewett and Swartz, 2004b). Unexpectedly, we also observed that protein synthesis in the Cytomim system continued and ATP concentrations were sustained after pyruvate (the only presumed energy substrate) was depleted (Jewett and Swartz, 2004b). These observations suggested that a new source of ATP production had been activated, which we hypothesized was oxidative phosphorylation. Until now, however, the fundamental mechanism has remained unknown.
In this study, we use a systems approach to quantitatively assess active biochemical reactions in the Cytomim system. Remarkably, our metabolite and biochemical inhibitor analysis provides evidence for co-activation of central metabolism, oxidative phosphorylation, and protein synthesis. These data reveal that our previously designed Cytomim system provides an unprecedented level of activation of biological networks in vitro (Figure 1). We also demonstrate, for the first time, that glutamate alone can be used as a natural, non-phosphorylated homeostatic energy substrate and, based on this observation, describe a more cost effective and productive Cytomim system. We believe that these results establish the new Cytomim system as a fertile platform for a broad range of imaginative cell-free synthetic biology projects.
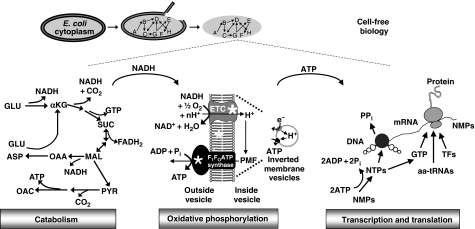
Figure 1.
Diagram of the molecular subsystems shown to be active in the Cytomim cell-free system. Glutamate (GLU) is used as a robust energy source in a natural chemical environment to produce reducing equivalents, primarily in the form of NADH, through the TCA cycle. NADH fuels oxidative phosphorylation in which oxygen serves as the final electron acceptor. Membrane-dependent respiration provides a stable supply of ATP, which is coupled to combined transcription and translation followed by protein folding. To prove the existence of oxidative phosphorylation in the Cytomim cell-free system, inhibitors were used to inactivate the electron transport chain (ETC), dissipate the proton motive force (PMF), and inactivate the F1FO-ATPase enzyme complex (each is denoted with an *) (αKG, alpha-ketoglutarate; SUC, succinate; MAL, malate; PYR, pyruvate; OAC, acetate; OAA, oxaloacetate; ASP, aspartic acid; Pi, inorganic phosphate; TFs, translation factors; aa-tRNAs, aminoacylated-tRNAs).
Results
CFPS is sustained without pyruvate
Early results using the Cytomim system with 33 mM pyruvate as the energy source and with initially tri-phosphorylated nucleosides (Cytomim/pyruvate/nucleoside triphosphate (NTP)) suggested that a new energy production pathway had been activated (Jewett and Swartz, 2004a, 2004b). To identify this pathway, we assessed metabolism in the Cytomim system. First, we confirmed that pyruvate metabolism was not the sole energy source. CFPS batch reactions were incubated with or without pyruvate for 6 h and chloramphenicol acetyl transferase (CAT) synthesis was assessed by 14C-leucine incorporation into trichloroacetic acid-precipitable protein. The production of 500 μg/ml of CAT was observed without pyruvate, and NTP concentrations were sustained (Figure 2A and B; Supplementary Figure 1). Autoradiograms confirmed that only CAT was produced as the T7 RNA polymerase limited transcription only to the T7 promoter on the plasmid added to the reaction.
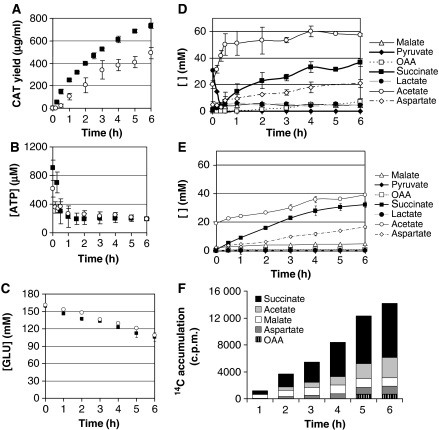
Figure 2.
Protein product, ATP, and metabolite kinetics for the Cytomim system indicate that central metabolism has been co-activated with protein synthesis. (A) Synthesis of chloramphenicol acetyl transferase (CAT) as determined by 14C-leucine incorporation in the Cytomim system (15 μl batch reactions) with 33 mM pyruvate (Cytomim/pyruvate/NTP—closed squares), and without pyruvate (Cytomim/glutamate/NTP—open circles). Control transcription and translation reactions without plasmid DNA were used to assess background protein production levels. These reactions demonstrated negligible background incorporation. (B) ATP concentration kinetics during protein synthesis (15 μl batch reactions) confirm the presence of an additional energy substrate besides pyruvate. Cytomim system with 33 mM pyruvate (Cytomim/pyruvate/NTP—closed squares). Cytomim system without pyruvate (Cytomim/glutamate/NTP—open circles). (C) Glutamate (GLU) depletion during protein synthesis (15 μl batch reactions) suggests that glutamate is fueling energy production. Cytomim system with 33 mM pyruvate (Cytomim/pyruvate/NTP—closed squares). Cytomim system without pyruvate (Cytomim/glutamate/NTP—open circles). (D) Organic acid formation kinetics in the Cytomim/pyruvate/NTP system indicate the accumulation of TCA cycle intermediates (OAA, oxaloacetate). As expected, acetate concentrations increased rapidly as a result of pyruvate catabolism through pyruvate dehydrogenase during the first 30 min of the reaction. (E) Organic acid formation kinetics in the Cytomim/glutamate/NTP system indicate the accumulation of TCA cycle intermediates (OAA, oxaloacetate). (F) Accumulation of radioactivity in metabolites of the Cytomim/glutamate/NTP cell-free reaction. A mixture of uniformly labeled 14C-glutamate and non-labeled glutamate was added at the start of the reaction. (A, B) Results are the average of n=6 experiments. (C–E) n=4. (F) n=3. Error bars=1 s.d.
