Abstract
Presenilin 1 (PS1) expression is repressed by the p53 tumor suppressor. As shown herein, wild-type PS1 is an effective antiapoptotic molecule capable of significantly inhibiting p53-dependent and p53-independent cell death. We analyzed, at the functional and molecular levels, the brains of p53 knockout mice. Surprisingly, we found that lack of p53 expression induces apoptotic brain lesions, accompanied by learning deficiency and behavioral alterations. p53-deficient mice show an unexpected overexpression of p21waf1 with subsequent down-regulation of PS1 in their brains. This process is progressive and age-dependent. These data indicate that the p53 pathway, besides affecting tumor suppression, may play a major role in regulating neurobehavioral function and cell survival in the brain.
Presenilin 1 (PS1) was characterized as a predisposition gene mutated in early-onset familial Alzheimer's disease (1). Increased evidence indicates that mutations in PS1 accelerate neurodegeneration and facilitate apoptosis (2–4). Furthermore, PS1 knockout mice have massive neuronal loss, skeletal defects, and severe hemorrhages and die shortly after birth (5, 6). Mutant PS1 transgenic, knockin mice, as well as transfected cells, show accelerated neurodegeneration or susceptibility to apoptosis (2–4). Experimental down-regulation of PS1 expression by antisense cDNA induces apoptosis in both hematopoietic and neuronal cells (7, 8), suggesting a conserved function in different cell types. PS1 interacts with a series of proteins among which are Notch and a c-Jun-interacting factor (9, 10). Repression by PS1 of c-Jun function through the specific interaction with QM/Jun-interacting factor-1 was also shown to inhibit apoptosis (10). Together, all these findings suggest that loss of PS1 function may contribute to apoptotic processes that underlie neurodegenerative diseases. Interestingly, PS1 is associated with the p53 signal transduction pathway (7, 11). Wild-type p53 down-regulates PS1 expression; a similar effect is exerted also by the p53 target gene product p21waf1 (12–15), raising the possibility that the effect of p53 on PS1 expression is attained indirectly through p21waf1 induction (7). In the present work, we investigated the effect of PS1 on apoptosis in both p53-dependent and p53-independent systems as well as the in vivo consequences of p53 deficiency on PS1 regulation, apoptosis in the brains, and neurobehavioral functions in p53 knockout mice.
Materials and Methods
Cells, Transfectants, Western Blot Analysis, Antibodies, and Terminal Deoxynucleotidyltransferase-Mediated UTP End Labeling (TUNEL) Assay.
LTR6 cells stably transfected with the temperature-sensitive p53Val135 mutant have been described (16). LTR6 cells were transfected by using Lipofectin with the full-length mouse PS1 cDNA subcloned in pcDNA3.1/Zeo (Invitrogen). Selection was performed with zeocin (50 μg/ml) for 6 weeks. All of the experiments were reproduced on at least two clones five consecutive times.
Polyclonal anti-PS1 antibodies (95/23) against the N-terminal fragment were kindly provided by C. L. Masters and G. Evin (University of Melbourne, Melbourne). For the detection of PS1, proteins were extracted adding 1% Nonidet P-40 and 1% Triton X-100, followed by the addition of SDS sample buffer with a final concentration of 8 M urea, heated for 20 min at 56°C, and loaded directly on the gel (10 μg of protein). Signals were detected with a secondary antibody coupled to peroxidase. Equal loading of proteins was analyzed by hybridizing the Western blots with anti-β-tubulin antibodies.
Human PS1 cDNA (2.7 kilobases) was cloned in the EcoRI site of the pBK-RSV vector (Stratagene), and US cells (17) were transfected with Lipofectin. Stable transfectants were obtained after 3 weeks of selection with 1.5 mg/ml of G418 (Sigma). Western blot analysis and TUNEL assay were performed as described above with anti-human PS1 antibodies (7). All of the experiments were reproduced on polyclonal cultures five consecutive times.
p21waf1 and antisense PS1 transfectants of U937 cells have been described (7, 18). For the detection of p21waf1, proteins were extracted by using standard conditions and sc-397 (Santa Cruz Biotechnology) as anti-p21waf1 antibody.
p53-Deficient Mice: Morris and Open Field Tests.
