Abstract
Systems biology relies on data sets in which the same group of proteins is consistently identified and precisely quantified across multiple samples, a requirement that is only partially achieved by current proteomics approaches. Selected reaction monitoring (SRM)—also called multiple reaction monitoring—is emerging as a technology that ideally complements the discovery capabilities of shotgun strategies by its unique potential for reliable quantification of analytes of low abundance in complex mixtures. In an SRM experiment, a predefined precursor ion and one of its fragments are selected by the two mass filters of a triple quadrupole instrument and monitored over time for precise quantification. A series of transitions (precursor/fragment ion pairs) in combination with the retention time of the targeted peptide can constitute a definitive assay. Typically, a large number of peptides are quantified during a single LC-MS experiment. This tutorial explains the application of SRM for quantitative proteomics, including the selection of proteotypic peptides and the optimization and validation of transitions. Furthermore, normalization and various factors affecting sensitivity and accuracy are discussed.
Keywords: mass spectrometry, MRM, proteomics, quantitative, SRM
Introduction
Biology in general and systems biology in particular increasingly require the detection and quantification of large numbers of analytes. Proteomic studies are commonly performed using a shotgun approach, in which the sample proteins are enzymatically degraded to peptides, which are then analysed by mass spectrometry (MS). Thereby, a subset of the peptides present in the sample is automatically and in part stochastically selected by the mass spectrometer in a process referred to as data-dependent precursor selection. Systems biology requires accurate quantification of a specified set of peptides/proteins across multiple samples derived from cells in differentially perturbed states (Ideker et al, 2001). This stringent requirement is driven by the long-term goal of systems biology to generate mathematical models that simulate the system and make specific predictions about its behaviour under different conditions. Although the comprehensive quantitative analysis of the transcriptome has become routine using microarray technology and other transcript profiling technologies (Katagiri and Glazebrook, 2004), quantitative proteomic analyses to a similar depth and consistency have not been achieved by the current shotgun proteomic approaches. Besides their limited sensitivity, a main shortcoming of these methods is poor reproducibility of target selection, which results in the identification of only partially overlapping sets of proteins from substantially similar samples. Such fragmentary data sets are also unsatisfactory for applications apart from systems biology, in which complete quantification profiles for each of the quantified proteins are required. Therefore, new approaches are needed to deliver precise quantitative data from defined sets of proteins reliably, across multiple samples.
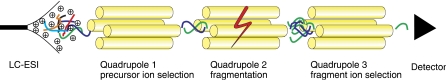
Selected reaction monitoring (SRM) has the potential to overcome, at least in part, the shortcomings of current shotgun proteomic approaches (See Box I for an overview of MS-based quantification methods). SRM exploits the unique capabilities of triple quadrupole (QQQ) MS for quantitative analysis. In SRM, the first and the third quadrupoles act as filters to specifically select predefined m/z values corresponding to the peptide ion and a specific fragment ion of the peptide, whereas the second quadrupole serves as collision cell (Figure 1). Several such transitions (precursor/fragment ion pairs) are monitored over time, yielding a set of chromatographic traces with the retention time and signal intensity for a specific transition as coordinates. The two levels of mass selection with narrow mass windows result in a high selectivity, as co-eluting background ions are filtered out very effectively. Unlike in other MS-based proteomic techniques, no full mass spectra are recorded in QQQ-based SRM analysis. The non-scanning nature of this mode of operation translates into an increased sensitivity by one or two orders of magnitude compared with conventional ‘full scan' techniques. In addition, it results in a linear response over a wide dynamic range up to five orders of magnitude. This enables the detection of low-abundance proteins in highly complex mixtures, which is crucial for systematic quantitative studies.
MS-based techniques for quantitative analyses.
The simplest method to quantify analytes by LC-MS is the use of eXtracted Ion Chromatograms (XIC). Data are collected in full MS scan mode and processed post-acquisition, to reconstruct the elution profile for the ion(s) of interest, with a given m/z value and a tolerance. XIC peak heights or peak areas are used to determine the analyte abundance.
Selected ion monitoring (SIM) is performed on scanning mass spectrometers, by restricting the acquisition mass range around the m/z value of the ion(s) of interest. The narrower the mass range, the more specific the SIM assay. SIM experiments are more sensitive than XICs from full scans because the MS is allowed to dwell for a longer time over a small mass range of interest. Several ions within a given m/z range can be observed without any discrimination and cumulatively quantified; quantification is still performed using ion chromatograms.
Selected reaction monitoring (SRM) is a non-scanning technique, generally performed on triple-quadrupole (QQQ) instruments in which fragmentation is used as a means to increase selectivity. In SRM experiments, two mass analysers are used as static mass filters, to monitor a particular fragment ion of a selected precursor ion. The selectivity resulting from the two filtering stages combined with the high-duty cycle results in quantitative analyses with unmatched sensitivity. The specific pair of m/z values associated with the precursor and fragment ions selected is referred to as a ‘transition' (e.g., 673.5/534.3).
The term SRM or ‘pseudo SRM' is occasionally used also to describe experiments conducted in LITs or QqTOFs instruments where, upon fragmentation of a precursor ion, MS/MS data are acquired on a partial mass range centred on a fragment ion. Although this scan mode resembles an SRM experiment, it is based on the ‘electronic' extraction of the fragment ion signal(s) and can thus be essentially viewed as the SIM of fragment ion(s). The full potential of SRM as described in this review is only tapped when the experiment is performed on QQQ MS.
Figure 1.
SRM/MRM analysis on QQQ MS. Several analytes are coeluting from the chromatographic system. The specific m/z selection in the first quadrupole filters out most coeluting ions. However, owing to identical mass, one interfering ion (blue) remains. In quadrupole 2, the analytes are fragmented. The m/z selection in the third quadrupole filters out all the fragments of the blue analyte and leaves only a particular fragment of the green analyte for specific detection.
Selectivity, sensitivity and dynamic range make SRM on a QQQ MS ideally suited to address a major proteomic challenge in systems biology: the accurate quantification of predefined sets of proteins spanning the whole range of the cellular proteome in a reproducible manner.
Establishing a proteomic SRM experiment
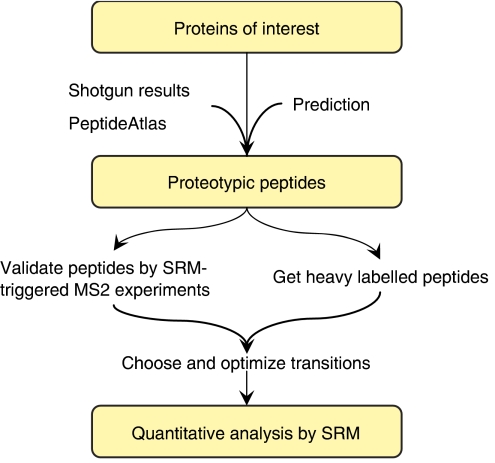
In contrast to conventional shotgun proteomic studies, SRM measurements are quantitative analyses strictly targeting a predetermined set of peptides and depend on specific SRM transitions for each targeted peptide. Previous information is required to define these transitions. Specifically, three types of information are of critical importance. First, the proteins that constitute the targeted protein set have to be selected. Second, for each targeted protein, those peptides that present good MS responses and uniquely identify the targeted protein, or a specific isoform thereof, have to be identified. Such peptides have been termed as proteotypic peptides (PTPs) (Mallick et al, 2007). Third, for each PTP, those fragment ions that provide optimal signal intensity and discriminate the targeted peptide from other species present in the sample have to be identified. These optimized transitions are the essence of an SRM assay. The time and effort required to establish these conditions is the price to pay for the excellent quantitative performance of SRM-based experiments. However, once established, such assays can be used indefinitely in any study that involves the particular targeted protein. The following sections guide the reader through the different steps of an SRM experiment (Figure 2).