Remarkably, this is the first time that CFPS has been reported without the addition of a secondary energy substrate, such as PEP, creatine phosphate, acetyl phosphate, glucose-6-phosphate, pyruvate, or other glycolytic intermediates.
Central metabolism is activated
As protein synthesis is sustained without pyruvate, we next sought to identify the secondary energy substrate. Primary candidates were the 20 amino acids and acetate. Using HPLC analyses and an enzymatic assay, we monitored consumption and formation of amino and organic acids in the cell-free system during CAT synthesis. Although the concentrations of most amino acids changed by at most 1–2 mM over the 6 h reaction (Supplementary Table II), a steady ∼9 mM/h decrease in the concentration of glutamate was observed (Figure 2C). We also detected the formation of succinate, acetate, malate, oxaloacetate, and aspartate (Figure 2D and E). These results suggest that glutamate is directed into the tricarboxylic acid (TCA) cycle to form α-ketoglutarate, likely through glutamate dehydrogenase, the levels of which are high in cells grown on glucose (a necessary condition for the Cytomim system; Jewett and Swartz, 2004b), and aspartate aminotransferase (McFall and Newman, 1996).
The carbon content of all amino and organic acids produced accounted for 82±7 and 79±6% of the consumed carbon with and without pyruvate, respectively (Supplementary Table II). This calculation does not, however, include carbon dioxide evolution. Although our efforts to capture CO2 were unsuccessful (likely a consequence of small reaction scales), we estimated CO2 production to a first-order approximation on a stoichiometric basis through α-ketoglutarate dehydrogenase (from glutamate consumed) and through pyruvate dehydrogenase (from acetate produced). This estimation resulted in carbon balances of 103±7 and 101±6% with and without pyruvate, respectively.
To verify formation of TCA cycle intermediates and side products through glutamate catabolism, radioactive tracing experiments with 84 μM L-[U-14C]glutamate added to Cytomim/glutamate/NTP reactions were performed (∼160 mM unlabeled glutamate is present initially). The accumulation of radioactive metabolites was determined by scintillation counting of fractions after HPLC separation (Figure 2F). Consistent with data presented in Figure 2E, labeled carbon appears in succinate, acetate, malate, aspartate, and oxaloacetate. Estimating CO2 evolution as above, we expected a 9% loss of the labeled carbon. The actual loss was 10±2%.
Protein synthesis is oxygen dependent
Although central metabolism was active, the new energy regeneration pathway was still undetermined. We hypothesized that reducing equivalents from glutamate catabolism and the TCA cycle could fuel ATP generation if they were linked to oxidative phosphorylation. To evaluate the possibility that inner membrane vesicles were competent for respiration, we tested the effect of oxygen availability on the Cytomim system. CAT synthesis yields and ATP concentrations under anaerobic conditions were significantly decreased relative to aerobic conditions (Figure 3A). Larger scale (>30 μl), aerobic batch reactions also produced less product than standard 15 μl reactions, presumably due to a decrease in the air/liquid surface area (required for oxygen transfer into the reaction mixture) to the reaction volume ratio (data not shown). More strikingly, both the dissolved oxygen and ATP concentrations rapidly dropped to zero and protein synthesis terminated when the pure oxygen feed to a 2 ml stirred CFPS reaction (Supplementary Figure 2) was interrupted (Figure 3B). As vigorous oxygen supply is directly linked to protein production, these data suggest that the new energy generation pathway is oxygen dependent and that oxygen consumption occurs at a relatively high rate.
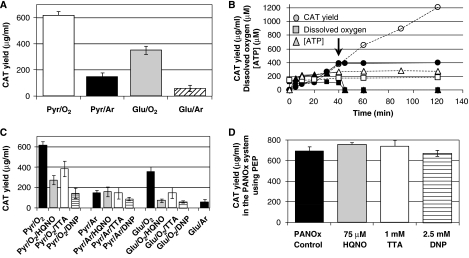
Figure 3.
Oxygen-dependent energy production in the Cytomim system is caused by oxidative phosphorylation. (A) Here, 20 μl cell-free batch reactions were carried out for 5 h. CAT production yields in the Cytomim/pyruvate/NTP system (Pyr) or in the Cytomim/glutamate/NTP system (Glu) in the presence of oxygen (O2) or argon (Ar). In (B), 2 ml stirred tank cell-free reactions using the Cytomim/glutamate-phosphate/NMP system are shown (see Materials and methods). Total CAT yield (circles), ATP concentration (triangles) and dissolved oxygen concentration (squares) are plotted versus time. After 40 min (see arrow), the oxygen feed was stopped (closed shapes). This resulted in complete consumption of available oxygen, depletion of ATP, and termination of protein synthesis. A control reaction (open shapes) is shown for comparison. After 6 h, 1447±64 mg/l of CAT is synthesized when oxygen is present. In (C), 20 μl cell-free batch reactions carried out for 5 h are shown. CAT production yields in the Cytomim/pyruvate/NTP system (Pyr) or in the Cytomim/glutamate/NTP system (Glu) in the presence of oxygen (O2) or argon (Ar). The reduction in protein synthesis after addition of either 75 μM 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO, an inhibitor of electron transport), 1 mM thenoyltrifluoroacetone (TTA, an inhibitor of electron transport) or 2.5 mM 2-4-dinitrophenol (DNP, an uncoupling agent) indicates that oxygen-dependent protein synthesis relies on energy derived from oxidative phosphorylation (e.g. Pyr/O2 versus Pyr/O2/HQNO). Oxygen-independent CAT synthesis is unaffected (e.g. Pyr/Ar versus Pyr/Ar/HQNO). In (D), 20 μl cell-free batch reactions using the conventional PANOx/PEP/NTP system (not the Cytomim system), carried out for 5 h, are shown. Consistent with previous results (Kim and Swartz, 2001), inhibitors of oxidative phosphorylation do not affect protein biosynthesis in this case (conducted in the presence of oxygen). (A, C, D) Results are the average of n⩾6 experiments. (B) n=3. Error bars=1 s.d.
