Abstract
Small non-coding RNAs (sRNAs) have important functions as genetic regulators in prokaryotes. sRNAs act post-transcriptionally through complementary pairing with target mRNAs to regulate protein expression. We use a quantitative approach to compare and contrast sRNAs with conventional transcription factors (TFs) to better understand the advantages of each form of regulation. In particular, we calculate the steady-state behavior, noise properties, frequency-dependent gain (amplification), and dynamical response to large input signals of both forms of regulation. Although the mean steady-state behavior of sRNA-regulated proteins exhibits a distinctive tunable threshold linear behavior, our analysis shows that transcriptional bursting leads to significantly higher intrinsic noise in sRNA-based regulation than in TF-based regulation in a large range of expression levels and limits the ability of sRNAs to perform quantitative signaling. Nonetheless, we find that sRNAs are better than TFs at filtering noise in input signals. Additionally, we find that sRNAs allow cells to respond rapidly to large changes in input signals. These features suggest a ‘niche' for sRNAs in allowing cells to transition quickly yet reliably between distinct states. This functional niche is consistent with the widespread appearance of sRNAs in stress response and quasi-developmental networks in prokaryotes.
Keywords: biophysics, genetic networks, signal processing, small RNA
Introduction
It is now clear that small non-coding RNAs (sRNAs) have a crucial function in prokaryotic gene regulation as both positive and negative regulators. sRNAs are involved in many biological functions, including quorum sensing (Fuqua et al, 2001; Lenz et al, 2004), stress response and virulence factor regulation (Gottesman, 2004; Storz et al, 2004, 2005; Majdalani et al, 2005; Gottesman et al, 2006), and the regulation of outer membrane proteins (Guillier et al, 2006; Vogel and Papenfort, 2006). One major class of prokaryotic sRNAs (antisense sRNAs) negatively regulates proteins by destabilizing the target protein's mRNA (Figure 1). These ∼100 bp antisense sRNAs prevent translation by binding to the target mRNAs in a process mediated by the RNA chaperone Hfq (Gottesman, 2004; Lenz et al, 2004). On binding, both the mRNAs and sRNAs are degraded (Gottesman, 2004), suggesting that prokaryotic sRNAs—unlike their eukaryotic counterparts—act stoichiometrically on their targets. Other antisense sRNAs positively regulate protein expression by promoting ribosome binding to target mRNAs, also in a stoichiometric manner (Gottesman, 2004).
Figure 1.
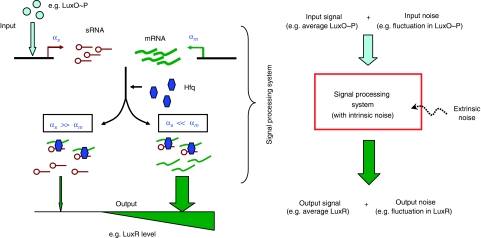
Genetic regulation through sRNAs. Left: small non-coding RNAs (sRNAs) regulate protein expression as part of a larger genetic network with a specific biological task (e.g. quorum sensing in Vibrio bacteria; Lenz et al, 2004). The sRNAs (stem loops) regulate target proteins by destabilizing target protein mRNAs (wavy lines), a stoichiometric process mediated by the RNA chaperone Hfq (hexagons). When the rate of sRNA transcription αs greatly exceeds the rate of mRNA transcription αm, i.e. when, αs≫αm, nearly all the mRNAs are bound by sRNAs and cannot be translated. By contrast, when αm≫αs, there are many more mRNAs than sRNAs, and protein is highly produced. Right: the stochasticity (randomness) of cellular processes results in noise—statistical fluctuations in the molecular numbers. It is helpful to classify the total noise in the output (output noise) into (i) input noise—noise in the input signal from upstream components in the gene circuit, (ii) intrinsic noise—noise from stochasticity inherent in gene regulation through sRNAs, and (iii) extrinsic noise—all other sources of noise impinging on the signal processing system.
Although transcription factor (TF)-based regulation is ubiquitous in prokaryotic gene circuits (Ptashne and Gann, 2001), thus far sRNAs have largely been found in circuits responding to strong environmental cues (e.g. extreme nutrient limitation). This leads to a natural question: are transcriptional regulation by TFs and post-transcriptional regulation by sRNAs distinctly well suited for different biological tasks?
To address this question, we report a quantitative comparison of the signaling properties of TF- and sRNA-based gene regulation. In general, a signaling system can be characterized by how it processes different types of inputs. We therefore treat TF- and sRNA-based regulation as signal processing systems with an input signal—the average concentration of the TFs controlling RNA transcription rates—and an output signal—the average level of the regulated protein (Ptashne and Gann, 2001)—and calculate engineering properties of the system such as the steady-state behavior, noise properties, frequency-dependent gain (amplification), and dynamical response to large input signals (Detwiler et al, 2000) (see Figure 1).
Results
Here, we focus on the case where sRNAs negatively regulate a target mRNA. Positive regulation by sRNAs is discussed in the Supplementary information. Post-transcriptional regulation through sRNAs is modeled using mass action equations with three molecular species: the number of sRNA molecules s, the number of target mRNA molecules m, and the number of regulated protein molecules p (Elf et al, 2003; Lenz et al, 2004; Levine et al, 2007; Mitarai et al, 2007; Shimoni et al, 2007). The effect of intrinsic noise is modeled by Langevin terms, 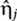 , that describe the statistical fluctuations in the underlying biochemical reactions (van Kampen, 1981). The kinetics of the various species are described by the differential equations
, that describe the statistical fluctuations in the underlying biochemical reactions (van Kampen, 1981). The kinetics of the various species are described by the differential equations
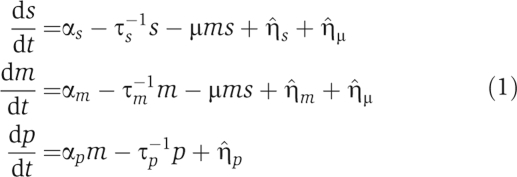 |
The terms can be interpreted as follows. sRNAs (mRNAs) are transcribed at a rate αs (αm), and are degraded at a rate τs−1 (τm−1). Additionally, both sRNAs and mRNAs are stoichiometrically degraded by pairing through Hfq at a rate that depends on the sRNA–mRNA interaction strength μ. Proteins are translated from mRNAs at a rate αp and are degraded at a rate τp−1.