Figure 2.
Workflow of SRM-based proteomic experiments.
Selection of a target protein set
The first step of a targeted proteomic experiment is the selection of a set of proteins of interest. Depending on the sensitivity and accuracy required, hundreds and eventually up to 1000 proteins can be targeted in a single LC-MS analysis after the transitions have been optimized (see Box II). The selection of the protein set might be on the basis of previous experiments or the scientific literature. In addition, multiple information resources on the Web can help to reveal potentially relevant proteins, for example, gene expression and protein expression data, protein–protein interaction data or sets of functionally related proteins derived from gene ontology groups or based on the KEGG database. Alternatively, network expansion can be used to complement an initial set of proteins that might be discovered in quantitative proteome screens (see Table I for links and citations).
Quantification of multiple proteins by SRM.
When targeting many proteins requiring numerous transitions in a single experiment, the dwell time of the individual transition is reduced. Therefore, the total number of targeted peptides that are reliably quantifiable within an LC run is limited and depends on the requirements for the limit of detection to be achieved. Practical dwell-time settings range between 10 ms for good sensitivity and 100 ms for excellent sensitivity. To ensure precise LC-MS quantification, at least eight data points should be acquired across the chromatographic elution profile of a peptide. Assuming a peak width of 20 s at 10% peak height, a cycle time of 2 s is required. This translates into 200 transitions of 10 ms dwell time. If every protein should be quantified by 2 peptides with 2 transitions each, 50 proteins can be quantified in total. In scheduled SRM, the transitions for a particular peptide are only acquired in a time window around the expected retention time, significantly increasing the number of peptides/proteins that can be detected and quantified in a single LC-MS experiment. More than 1000 transitions may be quantified with high sensitivity and reproducibility using scheduled SRM.
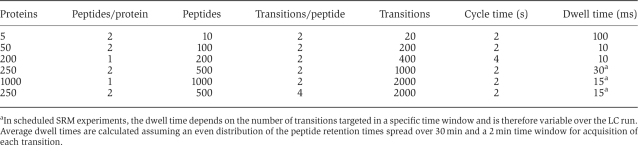
Table 1.
Online information resources relevant to selection of a set of proteins of interest
| Gene expression | GEO | http://www.ncbi.nlm.nih.gov/geo/ | Barrett et al (2007) |
| Protein expression | ProteinAtlas | http://www.proteinatlas.org/ | Uhlen and Ponten (2005) |
| Gene ontology groups | GO | http://www.geneontology.org/ | Ashburner et al (2000) |
| Functional groups | KEGG | http://www.genome.jp/kegg/ | Kanehisa and Goto (2000) |
| Protein–protein interactions | IntAct | http://www.ebi.ac.uk/intact/ | Kerrien et al (2007) |
| Protein–protein interactions | MINT | http://mint.bio.uniroma2.it/ | Chatr-aryamontri et al (2007) |
| Network expansion | PhosphoPep | http://www.phosphopep.org/ | Bodenmiller et al (2007) |
In addition to the proteins of interest, several ‘housekeeping' proteins should be selected as an invariant reference set to correct experimental variability such as uneven total protein amount per sample (see below). As the selection of the target protein set depends on the objective of each study and is therefore subjective, it is not further discussed here.
Peptide selection
Each of the targeted proteins yields tens to hundreds of peptides upon tryptic digestion (Picotti et al, 2007). Typically, only a few representative peptides per protein are targeted to infer the presence of a protein in a sample and to determine its quantity. The careful choice of the targeted peptides is essential for the success of the SRM experiment. Several factors affecting this choice are discussed in the following section. Additional useful empirical rules for the selection of peptides for SRM have been described earlier (Bronstrup, 2004; Kirkpatrick et al, 2005).
MS properties
It has been reported that among the peptides generated by tryptic digestion of a protein, only a (small) subset is routinely observed (Kuster et al, 2005). In principle, all possible peptides could be systematically tested. In practice, the time required for assay development is significantly reduced if previous information is used to select those peptides that are most likely observed in the experiment and provide the strongest specific signals. Previous information derived from the combination of multiple shotgun proteomics experiments can be used in two ways: the direct observation of highly detectable peptides and the inference of rules that predict MS observability. For a growing number of organisms, a significant number of MS experiments has been performed and the data have been deposited in online repositories like the ‘PeptideAtlas', ‘Human Proteinpedia', ‘GPM Proteomics Database' or ‘PRIDE', which support the retrieval of frequently observed peptides for the identified proteins (see Table II for links and citations). The recently released ‘ISPIDER Central' enables querying across several repositories. An intrinsic challenge of repositories is the difficulty to guarantee high data quality. In particular, the compilation of many similar experiments without researching the original data may increase the false discovery rates dramatically and therefore mislead in the selection of peptides.
Table 2.
Online repositories for proteomics data sets
| PeptideAtlas | http://www.peptideatlas.org/ | Desiere et al (2006) |
| Human proteinpedia | http://www.humanproteinpedia.org/ | Mathivanan et al (2008) |
| GPM Proteomics database | http://gpmdb.thegpm.org/ | Craig et al (2004) |
| PRIDE | http://www.ebi.ac.uk/pride/ | Jones et al (2008) |
| ISPIDER central | http://ispider.smith.man.ac.uk/ | Siepen et al (2008) |
The same data resources have also been used to train computational tools that attempt to predict the most likely MS-observable peptides from proteins so far not covered in the databases (Tang et al, 2006; Mallick et al, 2007). In combination, previous detection and predicted observability identify the most promising targets for SRM, even though observability and signal intensity in SRM experiments do not correlate perfectly. The observability in shotgun experiments is largely influenced by the fragmentation characteristics. Indeed, large peptides with many fragments of similar intensities are identified with high confidence. In contrast, more intense SRM signals are obtained from peptides yielding a few predominant fragments. Therefore, shorter peptides and those containing proline residues are better targets for SRM, even though their observability in shotgun experiments might be lower than longer peptides ionized with similar efficiency.
Uniqueness
By selecting peptides for targeted MS analysis, it is essential to ensure that the peptides selected uniquely identify the targeted protein or one isoform thereof. Additionally, it might be important to select peptides that can distinguish different splice isoforms or single nucleotide polymorphisms (SNP's). Both Ensembl (www.ensembl.org/) and NCBI (www.ncbi.nlm.nih.gov/sites/entrez, www.ncbi.nlm.nih.gov/projects/SNP/) databases provide information about splice variants and SNPs. The PeptideAtlas (www.peptideatlas.org/) helps to distinguish between multiple splice isoforms and distinct genes by reporting the number of genome locations for observed peptides and provides an interface to visualize the peptide–protein relationship by cytoscape (Shannon et al, 2003).
Post-translational modifications
Modified peptides cannot be detected by SRM unless specifically targeted. Quantitative differences observed in the analysis of an unmodified peptide reflect either a true change in abundance of the targeted protein in the samples or the (partial) modification of the peptide. Therefore, for reliable quantification, at least two peptides should be monitored for each targeted protein. If two peptides from the same protein reproducibly show divergent regulations, this is indicative of (post-translational) modification or processing of one of the peptides. SRM can also be applied to specifically target and quantify peptides with post-translational modifications like phosphorylation (Unwin et al, 2005; Williamson et al, 2006), ubiquitination (Mollah et al, 2007) or acetylation (Griffiths et al, 2007), provided that transitions for those peptides are established.