Biochemical evidence for oxidative phosphorylation
Specific inhibitors of oxidative phosphorylation provided conclusive evidence that respiration occurs in the Cytomim system and fuels protein synthesis. When aerobic cell-free reactions were incubated with four biochemical inhibitors of the electron transport chain and the F1FO-ATPase and with two membrane gradient uncouplers, significantly less protein was produced relative to control reactions (Figure 3C; Supplementary Figure 3). This occurred in both Cytomim/pyruvate/NTP and in Cytomim/glutamate/NTP reactions (note that glutamate is also present in the former). To rule out a general effect on protein synthesis imposed by these inhibitors, reactions were also conducted anaerobically. In these reactions, protein synthesis was unaffected by the inhibitors (Figure 3C; Supplementary Figure 3). In addition, protein production with conventional cell-free reactions that use PEP as an energy source (PANOx/PEP/NTP) was unaffected by the inhibitors (Figure 3D; Supplementary Figure 3).
When surfactants such as Triton X-100, a low CMC detergent that dissolves lipid bilayers, were added to the Cytomim/pyruvate/NTP system, aerobic protein synthesis yields were similar to those obtained under anaerobic conditions (∼60 μg CAT/ml reaction). These results are consistent with our earlier discovery that omitting PEG is critical for Cytomim system activation (Jewett and Swartz, 2004b).
The impact of purified vesicle addition
Results with the inhibitors and uncouplers support our interpretation that membrane vesicles had been activated. We next wanted to assess whether respiration or protein synthesis yields could be increased by vesicle supplementation. Using sucrose gradients (Wuu and Swartz, 2008), we isolated IMVs from our E. coli extracts (approximately 20% of the total lipid concentration) and first validated their NADH dehydrogenase, proton pumping, and ATPase activities. NADH should be directly accessible to NADH hydrogenases on the external surface of IMVs. By following NADH oxidation over time, we determined the rate of NADH consumption to be 1.2±0.1 μmol NADH/g lipid/min. To assess proton pumping, we used a self-quenching dye, quinacrine, and showed that the interior pH of the IMVs drops after initiating proton pumping with the addition of NADH or ATP (Figure 4A and B). Finally, we measured the ATP generation of purified IMVs to be 2.7±0.36 μmol ATP/g lipid/min, which is consistent with our NADH oxidation measurements assuming ∼2.5 ATP per NADH. Because the Cytomim system more faithfully reflects cytoplasmic conditions than the traditional biochemical assays used to assess vesicle activity, activity is likely higher in the more natural system. We have recently shown, for example, that the characterization of individual enzymes or purified components may not always predict the performance of multi-component cell-free systems with complex interdependencies (Jewett et al, 2008). Nevertheless, these data conservatively suggest that 0.052 mg/ml of IMVs in the extract can produce ∼0.15 μM ATP/min. Assuming protein synthesis requires four ATPs per peptide bond (Stryer, 1995), the ATP generation rate is sufficient to produce 247 μg protein/ml/h, a rate consistent with the data shown in Figure 2A. This does not consider additional ATP generation mechanisms (e.g. acetate kinase (ATP) and succinyl-coenzymeA (CoA) synthetase (GTP)) or ATP consumption independent of protein synthesis.
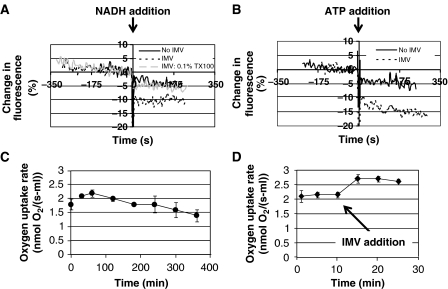
Figure 4.
The Cytomim system provides a chemical environment conducive for oxidative phosphorylation. Quinacrine, a self-quenching fluorescent dye that distributes according to pH, was used to indicate the proton pumping capability of IMVs. (A) When NADH is added to IMVs to initiate proton pumping, a 10% drop in fluorescence signal is observed, consistent with pH-induced quinacrine accumulation inside vesicles. This drop in fluorescence is not seen when vesicles are absent from the solution. As an additional control, the experiment was repeated with vesicles in the presence of 0.1% Triton X-100 detergent (TX100). Triton X-100 dissolves lipid bilayers and is shown here to prevent accumulation of protons or quinacrine. (B) Similar to NADH, we observe a drop in fluorescence when proton pumping through the F1FO-ATPase is initiated by ATP addition. (C) Oxygen uptake rate of the Cytomim/glutamate-phosphate/NMP system during synthesis of CAT. Results are the average of n=3 experiments. Error bars=1 s.d. (D) Addition of purified inner membrane vesicles to the Cytomim/glutamate-phosphate/NMP system during synthesis of CAT increases the oxygen uptake rate. At 10 min after the start of the reaction, 0.26 mg of purified inner membrane vesicles were added to the reaction, increasing the total vesicle concentration from 0.26 to 0.36 mg/ml. Results are the average of n=3 experiments. Errors bars=1 s.d.