The Langevin terms, 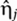 , model intrinsic noise by treating the birth and death processes of the various species in equation (1) as independent Poisson processes (van Kampen, 1981).
, model intrinsic noise by treating the birth and death processes of the various species in equation (1) as independent Poisson processes (van Kampen, 1981). 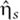 ,
, 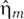 , and
, and 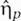 model the noise in the creation and degradation of individual sRNAs, mRNAs, and the regulated protein, respectively.
model the noise in the creation and degradation of individual sRNAs, mRNAs, and the regulated protein, respectively.  models sRNA–mRNA mutual degradation noise. The Langevin terms are characterized within the linear noise approximation by two-point time correlation functions (j=s, m, p, μ), which for steady states take the form
models sRNA–mRNA mutual degradation noise. The Langevin terms are characterized within the linear noise approximation by two-point time correlation functions (j=s, m, p, μ), which for steady states take the form
with σs2=αs+τs−1s̄, σm2=αm+τk−1m̄, σp2=2τp−1p̄, and σμ2=μm̄s̄ where s̄, m̄ and p̄ denote the mean number of sRNA, mRNA, and protein molecules, respectively. It is noted that we have separated the noise due to RNA production and degradation,  and
and 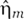 , from the noise due to the binary reaction between mRNAs and sRNAs, et̂aμ. This allows us to write equation (2) in terms of four independent Langevin terms while still capturing the cross-correlation between sRNAs and mRNAs.
, from the noise due to the binary reaction between mRNAs and sRNAs, et̂aμ. This allows us to write equation (2) in terms of four independent Langevin terms while still capturing the cross-correlation between sRNAs and mRNAs.
Recent evidence suggests that prokaryotic transcription may occur with RNA molecules being made in short intense bursts (Golding et al, 2005). The effects of transcriptional bursting can be incorporated into our model by allowing two states of gene activation, as reviewed below (for a detailed discussion, see Paulsson, 2005). Specifically, genes can be in a transcriptionally inactive ‘off' state or in a transcriptionally active ‘on' state. The average transcription rate of RNA, αj (j=m, s) in equation (1), is then related to the probability of the relevant gene being on, gjon, by
with αjon being the mean transcription rate of the relevant RNA when the gene is always on. We model the dynamics of a repressor-controlled gene using the equation
where k− and k+ are the unbinding and binding rates of the repressor and 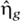 is a Langevin noise term. At steady state, it follows from the fluctuation dissipation theorem that
is a Langevin noise term. At steady state, it follows from the fluctuation dissipation theorem that 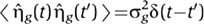 with σg2=2k+gon (Bialek and Setayeshgar, 2005). Thus, a full model that includes transcriptional bursting is described by equation (1) in conjunction with equations (3) and (4).
with σg2=2k+gon (Bialek and Setayeshgar, 2005). Thus, a full model that includes transcriptional bursting is described by equation (1) in conjunction with equations (3) and (4).
For completeness, we also briefly review the equations describing transcriptional regulation (Thattai and van Oudenaarden, 2001; Elowitz et al, 2002; Swain et al, 2002; Paulsson, 2004). The kinetics of transcription regulation is modeled using the Langevin equations
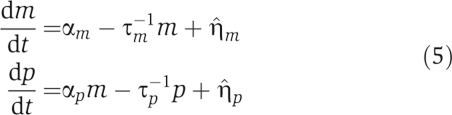 |
with m the number of mRNA molecules, p the number of proteins, αm the average rate of transcription, αp the average rate of translation, and τm−1 and τp−1 the first-order degradation rates of mRNA molecules and proteins, respectively. The two Langevin terms,  and
and  , model noise in the synthesis and degradation of the mRNA and protein, respectively (see Supplementary information) and obey the equations (j=m, p)
, model noise in the synthesis and degradation of the mRNA and protein, respectively (see Supplementary information) and obey the equations (j=m, p)
The effects of transcriptional bursting can also be included in this model using equations (3) and (4).
Mean steady-state protein number
The mean steady-state protein number for regulation through sRNAs can be approximated by ignoring the Langevin terms and setting the time derivatives to zero in equation (1) (see Supplementary information; Paulsson, 2004; Levine et al, 2007). The mean as calculated within this mean-field approximation may differ from the actual mean especially where noise is large. Nonetheless, the qualitative steady-state behavior of the mean can be understood within this approximation.
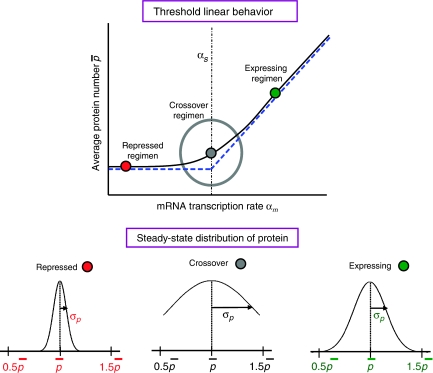
As shown in Levine et al (2007) and Elf et al (2005), the mean protein number exhibits a threshold linear behavior as a function of the mRNA transcription rate αm, with the threshold at αs (see Figure 2). This behavior should be contrasted with transcriptional regulation through TFs for which the mean protein number is a linear function of αm (Thattai and van Oudenaarden, 2001; Elowitz et al, 2002; Swain et al, 2002; Paulsson, 2004). For sRNA-based regulation, the mean steady-state protein number depends on RNA transcription rates only through the difference αm−αs, and this dependence can be characterized by three distinct regimens: repressed αs≫αm, expressing αs≪αm, and a crossover regimen αs≈αm. Increasing the sRNA–mRNA interaction strength μ results in a sharper crossover between the repressed and expressing regimens. The dashed line in Figure 2 depicts the μ → ∞ threshold linear behavior.