Chemically induced modifications
The introduction of artifactual chemical modifications due to sample processing is a potential source of error in quantitative MS experiments because a fraction of the targeted peptide might be converted into the modified form in an irreproducible and unpredictable manner. Therefore, care should be taken to avoid targeting peptides with a high propensity for artifactual modifications. In particular, peptides containing methionine or tryptophan residues should be avoided, as the side chains of these amino acids are prone to oxidation. Furthermore, peptides containing glutamine or asparagine residues may be chemically unstable and convert to glutamate or aspartate. The rate of conversion is dependent on the surrounding sequence, for example, asparagine followed by glycine or proline is particularly prone to deamidation (Piszkiewicz et al, 1970; Geiger and Clarke, 1987). In addition, N-terminal glutamine residues are quickly transformed to pyro-glutamate under acidic conditions.
Cleavage sites
Despite the high specificity and efficiency of trypsin, peptides with missed cleavages or non-tryptic cleavage sites are frequently observed in shotgun analyses (Picotti et al, 2007). Fortunately, for the most part, the ion current of such peptides is relatively low compared with the ion current of the true tryptic peptides. Such peptides should not be targeted for absolute quantification, as the extent of these events might be variable between samples. However, such peptides might be targeted as corroborating evidence for the presence of a protein in cases in which true tryptic PTPs are difficult to detect. In addition, peptides with two neighbouring basic amino acids at either cleavage site of the protein sequence (e.g., KRNGGGR or RNGGGKK) should be avoided, as those sites are predestined for a high rate of missed cleavages.
Selection of SRM transitions
The quantification of a peptide by SRM requires the selection of specific m/z settings for the first and third quadrupole, which, in combination, results in a highly sensitive and selective detection of the peptide (Box III). The combination of m/z setting for the first and third quadrupole is referred to as ‘transition'. The m/z value of the first quadrupole is determined by the mass and the predominant charge state of a peptide. In the third quadrupole, a particular fragment ion of the peptide is selected. The mass of the canonical fragment ions can be easily calculated. However, the intensities of individual fragments derived from one precursor ion differ substantially. To obtain a high-sensitivity assay, it is therefore essential to select transitions specific for the most intense fragments. Including transitions for all the major canonical fragments in the assay limits the analysis to a few peptides, as the total number of transitions per LC-MS run is limited (see Box II). Therefore, commonly only the best 2–4 transitions per peptide are selected for quantitative assays. The selection of these transitions might be either on the basis of data from shotgun experiments or experimentally determined on the QQQ instrument. Shotgun experiments can be exploited to derive information about the predominant precursor charge state and the MS/MS fragmentation pattern of a targeted peptide. However, one needs to be aware that the ionization conditions can affect the charge state distribution. Similarly, the distribution of relative fragment ion intensities is dependent on the type of instrument used and the operating parameters. This is particularly relevant when ion trap derived data are used to select transitions for a QQQ instrument. Owing to the different mode of collision-induced activation in ion traps compared with quadrupole collision cells, higher b-type ions and doubly charged fragments are usually less prominent or absent in the QQQ instrument mass spectra.
Alternatively, the fragment ion masses of the targeted peptide can be calculated and experimentally tested by SRM measurements on a QQQ instrument. This yields the most reliable selection of high-performing transitions. However, if two precursor charge states and several ion series are taken into account, more than 30 transitions per peptide might have to be measured. Therefore, the number of peptides that can be tested in a single LC-MS analysis is limited. However, the number of transitions analysed may be increased by using scheduled SRM (Stahl-Zeng et al, 2007). In such experiments, the transitions of a specific peptide are only acquired during a time window around its elution time. Therefore, using such time constraints, a much higher number of transitions may be tested to derive the best performing ones. To perform scheduled SRM experiments, the scheduling functionality needs to be supported by the instrument, and the retention times of the targeted peptides must be known. The retention times can be derived from previous experiments or they might be predicted by tools like SSRCalc (http://hs2.proteome.ca/SSRCalc/SSRCalc.html) (Krokhin et al, 2004) if the HPLC system is first calibrated. Alternatively, the retention time of the targeted peptides may be determined by analysing a limited number of transitions in a first run. This limited set of transitions may be selected on the basis of available MS/MS data, or singly charged y ions with m/z values above the m/z value of the precursor ions may be chosen blindly. Selecting 2–4 y-fragment ions from both doubly and triply charged precursor ions will usually result in at least one transition with reasonable performance to derive the retention time information for the scheduled experiments.
Limiting the final assay to 2–4 transitions per peptide enables the targeting of several hundred peptides in one LC-MS analysis (Box II). The smart selection of transitions as outlined ensures that this does not affect sensitivity or accuracy but that the highest sensitivity is obtained.
Validation of transitions
In spite of the high specificity of QQQ SRM analyses achieved by the two consecutive mass filtering stages, a particular precursor/fragment combination may not be specific for a peptide targeted in a complex sample. A typical case is illustrated in Figure 4. Unspecific signals may derive from other peptides with precursor/fragment ion pairs of similar masses. These peptides might have closely related sequences so that part of the transitions are identical. However, completely unrelated sequences might, by chance, result in mass pairs that are too close to be sufficiently filtered out in the quadrupoles. In particular, the often-not-considered additional peaks due to non-canonical fragments or the natural isotope distribution increase the likelihood for unspecific signals. Such signals will usually be of lower intensity than the optimized transitions. However, if SRM is used to target peptides that are several orders of magnitude less abundant than the most abundant peptides, such unspecific signals might still be well above the detection limit and often even more intense than the signal for the targeted peptide. As no full mass range spectra, neither on MS nor on MS/MS level, are acquired in the final SRM analysis, such signals might be easily mistaken as being derived from the targeted peptide and thus lead to mis-quantifications. Therefore, it is important to validate the initial set of transitions to ensure that the quantified signals indeed derive from the targeted peptide.
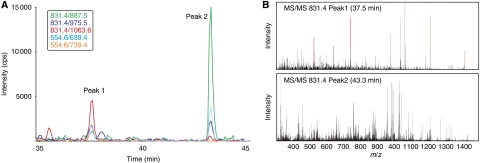
Figure 4.
Validation of transitions. SRM-triggered MS/MS experiment for the validation of transitions for peptide VFAQFSSFVDSVIAK. (A) SRM traces of five transitions. Two peaks with co-eluting transitions are apparent at 37.5 and 43.3 min. (B) MS/MS spectra triggered at the apex of SRM peaks 1 (upper panel) and 2 (lower panel). Peaks matching the respective y ions are coloured in red. Even though at 43.3 min, SRM transition intensities are higher, the MS/MS spectra clearly show that the targeted peptide is eluting at 37.5 min. Utilizing transition intensities at 43.3 min without validation would lead to false quantification values for the targeted peptide.
The first step of validation is the parallel acquisition of several transitions for a targeted peptide. At the time of peptide elution, such transitions yield a perfect set of ‘co-eluting' intensity peaks if they are derived from the same peptide. With each additional transition, the probability for a random match is markedly reduced, provided that perfect co-elution is observed. However, abundant non-target peptides with similar precursor m/z may generate a broad range of low-intense non-canonical fragment ions that could be detected by several transitions and lead to false quantitative data (see Figure 4B, peak 2 for an example). Therefore, it is highly advised to complement the parallel acquisition of several transitions by other means of validation.