To test whether the addition of IMVs to Cytomim CFPS reactions increased respiration, we used a stirred tank reactor sparged with oxygen (Supplementary Figure 2). The oxygen uptake rate for the Cytomim system, without IMV supplementation, is approximately 40 nmol O2 per mg of lipid in IMVs per second (Figure 4C). Strikingly, this is on the same order as for intact E. coli cells. The specific oxygen uptake rate for E. coli is about 110 nmol O2 per mg of cytoplasmic membrane lipid per second (Neijssel et al, 1996). When purified IMVs were added to Cytomim reactions to increase the total vesicle concentration by about 40%, the oxygen uptake rate increased by about 25% (Figure 4D). The disproportionate increase may reflect partial vesicle inactivation or a limitation in the supply of reducing equivalents. Nonetheless, this observation verifies that the Cytomim cell-free platform provides conditions conducive to oxidative phosphorylation. Addition of purified vesicles to Cytomim CFPS reactions did not enhance protein synthesis yields, however (data not shown). This is most likely because ATP concentrations were already well above the KmATP for the system (KmATP=27±4 μM; Jewett et al, 2008).
Enhancing protein synthesis yields and reducing costs
Addressing one of the key limitations of cell-free biology, our systems analysis demonstrates that catabolism, oxidative phosphorylation, and protein synthesis can be co-activated in the integrated Cytomim platform. Next, we sought to broaden the utility of this technology by enhancing protein synthesis yields and lowering costs. To increase protein synthesis yields in the Cytomim/glutamate/NTP system, we increased phosphate concentrations (Calhoun and Swartz, 2005a). It has been reported that low concentrations of phosphate limit phosphorylation reactions (e.g. nucleotide regeneration) (Calhoun and Swartz, 2005a). Production yields of CAT were enhanced approximately 33%, from 584±82 μg CAT/ml reaction to 802±48 μg CAT/ml reaction, when the reaction was augmented with 10 mM phosphate (the experimentally determined optimum). Figure 5 shows CAT accumulation over time for the Cytomim/glutamate-phosphate/NTP system. The soluble and active fraction of CAT was approximately 70±3% of the total yield. It is unknown whether the addition of phosphate enhances oxidative phosphorylation, inhibits phosphatase reactions, or both.
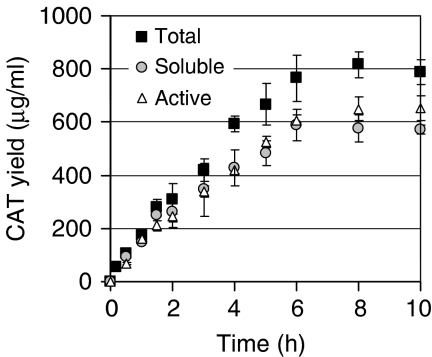
Figure 5.
CAT synthesis over time using the Cytomim/glutamate-phosphate/NTP system. Reactions were carried out for 10 h at 37°C and CAT synthesis was determined by 14C-leucine incorporation and enzymatic activity assay. Here, 10 mM phosphate was supplemented to the cell-free reaction without pyruvate and 15 μl reaction mixtures were prepared in a different tube for each time point. Results are the average of n=6 experiments. Error bars=1 s.d. Total yield of CAT as determined by 14C-leucine incorporation (closed squares). Soluble yield of CAT as determined by 14C-leucine incorporation (gray circles). Active yield of CAT as determined by enzymatic assay (open triangles).
High-energy phosphate-bonded substrates (e.g. PEP and nucleotides) have traditionally dominated production costs in cell-free systems (Swartz, 2006). Replacing NTPs with nucleoside monophosphates (NMPs) significantly reduces expenses in cell-free reactions energized by glucose (Calhoun and Swartz, 2005b). We found that NMPs are also effective nucleotides for the Cytomim/glutamate-phosphate system. While the overall CAT yield is reduced to 640±64 μg/ml (Supplementary Figure 4), the amount of protein produced per dollar of substrate costs is increased by more than an order of magnitude (Table I). This cost–benefit is even more dramatic when the Cytomim/glutamate-phosphate/NMP system is compared with the conventional PANOx/PEP/NTP reaction (Table I).
Table 1.
The Cytomim system is cost-effective
| System | Reaction size (μl) | Energy source and nucleotides | Synthesis duration (h) | CAT produced (mg/ml) | Energy source and nucleotides cost (US$/ml reaction) | mg protein/ (US$ energy source and nucleotides) |
|---|---|---|---|---|---|---|
| PANOx | 15 | PEP, NTPs | 3 | 0.74±0.02 | 1.64 | 0.45 |
| PANOx | 15 | Pyruvate, NTPs | 3 | 0.06±0.02 | 0.52 | 0.12 |
| Cytomim | 15 | Pyruvate, NTPs | 6 | 0.73±0.04 | 0.52 | 1.40 |
| Cytomim | 15 | Glutamate, NTPs | 6 | 0.51±0.05 | 0.51 | 1.00 |
| Cytomim | 15 | Glutamate, phosphate, NTPs | 6 | 0.85±0.05 | 0.51 | 1.65 |
| Cytomim | 15 | Glutamate, phosphate, NMPs | 6 | 0.62±0.06 | 0.004 | 155 |
Relative product yields in Eppendorf batch reactors (mg protein/US$ energy source and nucleotides cost) demonstrate a significant increase for the new Cytomim system when NMPs and phosphate are used. Reagent prices are from manufacturer's online catalog in August 2008.
As a further attempt to increase yields, we added up to 30 mM NADH to the Cytomim/glutamate-phosphate/NTP system (the concentration of the nicotinamide adenine dinucleotide (NAD)/NADH couple without supplementation is 0.3 mM). This, however, did not increase protein synthesis, likely because ATP concentrations were already well above the KmATP for the system (Jewett et al, 2008). Because glutamate salts were used to construct the required physicochemical environment, other avenues of TCA cycle activation linked to oxidative phosphorylation (e.g. α-ketoglutarate) were not pursued.