The experimental animals (aged 49–63 days) consisted of 12 female and 11 male p53−/− mice of the C57BL/6 genetic background (Taconic Farms) and 12 male and 12 female wild-type aged-matched control mice (C57BL/6). Mice were tested in a Morris swimming pool; the circular swimming pool (70 cm in diameter and 30 cm high) was filled with water (22°C) made opaque with the Opacifier 631. Dropped into the water from a different quadrant on each trial, mice had to learn to navigate to the invisible platform by using the spatial cues available in the room. After a training session to learn the procedural component of the task, mice were given three consecutive trials a day, for 4 days. After the third trial of the last session, mice were submitted to a probe test for the spatial bias. The platform was removed, and the mouse, starting from the opposite quadrant, was allowed a 1-min search for the platform. The path was recorded on a video tape, and a spatial bias index was computed as the number of crossings of an 8-cm diameter annulus surrounding the former location of the platform minus the mean crossings of the three annuli symmetrically laid out in the quadrants where the platform had never been. The experimenter was blind to the genotype and age of animals.
The same animals used in the Morris test were used in the Open Field test. Mice were introduced individually in a gray poly(vinyl chloride) cylinder (40 cm in diameter and 30 cm in height). This device, lit by six white bulbs (125 lux) diffusing through a frosted glass, represents a moderately anxiogenic situation. The relative number of central sector crossings corresponds to the number of crossings of a 15-cm diameter, centrally located annulus expressed as a percentage of the overall locomotor activity. Behavioral measurements were done over 5 min, providing a measurement of emotional reactions to absolute novelty.
Statistical Analysis.
A two-way Genotype X Sex ANOVA was used to analyze behavioral data. The squared correlation (r2) was computed to explain the part of variance accounted for by the effect of the knockout of the p53 gene. Scheffé post hoc tests were performed to analyze the group differences.
Northern Blotting.
Northern blot analysis was performed with 2 μg of mRNA. Hybridization was done with random-primed 32P-labeled probes for the fingerprint of the differentially expressed genes, p21waf1, PS1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Histopathological Analysis and TUNEL Assay on the Brains.
Four knockout mice and two controls were anesthetized and perfused transcardially with 10% (vol/vol) formaldehyde. After removal of the brains, 1-mm coronal blocks were embedded in paraffin, and 7-mm-thick sections were obtained and stained by hematoxylin/eosin and Bodian silver stain. Apoptotic nuclei were labeled by TUNEL according to the manufacturer's instructions (Roche Molecular Biochemicals). All of the slides were counted independently by at least two investigators with a double blind approach.
Results and Discussion
PS1 Is Antiapoptotic.