A proven method to validate transitions is to acquire MS/MS spectra, which are subjected to sequence database searching to confirm that the detected signals derive from the targeted peptide. This operation is ideally performed on the QQQ instrument used for the actual SRM experiment using a protocol referred to as SRM-triggered MS/MS scanning (Unwin et al, 2005). In such experiments, the instrument is programmed to acquire a full fragment ion spectrum whenever a signal for a particular transition is detected. The acquired MS/MS spectra are then compared with the predicted peptide fragments to assure that the major MS/MS peaks are matched (Figure 4). This effectively confirms that the detected SRM signals indeed derive from the targeted peptide. Although this protocol resembles a classical data-dependent acquisition mode, the triggering by SRM traces instead of full-scan MS spectra increases specificity and thereby allows the identification of peptides of low abundance in complex mixtures, which would not be detectable in data-dependent acquisition mode (Lange et al, 2008). However, as SRM is inherently more sensitive than full MS/MS spectrum acquisition, the validation of transitions for low-abundance proteins that might be detectable by SRM remains a challenge, even if SRM-triggered MS/MS scanning is applied.
The need to acquire MS/MS spectra for the validation of transitions can be obviated if heavy isotope-labelled peptides are spiked into the sample before analysis, which match the sequence of the targeted peptide. Heavy isotope-labelled peptides (with incorporated 15N and 13C) will exactly co-elute with the non-labelled target peptides. In addition, the signal intensity ratios of the transitions are the same for the labelled and unlabelled peptides. This allows distinguishing the detection of low-abundant peptides from unspecific signals that by chance elute at the exact same retention time. However, the costs for such heavy labelled peptides might exceed the resources for projects targeting a large number of proteins. In such cases, validating peptides by MS/MS spectra acquisition is a less expensive but more time-intensive alternative. Even though validation by MS/MS spectrum acquisition cannot quite reach the excellent sensitivity obtained by spiking heavy labelled peptides, it has been shown that it allows precise and reliable quantification of peptides that are below the detection limit of conventional shotgun approaches (Lange et al, 2008).
Software
The steps described that are required to set up an SRM assay for multiple proteins are fundamentally different from shotgun proteomics experiments where proteins are identified from a sample on the basis of matching MS/MS spectra to databases. Consequently, the different methods also have different software requirements. Ideally, a software system to support the set up of SRM assays would guide the user through the sequential steps outlined above. Specifically, the system should support the following:
Selection of a protein target set.
Selection of PTPs representing the target proteins, using a combination of available data and predictions.
Selection of transitions. Integration of diverse data types to derive transitions from MS/MS spectra. Calculation of canonical fragment transitions and support for transition selection on the basis of SRM runs.
Validation of transitions by MS/MS spectra if heavy labelled peptides are not available.
Integration of the selection and validation data to support the flexible export of sets of validated transitions for quantitative assays.
The selection of protein target sets is highly dependent on the individual project; a generic solution is thus difficult to envisage. Several Web-based tools as outlined above may assist the scientist in the target set selection. Starting with a target protein set, the recently released software suite TIQAM (Targeted Identification for Quantitative Analysis by multiple reaction monitoring (MRM)) supports the workflow of setting up an SRM experiment (Lange et al, 2008). TIQAM integrates proteomic data from local experiments and from the PeptideAtlas database to prioritize PTPs. The selection of transitions on the basis of proteomics data is not yet implemented. However, TIQAM can generate SRM transition lists and identify the best performing transitions from SRM pre-experiments. In addition TIQAM provides a viewer to validate transitions by SRM-triggered MS/MS experiments. All the peptide and transition information is stored in a database to enable smart retrieval of the validated transitions for quantitative analysis. TIQAM is freely available for download at http://tools.proteomecenter.org/TIQAM/TIQAM.html.
In addition to TIQAM, several commercial solutions have been announced supporting the setup of SRM assays for proteomics. These platform-specific tools include MRMPilot (Applied Biosystems), SRM Workflow Software (Thermo Scientific), VerifyE (Waters) and Optimizer (Agilent Technologies).
Quantification by SRM
SRM assay design
The validated and optimized transitions for a specific peptide constitute a robust assay that can be used to quantify the targeted protein(s) by SRM analysis in multiple samples. Ideally, one could target all peptides of the proteome in a single LC-MS analysis. However, there is a limit in the number of transitions that may be quantified with high sensitivity and accuracy in a single LC-MS analysis. In SRM mode, the instrument repeatedly cycles through a list of transitions spending a defined time, the dwell time, on each transition. For example, targeting 10 peptides with five transitions, 20 ms dwell time, results in a cycle time of 1 s (10 × 5 × 20 ms). Thus, in this situation, every second, an intensity value is recorded for each of the transitions. The number of transitions measured per cycle and the dwell time are mutually dependent at a fixed cycle time. To achieve high sensitivity, the dwell time has to be long enough to accumulate sufficient signal. Alternatively, the cycle time might be increased to analyse a higher number of transitions at a fixed dwell time. However, if too few data points are recorded over the chromatographic elution time of a peptide, the accuracy is diminished, as the peak cannot be correctly reconstructed with too few data points. To ensure precise LC-MS quantification, at least eight data points should be acquired across the elution profile. Therefore, the best compromise between dwell time and cycle time depends on the peak width, which is obtained from the chromatographic setup. The effect on the quantitative accuracy as a function of dwell time and cycle time is illustrated in Figure 5.
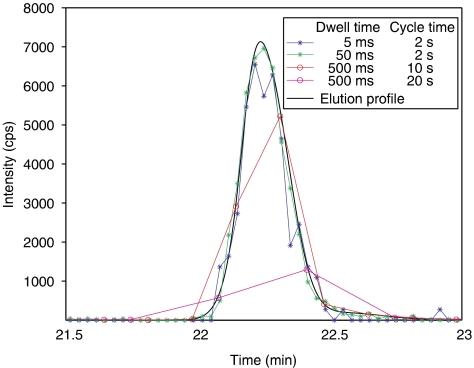
Figure 5.
Quantitative accuracy as a function of dwell time and cycle time. Quantification of a peptide using different settings for dwell time and cycle time. Reducing the dwell time from 50 to 5 ms decreases accuracy. With cycle times of 10 or 20 s, the peak height cannot be estimated correctly even though the accuracy of the individual data points is excellent at 500 ms dwell time. Changes in dwell time do not affect absolute signal intensity as it is plotted normalized as ‘counts per second'.
To avoid a loss of sensitivity in experiments in which a large number of transitions is measured, two main strategies have been used. First, the dwell times for each transition may be specifically adjusted to the expected concentration of targeted peptides, whereby a larger fraction of time is spent on lower abundance peptides, and shorter dwell times are chosen for peptides of higher abundance. A complication with this strategy is that the concentration of a peptide may not be known a priori and that, in addition, the relationship between peptide concentration and signal intensity is poorly understood. Another successful strategy to increase the number of targeted peptides without decreasing actual dwell times, accuracy and sensitivity is to restrict the acquisition of particular transitions to a window around the elution time of the corresponding peptide. By scheduling transitions, the full cycle time is deployed to detect and quantify the peptides expected to elute in a given time window. Depending on gradient length, elution peak width and reproducibility of chromatography, the number of transitions could be increased by a factor of 5–20 without a decrease in dwell time and sensitivity (Stahl-Zeng et al, 2007). See Box II for some examples discussing the number of proteins that may be quantified in a single LC-MS analysis.
The detection limit reached will highly depend on the LC conditions, ionization efficiency and instrument used. Wolf-Yadlin et al (2007) reported a limit of quantification of below 3 amol peptide loaded on column applying a dwell time of 100 ms. As a rule of thumb, a detection limit of 10–50 amol injected on column should be expected for a well-ionizing peptide with nano-LC and state-of-the-art QQQ instruments.
Normalization
The final goal of an SRM experiment is the precise quantification of a set of target proteins in a number of biological samples. In many cases, it is sufficient to determine the relative changes of protein amounts. Examples include the quantification of differences between healthy and disease or to determine protein abundance changes in response to changes in the environment, stress, pathway activation and so on. Such relative quantification might be performed on the basis of the absolute signal intensity of the individual samples. Alternatively, quantification may be based on the relative intensities of the sample analytes versus an internal standard labelled with stable isotopes. The pros and cons of ‘label free' versus ‘isotope labelled' quantification are discussed below.