Finally, to demonstrate the utility of the Cytomim system, we increased the production scale to 5 ml. Here, 205±28 nmol of CAT (>1 mg/ml) was produced during an 8 h incubation and 129±4 nmol were active (Supplementary Figure 5). CAT was a dominant band on a SDS polyacrylamide gel, and autoradiography confirmed that 14C-leucine was only incorporated into the target protein with minor quantities of translational pause or degradation products. Similar to our previous work synthesizing 100 nmol quantities of active protein (Jewett and Swartz, 2004c), these data suggest the feasibility of high-throughput protocols for generating sufficient quantities and purity of proteins for structure determination by NMR or X-ray crystallography.
Discussion
CFPS systems have many advantages as a complement to existing protein production technologies for producing disease treatments and diagnostics that are difficult to make in vivo. This includes therapeutic proteins that (i) are toxic to cells, (ii) require in vitro selections, (iii) require sequential production, or (iv) require rapid product/process development (e.g. personalized medicines). CFPS methods, however, have been limited by their inability to supply the intense energy needs of protein synthesis without deleterious changes to the chemical environment. This limitation was convincingly demonstrated in the landmark experiments of Spirin et al (1988) when they dramatically enhanced product yields by continually feeding the energy substrates. Yet, two decades have elapsed without the development of a simple, durable, and cost-effective batch cell-free system. More broadly, integrating complex metabolic networks is an outstanding challenge in cell-free synthetic biology (Simpson, 2006).
Here, by evaluating biochemical reactions, we determine the new source of ATP production in the Cytomim CFPS system (Jewett and Swartz, 2004a). Remarkably, we reveal that central metabolism, oxidative phosphorylation, protein synthesis, and protein folding are simultaneously activated in a single integrated platform. The two intense centrifugation steps in the cell extract preparation procedure remove any intact cells remaining after the high-pressure homogenization step, but IMVs do not sediment. IMVs serve to convert reducing equivalents gained from glutamate catabolism and the TCA cycle into the ATP required to fuel high-level transcription and translation (Figure 1). Never before have this many complex enzyme systems been shown to be simultaneously activated without living cells. Notably, the anionic salt species glutamate is, for the first time, used as a secondary energy substrate. With respect to CFPS systems, this possibility was not previously appreciated and stems from a major frame of reference shift. As our analysis has shown, crude extract systems can now be thought of as a complex set of biochemical reactions that can be identified and controlled, rather than a complex, inscrutable ‘black box'.
It is striking to note that the Cytomim system closely mimics E. coli cellular metabolism. It is homeostatic in pH and [Pi], uses natural, non-phosphorylated energy substrates, provides a long-lasting ATP source, and fuels highly productive protein synthesis (up to 600 mg protein/l/h). In addition, each ribosome can polymerize approximately 10 500 amino acids (42 copies of CAT), indicating that the Cytomim system is not limited by enzyme turnover (e.g. only one protein, or fraction of a protein, produced per ribosome). Furthermore, the specific oxygen uptake rate in the Cytomim system is on the same order as intact E. coli cells. As an additional indication of robust cell-free metabolism, other experiments in our laboratory have shown that the CFPS system exhibits second-order kinetics, i.e. when we doubled the cell extract concentration by ultrafiltration during the course of protein synthesis, volumetric protein synthesis rates increased four-fold (Jewett MC and Swartz JR (2008), in preparation). In other words, the specific translation rate is proportional to the cell extract (ribosome and cofactors) concentration. Cell-free protein elongation rates determined using polysome analysis were consistent with this interpretation (Underwood et al, 2005). Therefore, the maximum batch protein synthesis rate we observe for the Cytomim system suggests that this complex catalytic system is functioning at approximately the rate expected after a 20-fold dilution from cytoplasmic concentrations. To put this in perspective, even though the cell-free specific elongation rates are lower, the cell-free volumetric productivity compares favorably to typical rates for live cell cultures since the cell-free system uses the entire reactor volume rather the much smaller intracellular volume fraction used in typical cultures.
As our crude extract system can provide integrated metabolic function, we believe that the Cytomim system lays the foundation for constructive cell-free synthetic biology projects. Past cell-free systems have typically lacked the ‘housekeeping functions' of the cell (Simpson, 2006), such as activated metabolic networks, sustained energy production, and highly productive protein synthesis. Building synthetic systems of the complexity shown here from the ground up (Forster and Church, 2006) may ultimately be a more powerful approach for designing synthetic systems. However, the Cytomim platform represents an economically viable platform that can be used now for understanding and using biology, while gaps in our ability to construct purified systems are filled. For example, the Cytomim platform may also be combined with vesicle bioreactors (Noireaux and Libchaber, 2004; Murtas et al, 2007) as a further step toward making an artificial cell.
Beyond construction efforts, the Cytomim system also lays the foundation for analyzing complex biomolecular systems that are difficult to manipulate in vivo without the risk of new enzyme synthesis in response to changing environments. For example, this system will allow a more careful examination of the effects of component localization on overall system performance. It has already enabled us to determine the dependence of protein synthesis rate on [ATP] and [GTP] (Jewett et al, 2008) and is expected to stimulate intriguing hypotheses that can then be tested in vivo. For example, our metabolic flux measurements support the concept that many bacteria use intracellular glutamate as an emergency energy source under aerobic conditions following external energy source exhaustion.