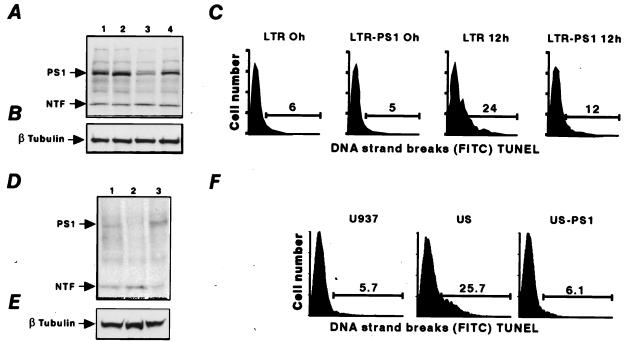
To assess a putative antiapoptotic function of PS1, we tested whether PS1 can protect cells against the apoptotic action of p53. The experimental system chosen for this purpose consisted of LTR6 cells (16). These cells are derived from the murine leukemic cell line M1 through stable transfection with the temperature-sensitive p53Val135 mutant. On shift from 37°C to 32°C, p53Val135 regains wild-type p53 function; this gain of function, in turn, results in apoptotic cell death (16). Of particular note, LTR6 cells show a pronounced down-regulation of PS1 expression after wild-type p53 activation (7, 11). Transfection of LTR6 cells with a PS1 expression vector resulted in only slightly more elevated levels of PS1 protein expression (Fig. 1A, lane 2) as compared with the control LTR6 (Fig. 1A, lane 1). After 12 h of wild-type p53 induction in the LTR6 cells at 32°C, the level of PS1 strongly decreased (Fig. 1A, lane 3). In contrast, the LTR6-PS1 transfectants in the same experimental conditions maintained a significantly higher level of PS1 (Fig. 1A, lane 4). These results indicate that transfection with the PS1 expression vector can compensate for the decrease in PS1 induced by p53. Importantly, induction of p53-mediated apoptosis at 32°C was markedly suppressed in these cells, which were allowed to maintain relatively high PS1 levels (Fig. 1C). This suppression by 50% of the program of cell death was also apparent after 24 h of incubation at 32°C, irrespective of whether apoptosis was measured by accumulation of cells with sub-G1 DNA content or by the TUNEL assay and whether LTR6 cells were transfected with a PS1 expression vector or infected with a PS1 retrovirus (data not shown). Hence, increased levels of PS1 protect against p53-induced apoptosis.
Figure 1.
PS1 is antiapoptotic. (A) Western blot analysis with anti-PS1 antibodies. (Lane 1) LTR6 cells at 37°C. (Lane 2) LTR6 cells transfected with PS1 (LTR-PS1) at 37°C. (Lane 3) LTR6 cells after 12 h at 32°C. (Lane 4) LTR-PS1 after 12 h at 32°C. PS1 indicates the full-length 50-kDa PS1 protein product, and NTF indicates the 30-kDa N-terminal fragment of PS1. (B) Western blot analysis with anti-β-tubulin antibodies. (C) Flow cytometry analysis of TUNEL assay in LTR6 cells and PS1 transfectants (LTR-PS1) at 37°C and after 12 h of incubation at 32°C. (D and E) Western blot analysis with anti-PS1 antibodies (D) or anti-β-tubulin antibodies (E) of protein extracts from U937 cells (lane 1), US cells (lane 2), and US cells transfected with PS1 (US-PS1) (lane 3). (F) Flow cytometry analysis of TUNEL assay in U937, US, and US-PS1 cells. Inhibition of apoptosis by PS1 is seen in both the LTR6-PS1 after 12 h and the US-PS1 transfectants.
This antiapoptotic function of PS1 was also confirmed in a p53-independent system (Fig. 1 D–F) in which US cells were transfected with PS1. These US cells (17), derived from the human monocytic cell line U937, express decreased levels of PS1 (7). As described previously, this down-regulation of PS1 in the US cells is due to increased levels of p21waf1 in the absence of p53 (7). Stable transfection of US cells with PS1 resulted in elevated levels of the 50-kDa full-length protein (Fig. 1D) with a drastic inhibition (75%) of apoptosis (Fig. 1F). These data show that increasing PS1 levels protects against apoptosis, whereas a decrease in PS1 facilitates apoptosis. Previous reports indicated that the C-terminal portion of PS2 (ALG3) protects against apoptosis (19, 20), whereas others reported increased apoptosis with mutant PS1 and PS2 (2–4, 19, 20). Furthermore, highly increased levels of wild-type human PS1 in transgenic rats were reported to be proapoptotic (21), perhaps because, in these systems, heterologous PS1 is recognized as a mutant one. Our data suggest that wild-type PS1 levels are tightly controlled within a narrow margin where PS1 can exert its antiapoptotic function.
Common Downstream Effector Genes Between p53-p21waf1 and PS1.