For particular questions regarding systems biology modelling, it is advantageous to determine the absolute abundance of proteins in samples, for example, as copies per cell or ng/ml. Absolute concentration cannot be inferred from the signal intensity directly, as the signal response is peptide dependent and cannot be predicted. Therefore, precise absolute quantification requires that accurately quantified isotopically labelled peptides or proteins be added to the samples. In contrast to the relative quantification approach, where a complex mixture is used as internal standard, only the peptides/proteins targeted for quantification are added.
Label-free quantification
Relative quantification can be based on the signal intensities of specific SRM transitions. Upon establishment of the SRM assay, the individual samples are processed and analysed by LC-MS. Even though the label-free approach seems conceptually and experimentally simple, precise label-free quantification is challenging due to variations in signal intensities from one LC-MS analysis to the other and also within one LC-MS analysis: A peptide spiked into different backgrounds will result in different intensities depending on the sample and time of analysis. The magnitude of the fluctuations depends on the peptide sequence, the background and several experimental factors that cannot be precisely controlled. These include fluctuations in ionization efficiency over time, and matrix effects leading to ion suppression or enhancement from co-eluting analytes. These effects are particularly problematic when many, potentially diverse, samples are analysed in a study. In an attempt to control for these effects, normalization is performed in label-free quantitative proteomics with the assumption that the majority of protein/peptide concentrations in a sample are not changing. However, such normalization can only correct for global shifts and do not compensate local suppression or enhancement effects on individual peptides. Despite these limitations, label-free quantification can be successfully performed if the sample processing is well controlled and the samples are closely related in background and protein composition. As matrix effects become more pronounced with higher total peptide amounts injected, the injected sample amount should be kept low. In addition, it is essential that several peptides for each protein are quantified to avoid false conclusion owing to these effects.
Stable isotope-based quantification
The addition of isotopically labelled internal standards effectively overcomes the problems of fluctuations in signal intensity. A common standard—for example, a mixture of the samples to be analysed—is labelled with heavy stable isotopes and spiked into the individual samples. Quantification is then not based any longer on the absolute signal intensity, but rather on the relative intensity of the analyte signals compared with that of the isotopically labelled internal standard. In such an approach, each peptide is quantified relative to the matching heavy labelled peptide. Selection and validation of transitions is usually performed on either a heavy or light sample. For the quantitative analysis, the list of transitions is supplemented with the corresponding light or heavy transitions resulting in a pair of light/heavy transitions for every precursor/fragment combination. The isotope labelling should introduce a sufficiently large mass difference of both the precursor and fragment ions to prevent cross talk to the light transitions. The mass difference required depends on the resolution of the quadrupoles and the charge state of the peptide. At a resolution of 0.7 Th FWHM, triply charged peptides should have a mass difference of at least 5–6 Da. To use isotope-labelling strategies with smaller mass differences, the quadrupole resolution needs to be tightened, which decreases sensitivity.
The isotopic label should be introduced in the processing workflow as early as possible to increase the number of steps that are being controlled and decrease the technical variability (Ong and Mann, 2005). Most labelling approaches applicable for quantitative shotgun proteomics, including ICAT (Gygi et al, 1999), SILAC (Ong et al, 2002), ICPL (Schmidt et al, 2005) and others (Ong and Mann, 2005) can similarly be used for SRM-based quantitative experiments if they introduce a sufficiently large mass shift. The main exception to this rule are isobaric reagents like TMT (Thompson et al, 2003) or iTRAQ (Ross et al, 2004) where all peptides share the same reporter ions and are only distinguished by the precursor mass. Therefore, peptides with similar precursor masses will interfere in quantification, and the advantages of SRM with regard to achieving superior selectivity by two-stage filtering are lost. Despite these limitations, iTRAQ has been successfully applied to the analysis of highly enriched samples of low complexity by SRM (Wolf-Yadlin et al, 2007). When choosing a labelling strategy, it should also be considered that the applied isotopes do not alter the retention time on reversed-phase chromatography. Therefore, labelling reagents based on isotopes of nitrogen, oxygen or carbon are preferred over those based on deuterium.
Isotope labelling increases the complexity and costs of an experiment with the benefit of more precise quantification. Matrix effects are largely corrected for by the isotope-labelled standard. Therefore, a higher sample amount can be loaded per LC-MS analysis, which increases the dynamic range of the analysis if the sample amount is not limiting. Taking everything into account, quantification using stable isotope labelling is the technically less challenging approach despite the additional preparation steps that are required.
Sample amount normalization
Quantification as discussed above yields precise quantitative values for the relative protein abundances in a set of samples. To be able to draw conclusions from these samples about a particular biological system, we have to ensure that the individual samples reflect the system adequately, for example, that each sample represents the same proportion of the system, the same number of cells and so on. When working with tissues, the sample amount is commonly based on tissue weight. This might distort the results when diseased, and normal tissues are compared with substantially different tissue structures. Non-adherent tissue culture cells could be counted. However, often the sample amount is based on the total protein mass as determined by assays, such as Lowry (Lowry et al, 1951), Bradford (Bradford, 1976) or BCA (Smith et al, 1985), which are often not particularly accurate. An alternative approach to this challenge of correct starting amounts is to normalize the samples on the basis of proteins, which are expected to be constant throughout the experiment. Usually, the more abundant ‘house keeping' proteins, which fulfil central cellular functions are good candidates. Even if a few of these presumably invariant proteins are changing in abundance, normalization is not compromised as long as the group is sufficiently large (>10 proteins), and outlier insensitive statistics (median) are applied to normalize the signal intensities. These proteins need to be included in the SRM assay and are quantified together with the target protein set. Normalization is performed after acquisition during the data analysis. For most experiments, such normalization on invariant proteins yields the most reliable data.
Absolute quantification
In particular cases, the relative quantification gained from experiments as outlined above is insufficient. For example, the determination of copy numbers per cell or the accurate concentration of a protein in blood requires that individual isotopically labelled reference peptides or proteins be spiked into the sample. As the amount of these peptides/proteins is precisely determined, the absolute amount of the target protein can be deduced from the relative intensity of the light/heavy transitions (Barr et al, 1996; Gerber et al, 2003; Kuhn et al, 2004; Kirkpatrick et al, 2005). However, it is important to make sure that there are no losses in the process before the peptides/proteins are added, for example, cell lysis conditions need to be well optimized to allow concluding about the copy numbers per cell. This also implies that peptides/proteins should be spiked in before any kind of separation or enrichment to control for losses during these processes. Spiking isotopically labelled proteins has the advantage that incomplete or unspecific digestion does not corrupt the results, as might be the case if isotope-labelled reference peptides are used.
Peptides/proteins should be spiked in sufficient amount to produce a relatively intense signal allowing high-precision quantification. However, excessive amounts might saturate the detector or bleed through into the transitions for the detection of the light peptide. In addition, stable isotope-labelled peptides might be contaminated with unlabelled peptides up to 1–2%. To test for bleed through or contamination, the heavy peptides should be injected alone and transitions for both the heavy and light peptides should be analysed. No signal should be detectable in the transitions for the light peptides. Ideally, the spiking amounts are derived from pre-experiments to determine the optimal amount for each individual peptide. We recommend as starting point to spike between 10 and 50 fmol peptide injected on column, but this is obviously dependent on instrument and setup.