Here, we have also engineered a more cost-effective, highly productive Cytomim system. High reagent costs, primarily energy substrates and nucleotides (amino acid costs are relatively minor), and low productivities have been a major deterrent for the commercial use of cell-free systems (Swartz, 2006). The new Cytomim/glutamate-phosphate/NMP batch system is more than two orders of magnitude more productive, on a cost basis, than the conventional PANOx/PEP/NTP system (Table I). This high-yielding (up to 1200 mg/l in 2 h), cost-effective protein synthesis technology is enabling us to produce complex pharmaceutical proteins, such as human insulin-like growth factor-1 at the 1-L scale (Voloshin and Swartz, 2007). This advance opens the door for commercial production of personalized medicines and for rapid synthesis of large quantities of vaccines and antidotes against bio-warfare attacks using cell-free factories.
Activating central metabolism in cell-free extracts also has utility for cofactor-dependent, multistep bioconversions. We have shown, for example, that the Cytomim system can convert inexpensive substrates, such as glutamate, to high-value products such as succinate (Ygs=0.68 C-mol succinate C-mol glutamate−1) (Figure 2) (Werpy and Petersen, 2004). More precise engineering of these pathways should lead to economically attractive bioconversions (Liao, 2004).
The reason for protein synthesis termination in the Cytomim system remains an unanswered question. As small molecular weight substrates appear to be in sufficient concentrations (Figure 2; Supplementary Figure 1; Supplementary Table II), inhibitory by-product accumulation or system degradation may be responsible.
In summary, although in vitro translation systems have been utilized for more than 50 years, demonstration that central metabolism, oxidative phosphorylation, and protein synthesis can be co-activated in a well-defined, robust, and customizable system is new. Given the capability to modify and control cell-free systems, the Cytomim platform will be a powerful tool for systems biology, for synthetic biology, and as a protein production technology.
Materials and methods
S30 crude extract preparation
S30 cell extract was prepared from E. coli strain NMR2, a derivative of K12 with mutations to stabilize arginine and tryptophan in the cell-free reactions (Michel-Reydellet et al, 2004), grown on a glucose and phosphate media (2 × YTPG) as described earlier (Swartz et al, 2004; Jewett and Swartz, 2004a). In this study, no measures were taken to inactivate hydrolytic enzymes in the cell extracts.
CFPS
Cytomim CFPS reactions were, in general, carried out in 1.5 ml Eppendorf tubes at 37°C (Supplementary Figure 2) (Jewett and Swartz, 2004a). The standard reaction mixture contained the following components: 1.2 mM ATP, 0.85 mM each of GTP, UTP and CTP, 34 μg/ml folinic acid, 170.6 μg/ml E. coli tRNA mixture (Roche, Indianapolis, IN), 13.3 μg/ml pk7CAT plasmid (encoding the gene for CAT; (Kim and Swartz, 2001)), 100 μg/ml T7 RNA polymerase, 5 μM L-[U-14C]leucine (Amersham Pharmacia Biotechnology, Uppsala, Sweden), 2 mM each of 20 unlabeled amino acids, 0.33 mM NAD, 0.26 mM CoA, 130 mM potassium glutamate, 10 mM ammonium glutamate, 8 mM magnesium glutamate, 1.5 mM spermidine, 1 mM putrescine, 4 mM sodium oxalate, and 0.24 volume of S30 extract. Reactions were supplemented with (the Cytomim/pyruvate/NTP system) or without (the Cytomim/glutamate/NTP system) 33 mM sodium pyruvate as specified in the text. T7 RNA polymerase was prepared as described earlier (Swartz et al, 2004). Unless otherwise specified, reaction volumes were 15 μl. The Cytomim/glutamate-phosphate/NTP system had the reaction mixture described above without pyruvate except that 10 mM potassium phosphate (pH=7.2 using glacial acetic acid) was also added. The Cytomim/glutamate-phosphate/NMP system simply exchanged NMPs for NTPs at the standard initial concentrations listed above. The conventional cell-free reactions using PEP as an energy substrate PANOx/PEP/NTP were carried out using previously described protocols (Jewett and Swartz, 2004a).
Here, 2 ml stirred tank reactions were performed in a reactor designed and fabricated in-house. Temperature was maintained through the use of a water jacket. Oxygen was delivered through one sparge point. Agitation was achieved through stirring with a micro magnetic bar at 100 r.p.m. Dissolved oxygen concentration was measured with a Clark Cell oxygen microprobe (Microelectrodes, Bedford, NH). Pluronic 31R1 antifoam agent was used at a concentration of 0.01 % v/v to prevent foaming.
Protein product, ATP, organic acid, and amino acid analyses
To quantify the amount of synthesized protein, reaction samples were analyzed by incorporation of 14C-leucine into trichloroacetic acid-precipitable radioactivity using a liquid scintillation counter following treatment with 0.1 N NaOH to stop translation or by enzymatic assay as described earlier (Swartz et al, 2004). ATP, organic acids, and amino acids were quantified using HPLC peak analysis as previously earlier (Jewett and Swartz, 2004b). Glutamate was analyzed using a reagent kit from R-Biopharm Inc. (Marshall, MI) that employs an enzymatic colorimetric method. Trichloroacetic acid (5%) at 4°C was added to the cell extract reaction mixture in a 1:1 volumetric ratio. Trichloroacetic acid precipitated samples were centrifuged at 12 000 g for 15 min at 4°C. The supernatant was collected and sodium hydroxide was added to bring the pH to between 7.8 and 8.0. The pH-adjusted supernatant was diluted to contain less than 0.07 g/l glutamate and concentrations were determined according to the manufacturer's recommendations.