Figure 2.
Steady-state behavior for gene regulation through sRNAs. For the regulated protein, the steady-state mean number p̄ exhibits an approximately threshold linear behavior as a function of the mRNA transcription rate αm. The threshold is set by the sRNA transcription rate αs. Protein expression can be classified into three regimens: repressed (αs≫αm), crossover (αs≈αm), and expressing (αs≪αm). In the repressed regimen, the average protein number is low. By contrast, the protein number increases almost linearly with αm in the expressing regimen. The typical behavior of the noise αp, the standard deviation of the protein number, is shown for the three regimens.
In the repressed regimen, on average, there are many more sRNAs transcribed than mRNAs. Consequently, almost all free mRNAs are quickly bound by sRNAs and degraded. This results in low levels of expression of the regulated protein. By contrast, in the expressing regimen, the average number of mRNAs greatly exceeds the number of sRNAs. The sRNAs degrade only a small fraction of the total mRNA population so mRNAs accumulate and are translated into proteins.
Signal transduction
To compare the signal-transduction properties of sRNA-based regulation with TF-based regulation, we consider the two regulation schemes as signal processing systems. Figure 1 depicts how sRNA-based regulation, e.g. in quorum sensing, can be viewed as a signal processing system (see also Supplementary information; Figure 3). In the context of quorum sensing, the input signal is the time-averaged number of phosphorylated LuxO (LuxO∼P) molecules in the cell, which, after a series of intermediate biochemical reactions, is converted into the output signal, the average number of LuxR molecules. Fluctuations in LuxO∼P and LuxR about their averages can be thought of as the input and output noise, respectively. The noise in the output is a combination of input noise (fluctuations in the input signal), intrinsic noise (stochasticity inherent in gene regulation), and extrinsic noise (other sources of noise impinging on the signal processing system not explicitly considered in the model, such as ribosome and RNA polymerase fluctuations).
Figure 3.
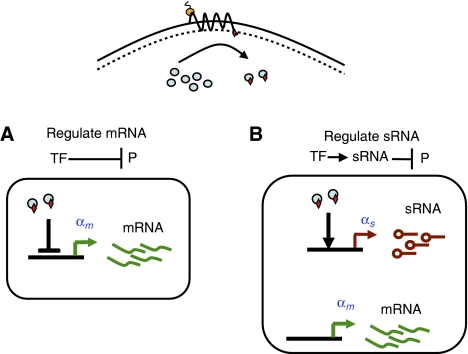
Schematic drawing showing our comparison of transcriptional and post-transcriptional sRNA-mediated regulation. We take as the input signal to both systems a protein regulator (blue discs) that either directly transcriptionally regulates the relevant gene by acting as a repressor or transcriptionally regulates an sRNA acting as an activator. The protein regulator is chosen to have identical kinetic properties in both cases.
The fidelity of a signaling system is ultimately limited by the output noise of the system. The output noise, defined as the ratio of the variance in the output protein number to the square of the mean output protein number, can be thought of as the square of the ‘percentage error' in the output. The higher the output noise, the poorer the signaling fidelity of a gene regulation scheme. Thus, examining the noise properties of sRNA-based and transcription factor gene regulation is important for comparing these two forms of gene regulation.
Gene regulation takes place as part of a larger genetic and biomolecular network, the purpose of which is to convert a measured signal into a concentration of the regulated protein. A simple but important observation is that sRNA-based regulation also requires protein regulators to couple to external signals. In particular, a protein regulator is necessary to vary the transcription rate of the sRNAs in response to an input. For this reason, we take as the input signal to both systems a protein that either transcriptionally regulates the relevant protein directly or else transcriptionally regulates the sRNAs. In the case of direct transcriptional regulation, the protein regulator acts as a repressor, whereas for post-transcriptional, sRNA-based regulation, it acts as an activator (see Figure 3). Furthermore, the kinetics of the protein regulator are chosen to be identical in both cases. The upstream components of the network that controls the level of the relevant protein regulator are also assumed to be identical. This allows for a principled comparison of the two regulatory schemes.
Intrinsic noise
Gene regulation is intrinsically noisy. In this paper, we define intrinsic noise as the fluctuations in the output protein number, given a fixed steady-state input, due to the stochastic nature of the underlying biochemical reactions. When calculating intrinsic noise, we neglect the contributions to output noise from fluctuations in the input and from extrinsic noise sources such as variations in the number of ribosomes and RNA polymerase molecules (see Figure 1).
We start by summarizing the noise properties of transcriptional regulation. For ordinary transcriptional regulation by a repressor, the intrinsic noise—defined as the variance in protein number divided by the mean protein number squared, σp2/p̄2, is given by (Thattai and van Oudenaarden, 2001; Elowitz et al, 2002; Swain et al, 2002; Paulsson, 2004, 2005; Golding et al, 2005; Supplementary information):
 |
where b=αpτm is the protein burst size (the average number of proteins made from an mRNA molecule) and pmax=αmαpτmτp is the mean protein level in the absence of repressor. The first term in equation (7) captures the noise due to translational bursting (the protein burst from each mRNA due to the translation of multiple proteins from each mRNA molecule) and the second captures the noise due to transcriptional bursting (the RNA burst while no repressor is bound). The transcriptional bursting contribution is typically much smaller than that of translational bursting as the unbinding rate of the repressor is generally much faster than the protein degradation rate, k−τp≫1. Consequently, the intrinsic noise for protein-based regulation is often approximated as σp2/p̄2≈(1+b)/p̄.