Several options exist to obtain such peptides/proteins as internal standards for absolute quantification. Custom-specified peptides with incorporated heavy labelled amino acids could be ordered by several suppliers. Accurate absolute quantification with these peptides, often referred to as AQUA peptides, relies on the accurate determination of the peptide concentrations, which is challenging. The most accurate procedure is amino-acid analysis (AAA) (Macchi et al, 2000), which relies on highly purified peptides. A drawback of these synthetic peptides is the relatively high cost, which may become an issue if large numbers of peptides are required.
Spiking isotopically labelled proteins is required when absolute quantification needs to be combined with separation and/or enrichment on the protein level, for example, enrichment of proteins by affinity enrichment or antibody capture (Brun et al, 2007). To obtain isotopically labelled proteins, the respective genes are cloned into expression vectors and expressed in in vitro expression systems with part of the amino acids isotope labelled. Alternatively, in vivo expression systems might be used with substituted amino acids or using metabolic labelling in Escherichia coli or yeast. Upon cleanup, the protein is quantified by AAA.
Beynon et al have described a recombinant genetic method to generate isotopically labelled reference peptides. Using gene synthesis, they create an artificial protein consisting of concatenated tryptic peptides, which is expressed as described for the synthesis of labelled proteins (Beynon et al, 2005; Pratt et al, 2006). One such construct may be assembled out of 30–50 peptides enabling the quantification of many proteins with this one artificial protein. As synthetic oligonucleotides are relatively inexpensive, such ‘QconCAT' proteins could be produced at a fraction of the cost of synthetic peptides.
A direct comparison of QconCAT derived and chemically synthesized peptides revealed the challenges of absolute quantification (Mirzaei et al, 2008). The accuracy of absolute quantification is dependent on the accuracy of quantification of the reference peptides. Incomplete digestion can distort the peptide amounts obtained from a QconCAT approach. Even when synthetic peptides are used, which have been quantified by AAA, losses of the peptides due to degradation or modification during storage or due to losses during lyophylization/re-solubilization may introduce a several-fold error (Mirzaei et al, 2008). Therefore, it is advisable to keep the peptides in solution after AAA and store aliquots at −80 °C.
Case studies
Case 1—yeast pathway analysis
Goal of the study
The aim of the study was to establish and test a mathematical model of the citrate acid cycle in the yeast Saccharomyces cerevisiae. All the enzymes that constitute the system were to be quantified by SRM in protein extracts from yeast cells grown under different metabolic conditions (unpublished results).
Selection of protein set
As the pathway has been extensively characterized, the target protein set of 20 proteins is well defined. The basic structure of the pathway is derived from the KEGG database, yeast build, and three additional proteins were included from literature reports.
Selection of targeted peptides
PTPs for the proteins of interest were derived from PeptideAtlas, yeast build (www.peptideatlas.org), which features more than 36 000 previously identified unique yeast peptides. On the basis of the number of reports in PeptideAtlas for each protein, the three most frequently observed peptides were selected.
Selection and validation of transition
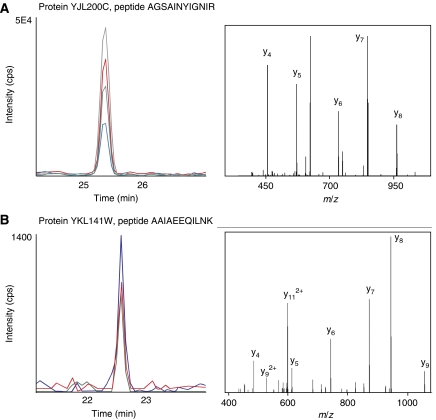
Five transitions per peptide were calculated. They corresponded to y-fragment ions and were prioritized on the basis of the fragment ions observed in the MS/MS consensus spectra stored in PeptideAtlas. The total of 300 transitions were split into three SRM-triggered MS/MS experiments, each performed using a 30-min chromatographic gradient. Each SRM transition was monitored with a 10 ms dwell time, followed by acquisition of two MS/MS spectra triggered by the two highest SRM signals. MS/MS data were searched against the S. cerevisiae protein database, and the analysis confirmed the identification of 11 proteins by at least two PTPs each and four by one PTP. Figure 6A shows validation of the SRM assay for the abundant protein YJL200C estimated to be expressed at ∼5000 copies per cell (Ghaemmaghami et al, 2003). For the remaining five proteins, which were not identified in the first pass, a second attempt was undertaken. Five peptides for each protein were targeted using six transitions for each of the two main charge states (doubly and triply charged). The 300 transitions, each acquired for 50 ms, were monitored in five MRM-triggered MS/MS runs using a 60-min gradient. Data analysis now confirmed peptides for four additional proteins. Figure 6B shows validation of the SRM assay for the low-abundance protein YKL141W estimated to be expressed at ∼250 copies per cell (Ghaemmaghami et al, 2003). The best transitions to be used in quantitative experiments were either selected on the basis of the MS/MS spectra acquired in the QQQ instrument or derived from an additional experiment conducted in scheduled MRM mode in which 20 transitions of the y-ion series with doubly and triply charged precursor were tested for each peptide, resulting in about 700 transitions. For each protein, the two best peptides, and for each peptide, the three transitions with the best signal-to-noise ratio were selected for quantitative analysis, resulting in 90 transitions (19 confirmed proteins, 30 peptides, 3 transitions each). For normalization purposes, 30 previously validated transitions corresponding to 10 moderately abundant yeast proteins were also included, which were expected to be invariant in the metabolic conditions used.
Figure 6.
Validation of transitions for yeast proteins. SRM-triggered MS/MS experiment for the validation of transitions for a high- (A) and a low-abundance (B) protein of S. cerevisiae. Owing to the low abundance of protein YKL141W (B), the SRM traces for the corresponding peptide AAIAEEQILNK were of low intensity. Nevertheless, the acquired MS/MS spectrum confirmed the identity of the peptide with high confidence.
Quantitative analysis
The validated SRM assays were used to measure the set of proteins of interest in six samples, that is, yeast grown in triplicate under glucose or under ethanol. Yeast cells grown under glucose were metabolically labelled, and the extracts from glucose and ethanol growth were combined. The protein set was quantified in a single analysis per replicate using scheduled SRM. Relative quantification was obtained by calculating the ratio between the height of the SRM peaks derived from the heavy and light version of each peptide. Differences in the total amount of protein loaded for each sample were corrected using the median of the abundance changes of the 10 normalization proteins. The analysis provided the relative abundance changes of the proteins involved in the citrate acid cycle under the two metabolic conditions studied. The data, optionally coupled to data sets of different type (e.g., metabolite concentration data), are used to validate and improve current mathematical models of the TCA cycle in S. cerevisiae.
Case 2—biomarker identification in plasma samples
On the basis of a set of candidate proteins, a collection of plasma samples was screened to identify potential novel biomarkers for a disease (unpublished results). To be able to quantify low abundance proteins in plasma, peptides with N-linked sugars were specifically enriched by solid phase capture (Zhang et al, 2003; Stahl-Zeng et al, 2007).
Selection of a target protein set
Preliminary results indicated 50 proteins as potential biomarkers for a disease. In addition, ten moderately abundant plasma proteins were selected for normalization.
Selection of peptides
On the basis of the number of observations in the human plasma build of the PeptideAtlas (www.peptideatlas.org), for each protein, one peptide with N-glycosylation site was selected. Peptides with non-tryptic or missed cleavages were not considered. In addition, peptides with potential chemical modifications were avoided whenever possible. The heavy isotope-labelled homologues of the 60 peptides were custom-synthesized and quantified.
Selection of SRM transitions
To optimize transitions, the peptides were mixed at high concentration (250 fmol/μl in 50% acetonitrile) in batches of 10 and analysed by infusion in nano-ESI mode (Box III). For each peptide, the three best performing transitions with associated parameters were selected for the final quantitative assay.
Factors affecting SRM transition efficiency.