To investigate further whether signal transduction by PS1 is related to p53 and p21waf1, we used a series of 15 previously characterized cDNAs as a molecular fingerprint of the p53 and/or p21waf1 transduction pathway (22). These genes have been named TSAP, for tumor suppression activated pathway, or TSIP, for tumor suppression inhibited pathway. Used as markers in Northern blot analysis, they have been shown to be differentially expressed in tumor suppression and/or apoptosis (22) and induced in p53-dependent or p53-independent biological model systems. As summarized in Table 1, the Northern blot analysis with these markers as a probe shows that there is a striking overlap in the differential expression of these genes between U937 cells transfected with p21waf1 and U937 cells transfected with antisense PS1. These results indicate that induction of p21waf1 or repression of PS1 triggers at least partially common downstream effector genes.
Table 1.
Characteristics of differentially expressed cDNA clones
| Cell type | cDNA clones
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSAP9 | TSAP10 | TSAP11 | TSAP12 | TSAP13 | TSAP14 | TSAP15 | TSAP16 | TSAP17 | TSAP18 | TSAP19 | TSAP20 | TSAP21 | TSAP22 | TSIP3 | ||
| U937 p21 | D | D | N | N | D | D | D | N | D | N | N | D | D | D | D | |
| U937 AS PS1 | D | D | N | N | D | D | D | N | N | D | N | D | D | D | D | |
D, differential expression by Northern analysis; N, no differential expression by Northern analysis. Homologies: TSAP 9, chaperonin containing t-complex polypeptide 1; TSAP 13, proteasome subunit p40.5 (Nas 7p); TSAP 21, SNARE syntaxin 11 (22).
p53-Deficient Mice Have Learning Deficiencies and Behavioral Alterations.
Because PS1 is a predisposition gene for early onset familial Alzheimer's disease and because its expression is regulated by p53, we evaluated the potential relevance of PS1 regulation by p53 by studying p53-deficient mice at the cognitive, behavioral, molecular, and pathological levels. p53 knockout mice have been widely studied for their increased cancer susceptibility (23). A significant portion of the female animals also have an abnormal closure of the neural tube resulting in exencephaly, leading to death in utero (24, 25). No abnormalities of the central nervous system were hitherto described in young or adult p53 knockout mice. For testing the memory and behavioral parameters, to avoid a possible interference of tumor formation with behavior, we studied mice during a disease-free time interval (49–63 postnatal days). All of the animals were verified by PCR to have a disrupted p53 gene (data not shown). Two tests were performed: the Morris water maze and the open field. The Morris water maze (26, 27), in which spatial learning and memory are evaluated, was used successfully in recent studies, including a mouse model of Alzheimer's disease (28). The open field test measures behavioral parameters in a mildly anxiogenic situation (29) and was used for mapping quantitative trait loci for fear-like behaviors in mice (30). A total of 23 p53 knockout mice and 24 control animals were tested.