The intrinsic noise of an sRNA-regulated protein differs significantly from that of a transcriptionally regulated protein. Noise in stoichiometrically coupled systems such as sRNA-based gene regulation has been studied earlier (Paulsson and Ehrenberg, 2001; Elf and Ehrenberg, 2003; Elf et al, 2003). It was found by Elf et al (2005) that the ultrasensitivity of stoichiometric systems in the crossover regimen necessarily gives rise to enhanced stochastic fluctuations. This ‘near-critical' behavior was related to the behavior at phase transitions where fluctuations also diverge (McNeil and Walls, 1974). We have extended these previous analyses to the context of gene regulation by sRNAs, and have calculated the intrinsic protein noise within the linear noise approximation (see Supplementary information; van Kampen, 1981; Elf and Ehrenberg, 2003), including the effects of transcriptional and translational bursting. We have checked our results using exact stochastic simulations (see Supplementary information; Supplementary Figures 2 and 3). The simulations confirm the existence of three regimens and verify that noise is enhanced in the crossover region due to critical fluctuations.
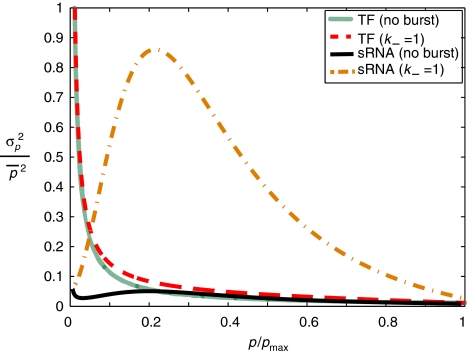
The full expressions for the intrinsic noise are lengthy and in the main text we present only our major findings. Figures 4 and 5 show typical intrinsic noise profiles as functions of the transcription rate ratio, αs/αm, and of the average protein level of the regulated protein, for various magnitudes of transcriptional bursting. For a given sRNA–mRNA interaction strength μ, the intrinsic noise increases with larger transcriptional bursts (smaller k−). Furthermore, for a fixed k−, the intrinsic noise increases with increasing sRNA–mRNA interaction strength μ, (see Supplementary Figure 1; Elf and Ehrenberg, 2003). The intrinsic noise is small in the repressed regimen αs≫αm, and shows a pronounced peak in the crossover region, αs≈αm (see Figure 4) as expected for a stoichiometric system. We have also obtained simplified, asymptotic expressions for the noise in the repressing and expressing regimens when τm≪τp, and there is no transcriptional bursting (see Supplementary information). The expressions for the intrinsic noise in the repressing and expressing regimens are given by, respectively:
Figure 4.
Protein noise with or without transcriptional bursting. Noise in protein expression σp2/p̄2 (variance divided by mean squared) as a function of the ratio of the sRNA and mRNA transcription rates, αs/αm, for different levels of transcriptional bursting. We have assumed that both the sRNAs and mRNAs are produced in bursts. The noise peaks in the crossover regimen, αs≈αm. A slower unbinding rate k− for the repressor proteins controlling sRNA and mRNA expression results in larger transcriptional bursts. Parameters are (in min−1): αm=3, αmon=10, αson=30, τm=10, τs=30, μ=0.02, αp=4, τp=30, and k+ is adjusted to set the mean protein levels (for a discussion of parameter dependence, see Supplementary information).
Figure 5.
Comparison of analytic expressions for the intrinsic protein noise for TF- and sRNA-based regulation. The intrinsic noise for sRNA-based regulation as a function of normalized average protein concentration, p̄/pmax, with and without transcriptional bursting, is shown. All parameters as in Figure 4.
(where pmax=αpτmαmτp and beff=b(p̄/pmax)≪b is the new ‘effective' protein burst size (see Supplementary information; Levine et al, 2007), and
We have written these expressions so that the contribution of sRNA–mRNA mutual degradation noise is contained entirely in the second term of equations (8) and (9).
Comparing the intrinsic noise of protein- and sRNA-based regulators in Figure 5, we observe that sRNA regulators are significantly less noisy than TFs in the repressed regimen. The dominant source of intrinsic noise for a TF-regulated protein, in the limit τm≪τp, is that proteins are made in bursts of average size b≫1. For an sRNA-regulated protein, the average size of a protein burst, beff, is much smaller (see equation (8)). This can be understood by noting that there are many more sRNAs than mRNAs in the repressed regimen, and therefore any free mRNA is quickly bound by an sRNA and degraded. This leads to a reduction in the effective mRNA lifetime and consequently a reduced beff (Levine et al, 2007). The reduction in effective mRNA lifetimes and intrinsic noise takes place even when mRNAs and sRNAs are produced in bursts.
The fidelity of a signaling system can be characterized by the output noise (σptotal)2/p̄2. In general, high-fidelity signaling requires (σptotal)2/p̄2≪1. Thus, from Figure 5 it is clear that over a large range of output protein levels, the large intrinsic noise due to transcriptional bursting makes it difficult for sRNAs to perform high-fidelity signaling.
One of the most striking features of Figure 5 is that sRNA-based regulation is much more sensitive to transcriptional bursting than protein-based regulation. For sRNAs, transcriptional bursting greatly enhances the near-critical fluctuations because the production of RNAs in bursts increases the anticorrelated sRNA–mRNA fluctuations in the crossover regimen (see Elf et al, 2003; Elf and Ehrenberg, 2003 for more details on the near-critical fluctuations). In contrast, for transcriptional regulation directly by a TF, the contribution of transcriptional bursting to the intrinsic noise is relatively small for most choices of parameters (see Figure 5). As recent experiments suggest that prokaryotic transcription may generically produce RNAs in bursts (Golding et al, 2005), this is likely to be a physiologically relevant effect for sRNA-based gene regulation.