The following parameters influence signal intensity.
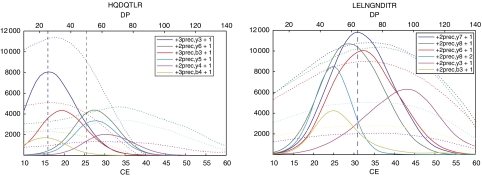
Ionization conditions. Precursor charge state: targeting the predominant charge state is essential for a sensitive detection. Charge states can be inferred from previous experiments on other ESI instruments. However, ionization devices and experimental conditions (flow rate, solvents, background) can influence charge state distributions. Ion source parameters: during the ionization process, single ions need to be generated. The process of dissolvation and dissociation of ion clusters is supported by a voltage potential referred to as ‘declustering potential' (DP), ‘fragmentor voltage' or ‘ion transfer capillary offset voltage' depending on the manufacturer. At too high DP, peptides are fragmented already in the source. From plotting many experimentally determined DP optima, a positive linear correlation of precursor m/z value and DP optimum was determined (unpublished results). As the DP displays a broad optimum, individual optimization usually does not result in a significant signal increase. Individual transitions of a peptide have the same DP optimum if they are derived from the same precursor charge state (Figure 3).
Fragmentation conditions. Fragment ion type: singly charged y ions are the predominant type of fragments generated by CID in a linear collision cell. Only small b ions are usually observed. Fragments with m/z values close to the precursor should be avoided as such transitions are usually noisy. Fragments with m/z values above the precursor generally display the highest selectivity, as the singly charged chemical background cannot result in fragments with higher m/z than the precursor. In contrast, tryptic peptide ions are predominantly doubly or triply charged with one charge at each terminus. Upon fragmentation, one charge is lost and therefore a part of the fragments has an m/z value bigger than the precursor value. Collision energy: with increasing collision energy, a larger part of the precursor ions is fragmented and fragment ion intensity increases until this increase is overcompensated by the losses due to secondary fragmentation events (Figure 3). The optimal collision energy is approximately linearly correlated with the precursor mass for a given charge state. However, particular peptides or fragments deviate considerably from the predicted value. We found that individually optimizing collision energy and declustering potential increased signal response two- to fivefold for every sixth transition (unpublished data).
The easiest and most systematic way of optimizing ionization and fragmentation conditions is to test possible transitions in direct infusion mode and ramp the parameters as in Figure 3. This process is partly automated by add-ons for the acquisition software. Less fine grained but often sufficient optimization may also be performed by LC-MS.
Quantification
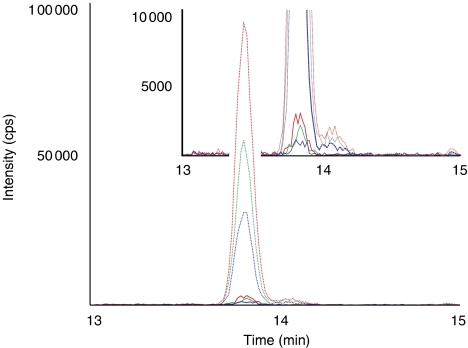
To derive the ideal spiking amount and the retention time, a representative sample processed by N-glycocapture was spiked with the isotopically labelled peptides to reach 200 fmol per labelled peptide injected on column. On the basis of the signal intensities observed, the peptides were spiked into 100 samples for the actual analyses at a reduced concentration to yield a signal intensity of 50 000 cps peak height (average of three transitions). The quantitative analyses were performed by scheduled SRM using a cycle time of 2 s with three transitions for each of the heavy and light peptides resulting in a total of 360 transitions (Figure 7).
Figure 7.
Quantification of proteins in plasma. A total of 360 transitions were acquired by scheduled SRM to quantify 60 peptides in plasma samples following glycocapture. Traces for one peptide in 1 out of 100 analysed samples are shown. On the basis of the heavy isotope-labelled peptide (dashed lines), the concentration of the endogenous protein (solid lines) was estimated to be 5 ng/ml.
Normalization
Quantification was based on the peak height, which we found more reliable for low-intense signals. For each peptide, the heavy/light transition pair with the best signal-to-noise ratio was selected for quantification. Two steps of normalization were performed: first, the ratio of light/heavy transition was calculated. Second, a normalization vector was calculated from the results of the 10 moderately abundant proteins. Each sample was corrected by the median of the normalized abundance of these 10 proteins to correct for global shifts in the individual samples.
Results
A total of 30 candidate biomarkers were successfully quantified over a set of 100 samples in this experiment. The remaining 20 candidates could not be detected in the plasma samples as the protein concentration was below the detection limit. Taking into account the estimated recovery of the glycocapture process and the spiked peptide amount, the concentration of the potential biomarker proteins in plasma was estimated, ranging from 5 to 100 ng/ml. Several candidates showed statistically significant differences between samples from healthy and diseased people.
Outlook
Shotgun proteomics has revolutionized the speed and depth of protein identification from complex mixtures. However, many biological questions remain unanswered by qualitative data alone, thus the demand for quantitative proteomic data has been steadily rising. Several strategies, including stable isotope labelling, were developed in the past decade to enable quantitative proteomic studies. Despite these advancements, technological limitations remain. First, in shotgun proteomics, only the more highly abundant part of the proteome is reproducibly identified. Less-abundant peptides are only sporadically identified resulting in missing data points in quantitative data sets. Second, sample fractionation, a key to increasing proteome coverage in proteomic studies, is more difficult to implement and requires prohibitive analysis time for larger quantitative studies that include numerous samples. The targeted SRM-based approach therefore ideally complements shotgun workflows. On the basis of a comprehensive map derived by shotgun proteomics, PTPs of protein sets of interest can be selected to generate specific SRM transitions. Upon validation, these proteins can be quantified reliably in multiple samples. This supports the growing demand by systems biology for consistent quantitative data sets of multiple samples challenged under varying conditions. It is also expected that SRM can bridge the gap between biomarker discovery, usually performed on few samples, and validation by antibody-based approaches, which are costly and slow to develop. When combining efficient enrichment strategies with SRM, detection limits in the low ng/ml range in plasma can be achieved under ideal conditions at a considerable throughput of 100 samples per week. If, however, higher throughput and/or higher sensitivity are required, antibody-based technologies should be considered.
Currently, setting up SRM assays for higher number of proteins remains challenging and requires substantial time investment despite the recently developed software solutions to support this process (Lange et al, 2008). Frequently, the same proteins are of interest for multiple projects, and the redundant transition selection in several laboratories is a waste of valuable resources. Thus, validated transitions should be stored in centralized databases, together with optimized experimental parameters, to be accessible to the proteomic community. Along these lines we have generated ‘MRMAtlas' (http://www.mrmatlas.org/), a publicly accessible database of validated SRM assays currently covering about 1500 proteins of S. cerevisiae (Picotti et al, 2008). Each validated SRM assay includes all the required parameters to quantify the targeted protein on a QQQ MS, such as the following: m/z of the precursor peptide ion, charge state, m/z of the fragment ions with the highest signal intensities, suggested collision energy and relative intensities of fragment ion signals, calculated hydrophobicity and observed elution times, both as a measure of relative retention time on reversed-phase separation. To ensure specificity of the assays, each peptide was validated by MS/MS spectra, which are accessible online. We intend to further extend the MRMAtlas to other species and encourage the submission of validated high-quality SRM assays. A remaining need is to concur on a standardized protocol to report normalized elution times, one of the critical parameters to be captured in such experiments. A set of common reference standards should serve as elution time landmarks and thus account for HPLC column and system variability.