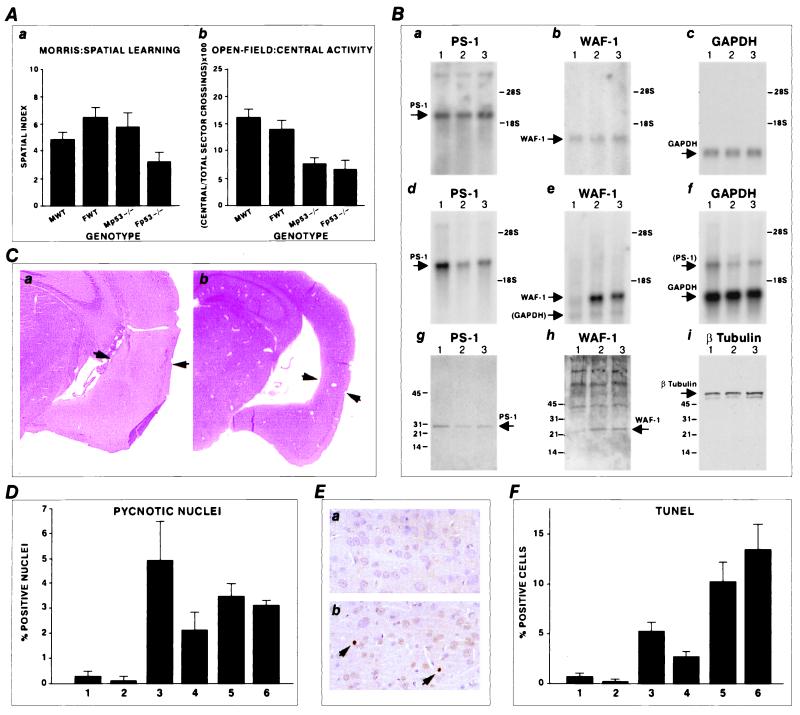
In the Morris water maze, mice are placed in a circular swimming pool and learn to escape to a hidden platform. To ensure that spatial learning occurred, a spatial probe test was given at the end of the learning session. Whereas p53−/− males did not differ from male and female wild-type controls, p53−/− females showed significantly inferior performance at the probe test, indicating poorer spatial learning and memory capacities (Fig. 2Aa). One of the traits measured by the open field test is the “wall-seeking” tendency (thigmotaxis). When individually introduced into a cylinder, both female and male p53−/− mice show a significantly lower percentage of relative central activity (Fig. 2Ab), which is an index of the ability to overcome the fear of leaving the wall to explore an unknown area. These results suggest that p53-deficient mice are more thigmotaxic than their normal counterparts. Moreover, from observing the breeding process of these animals, it becomes obvious that it is very hard to obtain any progeny from homozygous p53−/− mice. This difficulty is partially caused by the fact that p53−/− mice are less fertile, but also because they totally neglect their progeny and let them die. These observations together with the parameters obtained in the Morris and open field tests indicate that p53−/− animals suffer from a variety of neurological problems.
Figure 2.
Memory and behavioral alterations with apoptosis and deregulation of PS1 in p53 knockout mice. (A) Morris swim maze test. (a) Dropped into the water, mice had to learn to navigate to the invisible platform. After the end of the learning sessions, mice were submitted to a probe test for the spatial bias. F, female; M, male; WT, wild type. The two-way ANOVA revealed a significant genotypic X sex interaction [F(1,43) = 11.63; P = 0.001; r2 = 0.26]. Scheffé post hoc test: Fp53−/− versus FWT = 3.25; P = 0.01; Fp53−/− versus Mp53−/− = 2.77; P = 0.043. p53−/− females show significantly inferior performance. (b) Open field test. Behavioral measurements were done during the first 5 min after the introduction of each mouse in the open field. The relative central activity was the number of central sector crossings expressed as the percentage of the total sector crossings. ANOVA for relative central activity: F(1,43) = 44.93; P < 0.001; r2 = 0.52. p53−/− mice have a significantly lower percentage of relative central activity. (B) Northern blot analysis (a–f) of PS1, p21waf1, and GAPDH in the brains of young (a–c) and adult (d–f) mice. (Lane 1) Control mouse. (Lanes 2 and 3) p53 knockout female and male. a–c show the same Northern blot hybridized with the different probes. For the adult mice, hybridization was also performed on the same (d–f) Northern blot. The parentheses around GAPDH and PS1 indicate the residual signals after probe stripping in nonstringent conditions. A strong repression of PS1 (d) and induction of p21waf1 (e) are noticed in the adult p53-deficient mice. Western blot analysis of protein extracts from the brains (g–i) of the same adult animals with anti-PS1, anti-p21waf1, and anti-β-tubulin antibodies. (Lane 1) Control mice. (Lanes 2 and 3) p53 knockout animals. Increased p21waf1 and decreased PS1 expression is seen in the p53-deficient mice. (C) Hematoxylin and eosin stain of the isocortex in a control (a) and a knockout mouse (b). Note the thinning of the isocortex (arrows) in the knockout mouse (b). Original magnification = ×6.6. (D) Quantification of pyknotic nuclei in control mice (bar 1, male; bar 2, female) and p53 knockout mice (bars 3 and 4, males; bars 5 and 6, females). (E) TUNEL and counterstaining by Harris hematoxylin in control (a) and knockout mouse (b). Two nuclei are TUNEL-positive (arrows). Original magnification = ×132. (F) Quantification of the TUNEL assay in control mice (bar 1, male; bar 2, female) and p53 knockout mice (bars 3 and 4, males; bars 5 and 6, females).