The large intrinsic noise in the crossover regimen, αs≈αm can be understood by considering the special case αs=αm for very strong sRNA–mRNA binding, μ → ∞. In this limit, sRNAs and mRNAs, transcribed at the same average rate, quickly bind to each other and degrade and almost no protein is made. However, once in a while there is a fluctuation that produces more mRNAs than average. In this case, unless there is a corresponding fluctuation in sRNAs, the mRNAs cannot be degraded by sRNA–mRNA binding. The mRNAs produced in such a fluctuation will degrade by the usual slow degradation rate τm−1 resulting in a large burst of protein production, contributing to the large intrinsic noise. Transcriptional bursting further increases the magnitude of the aforementioned sRNA and mRNA fluctuations and consequently further increases the intrinsic noise in the crossover regimen.
Gain and filtering
We now consider, in the absence of noise, the change in output protein number about some steady state or ‘operating point' in response to a small, time-varying input signal. A small time-varying change from the steady-state value of the number of proteins controlling the sRNA transcription rate, δc(t)=c(t)−c̄, results in a corresponding time-varying change of the output protein number from its steady-state value, δp(t)=p(t)−p̄. For small enough signals, the dynamics are captured by linearized versions of the mass action equations (equation (1)) (see Supplementary information). In the frequency domain, the relationship between the output protein response at frequency ω and the input signal at frequency ω takes the simple form
where the frequency-dependent gain is given by
 |
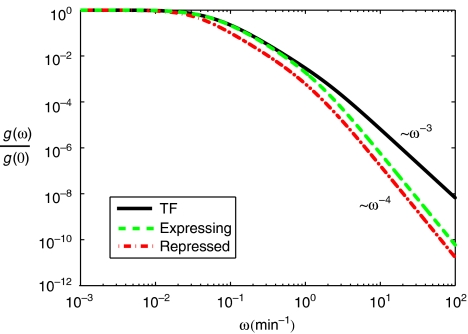
with τg=k−+k+ the characteristic time the sRNA gene is ‘on' and τ± two times related to—and of the same order of magnitude as––the mRNA and sRNA lifetimes (see Supplementary information for exact definition of τ±). Each term of the form (iω+τ−1)−1 can be interpreted as a low-pass filter with a cutoff frequency τ−1. The four low-pass filters in the frequency-dependent gain come from different intermediate steps: I from the binding–unbinding of the protein regulator (activator), II from the transcription of RNAs and the sRNA–mRNA interaction, and III from the translation of mRNAs into proteins. The amplitude of the frequency-dependent gain decreases rapidly ∝ω−4 at high frequencies. This can be compared with the gain in TF-based regulation, which has only three low-pass filters and falls of at high frequencies ∝ω−3 (see Figure 6; Supplementary information). Thus, we conclude that sRNA-based regulation is less sensitive to high-frequency input noise than TF-based regulation.
Figure 6.
Normalized frequency-dependent gain, g(ω)/g(0), as a function of the frequency, ω, for a small input signal for TF- and sRNA-based regulation in the repressed and expressing regimens. The amplitude of the frequency-dependent gain decreases rapidly ∝ω−4 at high frequencies for sRNAs compared with ∝ω−3 for TFs. Consequently, sRNA-based regulation is less sensitive to high-frequency input noise than TF-based regulation. Parameters as in Figure 4.
The underlying reason for the enhanced noise filtering properties of sRNAs is that sRNA-based regulation involves an additional step when compared with transcriptional regulation. Namely, the input signal from upstream components in the genetic network is transmitted to the mRNAs encoding the output protein through sRNAs, which corresponds to an additional noise filter. This extra filtering could also be achieved by introducing an additional layer of transcriptional regulation in the genetic network. However, adding an extra layer of transcriptional regulation also leads to a slower kinetic response of the signaling network to changes in the input signal because an additional protein regulator must be synthesized or degraded to transmit signals. This kinetic cost is much smaller for sRNA-based regulation (see below). Consequently, sRNA-based regulation allows for an extra layer of noise filtering without sacrificing the ability to respond quickly to changes in input.
The above results hold only when the input signal is coupled to the sRNAs. Small input signals can also modulate the transcription of the protein-coding mRNAs instead of the sRNAs. In this case, at high frequencies, the gain falls off as ∝ω−3 similar to TF-based regulation, as the input signal does not pass through the sRNAs (see Supplementary information). Thus, coupling the input signal to sRNAs instead of mRNAs is necessary to achieve the advantageous high-frequency filtering properties of sRNA-based gene regulation. This may explain why input signals are often found coupled to the sRNAs rather than to the mRNAs in sRNA-based regulatory circuits.
Fidelity of small signal response
Intrinsic noise limits the ability of a signaling system to faithfully respond to small signals. Typically, the ability of a system to transduce small signals is quantified by its gain (amplification factor) (Detwiler et al, 2000; Elf and Ehrenberg, 2003; Elf et al, 2003). A large gain is interpreted to mean the system can differentiate small changes in the input signal. However, even if the gain is large, if there is also high intrinsic noise—as is the case in sRNA-based regulation—it may be impossible to distinguish the output signal from the output noise (see Detwiler et al, 2000; Supplementary information). Furthermore, the gain often depends on how input and output signals are defined (e.g. logarithmic gain versus linear gain). For this reason, we consider an alternative measure to compare the small signal responses of sRNA- and protein-based regulators, namely the minimal signal that can be faithfully transmitted by the system (Detwiler et al, 2000).