The setup of SRM assays is largely simplified by isotopically labelled peptides. As these become routinely available at lower prices, their use in larger scale projects is conceivable. As many researchers, particularly in the biomarker field, will require the same sets of PTPs, suppliers may soon offer panels of specific isotopically labelled peptides in combination with optimized SRM parameters. As the quantities required as internal standards are several orders lower than the amount produced in one chemical synthesis, such peptide kits will become affordable. Alternatively, where unique peptides are required, the QconCAT strategy may, once commercialized, allow the production of personalized QconCAT proteins covering peptides for a full set of targeted proteins at the cost of a microarray experiment. Therefore, we expect the availability of isotopically labelled peptides to increase significantly by one way or another.
As SRM finds more widespread use in proteomics, several of the remaining obstacles will be solved. In addition to the quantification of protein abundance, SRM has a great potential for the reliable quantification of post-translational modifications. In conclusion, SRM strategies appear as an excellent match to the requirements of systems biology, not only allowing quantitative analysis of low-abundant proteins, but also delivering reliably quantitative data when proteins are analysed across multiple samples.
Figure 3.
Optimization of transitions. Plot of intensity versus collision energy (CE, solid lines) or declustering potential (DP, dotted lines) for the six best transitions of two peptides obtained by ramping CE and DP in constant infusion mode. The predicted CEs for the doubly (blue, CE=0.044 × m/z+5.5) and triply (green, CE=0.051 × m/z+0.5) charged precursors are marked by vertical dashed lines.
Acknowledgments
This work was supported in part by a grant from F Hoffmann-La Roche Ltd (Basel, Switzerland) through support for the Competence Center for Systems Physiology and Metabolic Disease, by the Swiss National Science Foundation under grant no. 31000-10767 to RA, by Federal funds from the National Heart, Lung and Blood Institute, National Institutes of Health, under contract no. N01-HV-28179 and by SystemsX.ch, the Swiss initiative in systems biology. PP is a recipient of a Marie Curie Intra-European fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JR, Maggio VL, Patterson DG Jr, Cooper GR, Henderson LO, Turner WE, Smith SJ, Hannon WH, Needham LL, Sampson EJ (1996) Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem 42: 1676–1682 [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R (2007) NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucl Acids Res 35: D760–D765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods 2: 587–589 [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, Panse C, Lee HK, Schlapbach R, Aebersold R (2007) PhosphoPep–a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol 3: 139 10.1038/msb4100182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bronstrup M (2004) Absolute quantification strategies in proteomics based on mass spectrometry. Expert Rev Proteomics 1: 503–512 [DOI] [PubMed] [Google Scholar]
- Brun V, Dupuis A, Adrait A, Marcellin M, Thomas D, Court M, Vandenesch F, Garin J (2007) Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics 6: 2139–2149 [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G (2007) MINT: the Molecular INTeraction database. Nucl Acids Res 35: D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC (2004) Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3: 1234–1242 [DOI] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R (2006) The PeptideAtlas project. Nucleic Acids Res 34: D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Clarke S (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem 262: 785–794 [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 100: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Griffiths JR, Unwin RD, Evans CA, Leech SH, Corfe BM, Whetton AD (2007) The application of a hypothesis-driven strategy to the sensitive detection and location of acetylated lysine residues. J Am Soc Mass Spectrom 18: 1423–1428 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Jones P, Cote RG, Cho SY, Klie S, Martens L, Quinn AF, Thorneycroft D, Hermjakob H (2008) PRIDE: new developments and new datasets. Nucleic Acids Res 36: D878–D883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Glazebrook J (2004) Overview of mRNA expression profiling using microarrays. Curr Protoc Mol Biol 22.4.1–22.4.8 [DOI] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B et al. (2007) IntAct—open source resource for molecular interaction data. Nucl Acids Res 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Gerber SA, Gygi SP (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35: 265–273 [DOI] [PubMed] [Google Scholar]
- Krokhin OV, Craig R, Spicer V, Ens W, Standing KG, Beavis RC, Wilkins JA (2004) An improved model for prediction of retention times of tryptic peptides in ion pair reversed-phase HPLC: its application to protein peptide mapping by off-line HPLC-MALDI MS. Mol Cell Proteomics 3: 908–919 [DOI] [PubMed] [Google Scholar]
- Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B (2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4: 1175–1186 [DOI] [PubMed] [Google Scholar]
- Kuster B, Schirle M, Mallick P, Aebersold R (2005) Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6: 577–583 [DOI] [PubMed] [Google Scholar]
- Lange V, Malmström JA, Didion J, King NL, Johansson BP, Schäfer J, Rameseder J, Wong C-H, Deutsch EW, Brusniak M-Y, Bühlmann P, Björck L, Domon B, Aebersold R (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics 7: 1489–1500 M800032-MCP800200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Macchi FD, Shen FJ, Keck RG, Harris RJ (2000) Amino acid analysis, using postcolumn ninhydrin detection, in a biotechnology laboratory. Methods Mol Biol 159: 9–30 [DOI] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125–131 [DOI] [PubMed] [Google Scholar]
- Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, Bjorling E, Blagoev B, Bose R, Brahmachari SK, Burlingame AS, Bustelo XR, Cagney G, Cantin GT, Cardasis HL, Celis JE et al. (2008) Human Proteinpedia enables sharing of human protein data. Nat Biotechnol 26: 164–167 [DOI] [PubMed] [Google Scholar]
- Mirzaei H, McBee JK, Watts J, Aebersold R (2008) Comparative evaluation of current peptide production platforms used in absolute quantification in proteomics. Mol Cell Proteomics 7: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollah S, Wertz IE, Phung Q, Arnott D, Dixit VM, Lill JR (2007) Targeted mass spectrometric strategy for global mapping of ubiquitination on proteins. Rapid Commun Mass Spectrom 21: 3357–3364 [DOI] [PubMed] [Google Scholar]
- Murray KK, Boyd RK, Eberlin MN, Langley GJ, LIANG Li L, Naito Y (2006) Standard definitions of terms relating to mass spectrometry: 3rd Draft Document, August 2006 (in press; http://www.iupac.org/web/ins/2003-056-2-500)
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M (2005) Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1: 252–262 [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R, Domon B (2007) The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics 6: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, Domon B, Aebersold R (2008) A database of mass spectrometric assays for the yeast proteome. Nat Methods, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piszkiewicz D, Landon M, Smith EL (1970) Anomalous cleavage of aspartyl-proline peptide bonds during amino acid sequence determinations. Biochem Biophys Res Commun 40: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ (2006) Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc 1: 1029–1043 [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kellermann J, Lottspeich F (2005) A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 5: 4–15 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepen JA, Belhajjame K, Selley JN, Embury SM, Paton NW, Goble CA, Oliver SG, Stevens R, Zamboulis L, Martin N, Poulovassillis A, Jones P, Cote R, Hermjakob H, Pentony MM, Jones DT, Orengo CA, Hubbard SJ (2008) ISPIDER Central: an integrated database web-server for proteomics. Nucleic Acids Res 36: W485–W490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics 6: 1809–1817 [DOI] [PubMed] [Google Scholar]
- Tang H, Arnold RJ, Alves P, Xun Z, Clemmer DE, Novotny MV, Reilly JP, Radivojac P (2006) A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics 22: e481–e488 [DOI] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75: 1895–1904 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Ponten F (2005) Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics 4: 384–393 [DOI] [PubMed] [Google Scholar]
- Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD (2005) Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol Cell Proteomics 4: 1134–1144 [DOI] [PubMed] [Google Scholar]
- Williamson BL, Marchese J, Morrice NA (2006) Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol Cell Proteomics 5: 337–346 [DOI] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA 104: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li XJ, Martin DB, Aebersold R (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol 21: 660–666 [DOI] [PubMed] [Google Scholar]