L-[U-14C]glutamate labeling
Cytomim CFPS reactions with 84 μM L-[U-14C]glutamate (0.02 μCi/ml of reaction; Amersham Pharmacia Biotechnology) were incubated at 37°C and samples were periodically taken for HPLC peak analysis. During the HPLC separation using the organic acid method described above, fractions were collected every 15 s for radioisotope analysis by liquid scintillation counting.
Aerobic/anaerobic CFPS reactions
After all reagents for CFPS were mixed together at 4oC (as specified above), the headspace of the reactor, an Eppendorf tube, was filled with either pure oxygen or argon. Sparging of the specified gas was performed for 2 min in a closed Eppendorf tube with vent holes to prevent pressure build-up. After 2 min, the top of the Eppendorf tube was sealed to cover the holes in the cap and prevent gas exchange. Control experiments demonstrated that reaction volumes did not decrease as a result of gas sparging.
CFPS with inhibitors for oxidative phosphorylation
CFPS (20 μl reactions) was carried out for 5 h after pre-equilibrating cell extract with an inhibitor of oxidative phosphorylation for 15 min at 25°C. Substrates and solutes for the cell-free translation reaction were added to the extract/inhibitor mixture for CAT biosynthesis to proceed. Inhibitors included: 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO, 75 μM; Cox et al, 1970), thenoyltrifluoroacetone (TTA, 1 mM; Tappel, 1960), 2-4-dinitrophenol (DNP, 2.5 mM; McLaughlin, 1972), cyanide (1 mM; Miller and Gennis, 1983), N,N′-dicyclohexylcarbodiimide (DCCD, 50 μM; Hare, 1975) and carbonyl-cyanide-m-chlorophenylhydrazone (CCCP, 100 μM; (Heytler and Prichard, 1962). HQNO and DCCD were solubilized in ethanol. TTA was solubilized in methanol. Cyanide and DNP were solubilized in water. CCCP was dissolved in DMSO. Negative controls performed with ethanol, methanol, and DMSO demonstrated that these solvents did not affect protein synthesis at concentrations used in this study.
Vesicle purification, characterization, and addition to CFPS reactions
Vesicles were purified following the protocol of Wuu and Swartz (2008). Detailed descriptions and methods used for vesicle characterization are described in the Supplementary information. Vesicle addition experiments were performed in 2 ml batch Cytomim/glutamate-phosphate/NMP reactions synthesizing CAT at 37°C. The initial lipid concentration due to vesicles in the extract was ∼0.26 mg/ml. At 10 min after the start of the reaction, 0.26 mg (lipid content) of purified inner membrane vesicles was added to the 2-ml reaction mixture, bringing the total vesicle concentration to 0.36 mg/ml.
Oxygen consumption rate measurements
To measure the oxygen consumption rate, the oxygen feed was terminated and the change in oxygen concentration in the cell-free reaction mixture was measured. The sensor was calibrated using a 30-min nitrogen sparge (0 mM DO2) and an air sparge (0.2 mM DO2). Both calibration points were performed at 37°C, the temperature of the CFPS reaction. The oxygen consumption rate was taken as the slope (linear section) of the oxygen depletion curve.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to C Yanofsky and C Khosla for critical reading of the paper. This study was supported by an NIH grant to JRS (RO1-GM60615). MCJ was a recipient of a predoctoral fellowship from the Stanford-NIH training program in biotechnology. KAC was supported by an NSF Graduate Research Fellowship.
References
- Calhoun KA, Swartz JR (2005a) Energizing cell-free protein synthesis with glucose metabolism. Biotechnol Bioeng 90: 606–613 [DOI] [PubMed] [Google Scholar]
- Calhoun KA, Swartz JR (2005b) An economical method for cell-free protein synthesis using glucose and nucleoside monophosphates. Biotechnol Prog 21: 1146–1153 [DOI] [PubMed] [Google Scholar]
- Cox GB, Newton NA, Gibson F, Snoswell AM, Hamilton JA (1970) The function of ubiquinone in Escherichia coli. Biochem J 117: 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktycz MJ, Simpson ML (2007) Nano-enabled synthetic biology. Mol Syst Biol 3: 125 10.1038/msb4100165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Church GM (2006) Towards synthesis of a minimal cell. Mol Syst Biol 2 45 10.1038/msb4100090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Church GM (2007) Synthetic biology projects in vitro. Genome Res 17: 1–6 [DOI] [PubMed] [Google Scholar]
- Goerke AR, Swartz JR (2008) Development of cell-free protein synthesis platforms for disulfide bonded proteins. Biotechnol Bioeng 99: 351–367 [DOI] [PubMed] [Google Scholar]
- Hare JF (1975) Purification and characterization of a dicyclohexylcarbodiimide-sensitive adenosine triphosphatase complex from membranes of Escherichia coli. Biochem Biophys Res Commun 66: 1329–1337 [DOI] [PubMed] [Google Scholar]
- Hertzberg EL, Hinkle PC (1974) Oxidative phosphorylation and proton translocation in membrane vesicles prepared from Escherichia coli. Biochem Biophys Res Commun 58: 178–184 [DOI] [PubMed] [Google Scholar]
- Heytler PG, Prichard WW (1962) A new class of uncoupling agents—carbonyl cyanide phenylhydrazones. Biochem Biophys Res Commun 7: 272–275 [DOI] [PubMed] [Google Scholar]
- Jewett MC, Miller ML, Chen Y, Swartz JR (2008) Continued protein synthesis at low [ATP] and [GTP] enables cell adaptation during energy limitation. J Bacteriol (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MC, Swartz JR (2004a) Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng 86: 19–26 [DOI] [PubMed] [Google Scholar]
- Jewett MC, Swartz JR (2004b) Substrate replenishment extends protein synthesis with an in vitro translation system designed to mimic the cytoplasm. Biotechnol Bioeng 87: 465–472 [DOI] [PubMed] [Google Scholar]