As discussed above, the noise in the output protein limits the detection of small input signals. For an input signal to be detectable, the corresponding output signal must be greater than the output noise (Detwiler et al, 2000). In particular, the power of the output signal must be greater than the power of the output noise. Consider a periodic input signal at a frequency ω0 and amplitude δcω0, δcω0, δcω0eiω0t. For small input signals, the output signal is related to the input signal by the frequency-dependent gain g(ω). Thus, the output signal is O(t)=g(ω0)δcω0eiω0t and the power of the output signal is by definition
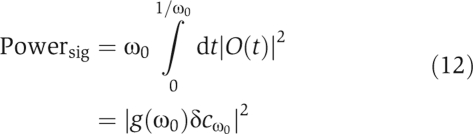 |
On the other hand, the power of the output noise is calculated by integrating fluctuations over all frequencies, and is given within the linear noise approximation by the expression
where δp(ω′) is just the fluctuation in the output protein level at a frequency ω due to intrinsic noise as calculated in the Supplementary information. For a signal to be detectable, we must have
 |
For a step input signal with amplitude δco (ω0 → 0 in the above expressions), the requirement that the output signal is larger than the noise sets a lower bound on the detectable input signal δcomin⩾σp/go (Detwiler et al, 2000). Of course, by time-averaging the output, one can reduce the output noise and hence detect smaller signals, but this does not affect our comparison. Therefore, we computed the minimum input signal without time-averaging for both sRNA- and TF-based regulation and found that, for even moderate amounts of transcriptional bursting, protein regulators are better than sRNAs at responding to small signals across the whole range of output protein levels. At low protein levels (repressed regimen), the minimum detectable signal for sRNA-based regulation is larger due to the lower gain for sRNA-based regulation than for TF-based regulation. At intermediate to high levels of output protein (crossover and expressing regimens), the minimum detectable signal for sRNAs is also larger due to the large protein noise σp2 arising from transcriptional bursting for sRNA-based regulation.
Consequently, contrary to previous speculations (Levine et al, 2007), results indicate that sRNA-based regulation is unlikely to be useful for amplifying small signals despite the large gain of sRNA-based regulation in the crossover region. Our results also imply that it is more advantageous to use TF-based regulation than sRNA-based regulation in genetic networks designed to respond to small changes in upstream components.
Large signal response
In nature, an organism may benefit from switching quickly between two different gene expression states in response to a large persistent input signal. We have compared here the rates at which a regulated protein can switch between ‘off' and ‘on' states in response to an input signal when its mRNA is directly regulated by a TF or indirectly regulated by an sRNA.
Figure 7 shows the time evolution of the average mRNA level for both sRNA- and TF-based regulation in response to a step change in the input. The response for sRNA-based regulation depends on the initial conditions, and can be tuned by changing the location in which the system is initially located in the repressed regimen. In particular, the effective mRNA degradation (and dilution) rate depends on the sRNA pool size and on the sRNA–mRNA interaction strength μ. However, our conclusions do not strongly depend on the choice of parameters (see Supplementary information).
Figure 7.
Large signal switching. Normalized mRNA level m/mmax, as a function of time, in response to step changes in the input, for both the sRNA- and TF-based regulation. Switching from high mRNA level (on state) to low mRNA level (off state) and vice versa. Switching from off to on state has a lag in the case of sRNA-based regulation, whereas the switching time from the on to off state for sRNAs is faster or comparable to that for TFs, depending on the choice of kinetic parameters. For sRNA-based regulation, αm=3.5 and αs goes from ∼0.35 to 4.5 for switching from low to high and vice versa for high to low. For TF-based switching, αm is such that both schemes have same steady states. Other parameters as in Figure 4.
We find that using sRNAs to switch protein expression on, i.e. going from low output protein number to high output protein number, is slower than direct TF regulation. This slower response is due to the sRNA pool that needs to be depleted before target mRNAs can accumulate. On the other hand, sRNA-based regulation can be faster than TF-based regulation when switching off expression of a protein—the large input signal rapidly increases the concentration of sRNAs resulting in fast degradation of target mRNAs (see Figure 7; Shimoni et al, 2007). The slower response of the sRNA-based regulation at turning on protein expression stems from the delay introduced by having an additional layer of sRNA regulation in the signal-transduction pathway when compared with protein-based regulation (see Figure 7). However, this delay is much smaller than that which would be introduced by having an additional layer of transcriptional regulation as the synthesis and degradation rates of proteins are much slower than those of RNAs (see Supplementary information for a discussion comparing our results with Shimoni et al, 2007).
Thus far, we have considered the case where a protein is negatively regulated by sRNAs. However, a protein can also be positively regulated by sRNAs (see Storz et al, 2004; Hammer and Bassler, 2007; Supplementary information), and in this case switching protein expression on using sRNAs can be faster than TF-based regulation. Typically, sRNAs positively regulate protein expression by preventing the formation of inhibitory secondary structures that occlude the ribosome-binding sites of the regulated mRNA. As there is generally a background pool of translationally inactive target mRNAs, a large input signal that produces sRNAs allows the target mRNAs to be quickly converted into the translationally active form.
Discussion
sRNAs have an important regulatory function in prokaryotic gene circuits. sRNAs are involved in a variety of critical physiological tasks such as quorum sensing, stress response, and the regulation of outer-membrane proteins. Yet sRNAs are not currently thought to be as common as TFs in prokaryotic gene regulatory circuits (at least based on our present knowledge), suggesting sRNAs may be well suited for certain biological tasks but not for others. This paper evaluates the suitability of sRNA-based regulation to particular biological tasks by treating gene regulation as a signal processing system.