Aged p53-Deficient Mice Exhibit Increased p21waf1 Expression and Down-Regulation of PS1 in the Brain.
We next investigated whether the perturbations in central nervous system function in p53 knockout mice are correlated with changes in PS1 and p21waf1 expression in the brain, because both genes are regulated by wild-type p53 and have been reported to modulate apoptosis (7, 12–15). Two groups were analyzed: young 6-week-old to 2-month-old mice and adult 6- to 7-month-old mice. The first group of young animals consisted of new p53−/− offspring. The second one consisted of the same series of p53−/− mice previously tested for the Morris and open field, except those that died in the meantime from cancer. We found striking age-related differences. Young p53−/− animals possess the same levels of PS1 and p21waf1 mRNA as control mice (Fig. 2 Ba–Bc). In contrast, adult p53−/− mice show a dramatic repression of PS1 and increase in p21waf1 mRNA expression (Fig. 2 Bd–Bf). As seen in Fig. 2 Bg–Bi, PS1 protein expression was markedly decreased in the brains of p53 knockout mice. We also tested cDNA from the p53 knockout mice for mutations in the coding region of PS1; none were found (data not shown). These observations suggest that older p53 knockout mice overcompensate for the lack of p53 by overexpressing p21waf1 with consequent PS1 repression. Our previous studies indicated that increased expression of p21waf1 resulted in repression of PS1 with induction of apoptosis (7) and that inhibition of PS1 production by antisense cDNA induces apoptosis (7, 8). These findings support the notion that the decreased PS1 expression in the brain may cause defects in central nervous system function through augmentation of apoptosis.
The fact that young p53 knockout mice already have learning, memory, and behavioral alterations before any measurable decrease in PS1 levels is similar to what is found in neurodegenerative diseases. Abnormalities in behavior can occur in Alzheimer's disease patients long before the terminal stage of the disease in which neurodegeneration is observed (31). Furthermore, we could have easily missed a decrease in PS1 that was confined to a small territory of the brain but that already could have resulted in consequences on learning, memory, and behavior. Moreover, the mutant PS1 transgenic mice show significant levels of apoptosis only in the aged, 13-month-old animals (4). These data suggest a slowly progressive process in which abnormalities in memory and behavior occur before the down-regulation of PS1 becomes measurable. Conceivably, additional as-yet unknown molecular deregulations induced by p53 deficiency may also contribute to this process.
Apoptosis in the Brains of p53-Deficient Mice.
Histopathological analysis, combined with a TUNEL assay to evaluate apoptosis, was performed on the older animals with the PS1 down-regulation described above. Three of four animals showed no striking gross pathological abnormality. One knockout mouse (female) had more than 50% thinning of the isocortex, with a highly enlarged ventricle (Fig. 2Cb), as compared with the control animals (Fig. 2Ca). Defects in neural tube closure were described exclusively in female p53 knockout mice (24, 25). Interestingly, we find that memory was also significantly affected only in females. The thinning of the isocortex is probably not related to the anencephaly in utero described in the p53−/− females, because, in the former case, apoptotic cells are still present suggesting a progressive neurodegenerative process. Quantification of the pyknotic nuclei and TUNEL-positive cells showed apoptotic lesions (Fig. 2 D–F) in the brains of all of the p53 knockout mice.