- Jewett MC, Swartz JR (2004c) Rapid expression and purification of 100 nmol quantities of active protein using cell-free protein synthesis. Biotechnol Prog 20: 102–109 [DOI] [PubMed] [Google Scholar]
- Kanter G, Yang J, Voloshin A, Levy S, Swartz JR, Levy R (2007) Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood 109: 3393–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F, Chang G, Kudlicki W (2005) The past, present and future of cell-free protein synthesis. Trends Biotechnol 23: 150–156 [DOI] [PubMed] [Google Scholar]
- Kim DM, Swartz JR (2001) Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol Bioeng 74: 309–316 [PubMed] [Google Scholar]
- Kim J, White KS, Winfree E (2006) Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol Syst Biol 2 68 10.1038/msb4100099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC (2004) Custom design of metabolism. Nat Biotechnol 22: 823–824 [DOI] [PubMed] [Google Scholar]
- McFall E, Newman EB (1996) Amino acids as carbon sources. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), 2nd edn, pp 358–379. Washington DC: ASM Press [Google Scholar]
- McLaughlin S (1972) The mechanism of action of DNP on phospholipid bilayer membranes. J Membr Biol 9: 361–372 [PubMed] [Google Scholar]
- Meyer A, Pellaux R, Panke S (2007) Bioengineering novel in vitro metabolic pathways using synthetic biology. Curr Opin Microbiol 10: 246–253 [DOI] [PubMed] [Google Scholar]
- Michel-Reydellet N, Calhoun K, Swartz J (2004) Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome. Metab Eng 6: 197–203 [DOI] [PubMed] [Google Scholar]
- Miller MJ, Gennis RB (1983) The purification and characterization of the cytochrome d terminal oxidase complex of the Escherichia coli aerobic respiratory chain. J Biol Chem 258: 9159–9165 [PubMed] [Google Scholar]
- Murtas G, Kuruma Y, Bianchini P, Diaspro A, Luisi PL (2007) Protein synthesis in liposomes with a minimal set of enzymes. Biochem Biophys Res Commun 363: 12–17 [DOI] [PubMed] [Google Scholar]
- Neijssel OM, Teixeira de Mattos MJ, Tempest DW (1996) Growth yield and energy distribution. In Escherichia Coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), 2nd edn, pp 1683–1692. Washington DC: ASM Press [Google Scholar]
- Nishio K, Iwamoto-Kihara A, Yamamoto A, Wada Y, Futai M (2002) Subunit rotation of ATP synthase embedded in membranes: a or beta subunit rotation relative to the c subunit ring. Proc Natl Acad Sci USA 99: 13448–13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg M (2004) Historical review: deciphering the genetic code—a personal account. Trends Biochem Sci 29: 46–54 [DOI] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH (1961) The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci USA 47: 1588–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Bar-Ziv R, Libchaber A (2003) Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci USA 100: 12672–12677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A (2004) A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA 101: 17669–17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T (2001) Cell-free translation reconstituted with purified components. Nat Biotechnol 19: 751–755 [DOI] [PubMed] [Google Scholar]
- Simpson ML (2006) Cell-free synthetic biology: a bottom–up approach to discovery by design. Mol Syst Biol 2 69 10.1038/msb4100104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB (1988) A continuous cell-free translation system capable of producing polypeptides in high yield. Science 242: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Stryer L (1995) Biochemistry, 4th edn. New York, NY: WH Freeman and Company [Google Scholar]
- Swartz JR (2001) A PURE approach to constructive biology. Nat Biotechnol 19: 732–733 [DOI] [PubMed] [Google Scholar]
- Swartz JR (2006) Developing cell-free biology for industrial applications. J Ind Microbiol Biotechnol 33: 476–485 [DOI] [PubMed] [Google Scholar]
- Swartz JR, Jewett MC, Woodrow KA (2004) Cell-free protein synthesis with prokaryotic combined transcription–translation. Methods Mol Biol 267: 169–182 [DOI] [PubMed] [Google Scholar]
- Tappel AL (1960) Inhibition of electron transport by antimycin A, alkyl hydroxy naphthoquinones and metal coordination compounds. Biochem Pharmacol 3: 289–296 [DOI] [PubMed] [Google Scholar]
- Underwood KA, Swartz JR, Puglisi JD (2005) Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol Bioeng 91: 425–435 [DOI] [PubMed] [Google Scholar]
- Werpy T, Petersen G (2004) Top Value Added Chemical from Biomass, Volume I: Results of Screening for Potential Candidate from Sugars and Synthesis Gas. August Report by the Pacific Northwest National Laboratory (PNNL) and the National Renewable Energy Laboratory (NREL). US Department of Energy. http://www.nrel.gov/docs/fy04osti/35523.pdf
- Wuu JJ, Swartz JR (2008) High yield cell-free production of integral membrane proteins without refolding or detergents. Biochim Biophys Acta 1778: 1237–1250 [DOI] [PubMed] [Google Scholar]
- Voloshin AM, Swartz JR (2007) Large-scale batch reactions for cell-free protein synthesis. In Cell-free Protein Synthesis: Methods and Protocols, Spirin AS, Swartz JR (eds), pp 207–235. Weinheim, Germany: Wiley-VCH [Google Scholar]
- Xie XS, Yu J, Yang WY (2006) Living cells as test tubes. Science 312: 228–230 [DOI] [PubMed] [Google Scholar]
- Yang J, Kanter G, Voloshin A, Michel-Reydellet N, Velkeen H, Levy R, Swartz JR (2005) Rapid expression of vaccine proteins for B-cell lymphoma in a cell-free system. Biotechnol Bioeng 89: 503–511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information