Our analysis shows that for a large (intermediate to high) range of output protein levels, the intrinsic noise for sRNA-based regulation is much larger than for TF-based regulation. However, even at a high level of transcriptional bursting, sRNA-based regulation is less noisy than TF-based regulation at low protein levels (in the repressed regimen) because a large pool of sRNAs shortens the effective mRNA lifetime and buffers against target mRNA fluctuations. Thus, in all cases, protein expression can be kept off much more reliably by sRNAs than by TFs (see Supplementary information for a discussion of the dependence on kinetic parameters). We also find (when the input signal is coupled to the sRNAs) that sRNAs are better filters of high-frequency input noise than TFs as they implement an extra low-pass filter when compared with TFs. This is likely to be physiologically relevant as sRNAs are often found in networks that couple to external signals (Majdalani et al, 2005). In such networks, high-frequency noise in the input could arise from noise in external concentrations or from the fast upstream protein modification reactions such as phosphorylation–dephosphorylation of a two-component system. sRNAs also allow cells to respond quickly to large changes in input signal. In particular, sRNAs can quickly turn off negatively regulated genes and quickly turn on positively regulated genes (Shimoni et al, 2007). This ability to filter high-frequency noise without compromising the ability to rapidly respond to input signals is a defining feature of sRNAs. The above characteristics make sRNA-based regulation useful for constructing genetic switches. In contrast, even for moderate levels of transcriptional bursting, sRNA-based regulatory circuits are worse than TFs at transducing small input signals, suggesting that TFs are likely better suited for quantitative adjustment of protein expression. Additionally, the use of sRNAs in more complex network motifs such as feed-forward loops is likely to give rise to new behaviors (Shimoni et al, 2007). Our results are summarized in Table I.
Table 1.
Summary of the advantages and disadvantages of sRNAs when compared with transcriptional protein regulators (transcription factors)
| Advantages of sRNAs | Advantages of protein regulators |
|---|---|
| • sRNAs are better than protein regulators at keeping proteins ‘off' because a large pool of sRNAs shortens the effective mRNA lifetime and buffers against target mRNA fluctuations | • The intrinsic noise for sRNA-based regulation is much larger than that for transcriptional regulation in a large (intermediate to high) range of expression levels of the regulated protein, especially in the presence of transcriptional bursting |
| • sRNAs can filter high-frequency noise without compromising the ability to rapidly respond to large changes in input signals | • Protein regulators are better than sRNAs at transducing small input signals |
| → sRNAs likely fill a ‘niche' in allowing cells to transition quickly yet reliably between distinct states | → Protein regulators are likely better suited for quantitative adjustment of protein expression than sRNAs |
Summary comparison of signaling properties of sRNAs and protein regulators.
Indeed, sRNAs are often found in genetic circuits that switch gene expression states in response to strong environmental cues. For example, under iron limitation, the sRNA RyhB rapidly shuts off synthesis of several iron-binding proteins, making iron available for essential proteins (Masse and Gottesman, 2002). In the quorum-sensing network of Vibrio harveyi and of the human pathogen V. cholerae, multiple sRNAs (Qrr1–5) switch the expression state of the cell in response to external cell density (Lenz et al, 2004). The fast dynamical response of sRNA-based regulation, accelerated by the presence of five sRNAs, may allow the pathogen V. cholerae to quickly switch expression states in response to a sudden change in the environment—for example, from a high bacterial cell density in a eukaryotic host to low cell density in the marine environment (Zhu et al, 2002; Hammer and Bassler, 2007). Recent modeling work by Shimoni et al (2007) suggests that the kinetic properties of sRNAs are crucial to understanding the behavior of Escherichia coli regulatory circuits involved in responding to osmotic stress. In both the iron metabolism and quorum-sensing circuits discussed above, the input signals, iron limitation and cell density are coupled to the expression of sRNAs and not to the target mRNAs (Lenz et al, 2004; Masse et al, 2007), suggesting that filtering input noise may also be an important consideration (see Figure 4).
We have considered the case where a single sRNA species regulates a single mRNA species. However, as in the Vibrio quorum-sensing circuit, multiple sRNAs may regulate multiple mRNAs (Repoila et al, 2003; Lenz et al, 2004; Mitarai et al, 2007). Even in such a case, mean steady-state protein numbers are expected to exhibit a threshold linear behavior with three distinct regimens. The main difference from the single sRNA/mRNA case is that the threshold occurs when the combined sRNA transcription rate exceeds the target mRNA transcription rate (Levine et al, 2007; Shimoni et al, 2007). This may allow sRNAs to prioritize usage of different target mRNAs (Levine et al, 2007; Mitarai et al, 2007).
There are additional considerations that may favor sRNA- or TF-based regulation. For example, TFs are likely to be better global regulators than sRNAs—as sRNAs degrade mRNAs stochiometrically, only a limited number of genes can be regulated by a given sRNA. Also, the cost in space on the genome is generally larger for sRNA-based regulation than for direct regulation by TFs because in the former it is necessary to encode for the sRNA in addition to the regulatory region of the regulatory TF coupling the sRNA to external signals (see Figure 3) (Semsey et al, 2006). Additionally, sRNAs and TFs are likely to respond differently to extrinsic noise. For example, one expects sRNA-based regulation to be less sensitive to global RNA polymerase fluctuations than TFs as sRNAs and their target mRNAs are affected identically by polymerase abundance (Paulsson and Ehrenberg, 2001). Finally, the metabolic costs of sRNA- and protein-based regulation may differ (Mitarai et al, 2007).
In this paper, we have considered gene regulation by non-coding RNAs in prokaryotes. Regulatory RNAs are also found in eukaryotes. In eukaryotes, these regulatory RNAs are believed to act catalytically, not stoichiometrically. Nonetheless, our analysis suggests that, even in eukaryotes, regulatory RNAs are better at keeping protein expression off than TFs, as in both cases, regulatory RNAs shorten the effective lifetime of their target mRNAs, thus reducing protein fluctuations. Furthermore, our analysis also suggests that regulatory RNAs in eukaryotes are likely better than TFs at filtering out high-frequency input noise in upstream signals.
Recently, it has been shown that noise in protein expression may exhibit a universal behavior (Bar-Even et al, 2006). However, our analysis for the intrinsic noise of an sRNA-regulated protein differs significantly from the proposed universal behavior in the presence of transcriptional bursting (see also Tkacik et al, 2008). It would be interesting to test our predictions for intrinsic noise experimentally by quantifying intrinsic cell-to-cell variation of a fluorescent protein (Elowitz et al, 2002) alternatively regulated by an sRNA or a TF, particularly with controllable transcriptional bursting (Blake et al, 2006).
The analogy between biochemical circuits and signal processing systems in engineering provides a general framework for characterizing the signal-transduction pathways found in biology (Detwiler et al, 2000). Different biological tasks place different requirements on signal-transduction circuits. For example, in chemotaxis, bacteria must respond quickly to changing input signals (Berg, 2003; Bialek and Setayeshgar, 2005; Keymer et al, 2006), whereas in quorum sensing or stress response, reliability may be more crucial than speed. One suspects that biological networks exhibit a harmony between network architecture and network function. For this reason, understanding the comparative advantages and disadvantages of different architectures is likely to yield new insights into biological function, as well as new schemes for synthetic circuits.
Materials and methods
The analyses were carried out using rate equation models extended to include stochastic fluctuations and our results were tested using Monte Carlo (Gillespie) simulations. The equations account for the concentration of each component in the circuit, and for noise around the means of these components. The dynamics of gene regulation was modeled using Langevin equations for the various species in the system: mRNAs, sRNAs, and proteins. Using this model, we analyzed the signaling properties of the two regulation schemes, focusing on gain, filtering, and switching times in response to large input signals. For further details, see the Supplementary information.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Bonnie Bassler, Matthias Kaschube, Anirvan Sengupta, Gasper Tkacik, Chris Waters, and Kerwyn C Huang for helpful discussions and suggestions on the paper. This study was partially supported by US National Institutes of Health (NIH) Grant PSO GM071508, the Defense Advanced Research Projects Agency (DARPA) under Grant HR0011-05-1-0057, and the Burroughs Wellcome Fund Graduate Training Program.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Berg HC (2003) E. coli in Motion. New York, USA: Springer-Verlag [Google Scholar]
- Bialek W, Setayeshgar S (2005) Physical limits to biochemical signaling. Proc Natl Acad Sci USA 102: 10040–10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ (2006) Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Detwiler PB, Ramanathan S, Sengupta A, Shraiman BI (2000) Engineering aspects of enzymatic signal transduction: photoreceptors in the retina. Biophys J 79: 2801–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Ehrenberg M (2003) Fast evaluation of fluctuations in biochemical networks with the linear noise approximation. Genome Res 13: 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Paulsson J, Berg OG, Ehrenberg M (2003) Near-critical phenomena in intracellular metabolite pools. Biophys J 84: 154–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Paulsson J, Berg OG, Ehernberg M (2005) Mesoscopic kinetics and its applications in protein synthesis. In Topics in Current Genetics: Systems Biology: Definitions and Perspectives, Alberghina L, Westerhoff HV (eds), pp 95–116. Berlin, Germany: Springer-Verlag [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35: 439–468 [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Gottesman S (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 58: 303–328 [DOI] [PubMed] [Google Scholar]
- Gottesman S, McCullen CA, Guiliier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, Fitzgerald PC, Sowa NA, Fitzgerald DJ (2006) Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol 71: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G (2006) Modulating the outer membrane with small RNAs. Genes Dev 20: 2338–2348 [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL (2007) Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 104: 11145–11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer JE, Andres RG, Skoge M, Meir Y, WIngreen NS (2006) Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci USA 103: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82 [DOI] [PubMed] [Google Scholar]
- Levine E, Kuhlman T, Zhang Z, Hwa T (2007) Quantitative characteristics of gene regulation by small RNA. PLoS Biol 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, Gottesman S (2005) Bacterial small RNA regulators. Crit Rev Biochem Mol Biol 40: 93–113 [DOI] [PubMed] [Google Scholar]
- Masse E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Salvail H, Desnoyers G, Arguin M (2007) Small RNAs controlling iron metabolism. Curr Opin Microbiol 10: 140–145 [DOI] [PubMed] [Google Scholar]
- McNeil KJ, Walls DF (1974) Nonequilibrium phase transitions in chemical reactions. J Stat Phys 10: 439–448 [Google Scholar]
- Mitarai N, Andersson MC, Krishna S, Semsey S, Sneppen K (2007) Efficient degradation and expression prioritization with small RNAs. Phys Biol 4: 164–171 [DOI] [PubMed] [Google Scholar]
- Paulsson J (2004) Summing up the noise in gene networks. Nature 427: 415–418 [DOI] [PubMed] [Google Scholar]
- Paulsson J (2005) Models of stochastic gene expression. Phys Life Rev 2: 157–175 [Google Scholar]
- Paulsson J, Ehrenberg M (2001) Noise in a minimal regulatory network: plasmid copy number control. Q Rev Biophys 34: 1–59 [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A (2001) Genes and Signals. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Repoila F, Majdalani N, Gottesman S (2003) Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 48: 855–861 [DOI] [PubMed] [Google Scholar]
- Semsey S, Andersson AMC, Krishna S, Jensen MH, Masse E, Sneppen K (2006) Genetic regulation of fluxes: iron homeostasis of Escherichia coli. Nucleic Acids Res 34: 4960–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H (2007) Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol 3: 138 10.1038/msb4100181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman N (2005) An abundance of RNA regulators. Annu Rev Biochem 74: 199–217 [DOI] [PubMed] [Google Scholar]
- Storz G, Opdyke JA, Zhang A (2004) Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol 7: 140–144 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A (2001) Stochastic gene expression in fluctuating environments. Proc Natl Acad Sci USA 98: 8614–861911438714 [Google Scholar]
- Tkacik G, Gregor T, Bialek W (2008) The role of input noise in transcriptional regulation. PLoS ONE 3: e2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kampen NG (1981) Stochastic Processes in Physics and Chemistry. Amsterdam: North-Holland [Google Scholar]
- Vogel J, Papenfort K (2006) Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9: 605–611 [DOI] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ (2002) Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA 99: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information