In conclusion, although our previous in vitro experiments indicated that down-regulation of PS1 by p53, p21waf1, or antisense PS1 induces apoptosis, we presently demonstrate the antiapoptotic function of wild-type PS1 in both p53-dependent and p53-independent systems. The analysis of p53 knockout mice reveals an array of neurological defects associated with apoptotic lesions in the brain. These alterations may involve a long-term age-dependent overcompensation of p53 deficiency by increased p21waf1 expression, which leads to down-regulation of PS1. This down-regulation, in turn, compromises the antiapoptotic effect of PS1. Our findings indicate that the loss of PS1 function, through either mutations as seen in early onset familial Alzheimer's disease or deregulation of expression as seen in p53-deficient mice, eventually leads to cell death. Together with the data showing developmental abnormalities in the brains of p53-deficient mice, our study indicates that p53, besides its role in cancer prevention, also plays a crucial role in maintaining the integrity of the central nervous system.
Acknowledgments
A.T. and R.A. dedicate this work to Howard Cann for constant support and critical advice. We thank Laurence Piouffre for excellent technical help. We are mostly grateful to Collin Masters and Geneviève Evin for providing the 95/23 anti-PS1 antibody. We are grateful to Harvey Lodish, Barbara Osborne, Joseph Schlessinger, Hermann Steller, Bert Vogelstein, and Andrew Wyllie for critical review of the work and encouragement. We thank Brent Passer, Tim Allsopp, Laurent Pradier, Christian Czech, Marianne Colle, and Nicolas Privat for stimulating discussions and expert advice. We thank Bernard Boursin, Elizabeth Savariav, and Robin Nancel at the photography department, Hôpital St. Louis. We are indebted to Cécile Rouzaud and Pascale Villedieu for efficiency and help.
Abbreviations
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 2.Guo Q, Sopher B L, Furukawa K, Pham D G, Robinson N, Martin G M, Mattson M P. J Neurosci. 1997;17:4212–4222. doi: 10.1523/JNEUROSCI.17-11-04212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Q, Fu W, Sopher B L, Miller M W, Ware C B, Martin G M, Mattson M P. Nat Med. 1999;5:101–106. doi: 10.1038/4789. [DOI] [PubMed] [Google Scholar]
- 4.Chui D H, Tanahashi H, Ozawa K, Ikeda S, Checler F, Ueda O, Suzuki H, Araki W, Inoue H, Shirotani K, et al. Nat Med. 1999;5:560–564. doi: 10.1038/8438. [DOI] [PubMed] [Google Scholar]
- 5.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J S, Trumbauer M E, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–291. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 6.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 7.Roperch J-P, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron M-C, Israeli D, Dausset J, et al. Nat Med. 1998;4:835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- 8.Hong C S, Caromile L, Nomata Y, Mori H, Bredesen D E, Koo E H. J Neurosci. 1999;19:637–643. doi: 10.1523/JNEUROSCI.19-02-00637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray W J, Yao M, Nowotny P, Mumm J, Zhang W, Wu J Y, Kopan R, Goate A M. Proc Natl Acad Sci USA. 1999;96:3263–3268. doi: 10.1073/pnas.96.6.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imafuku I, Masaki T, Waragai M, Takeuchi S, Kawabata M, Hirai S, Ohno S, Nee L E, Lippa C F, Kanazawa I, et al. J Cell Biol. 1999;147:121–134. doi: 10.1083/jcb.147.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amson R, Nemani M, Roperch J-P, Israeli D, Bougueleret L, Le Gall I, Medhioub M, Linares-Cruz G, Lethrosne F, Pasturaud P, et al. Proc Natl Acad Sci USA. 1996;93:3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 14.Michieli P, Chedid M, Lin D, Pierce J H, Mercer W E, Givol D. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 15.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 16.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 17.Nemani M, Linares-Cruz G, Bruzzoni-Giovanelli H, Roperch J-P, Tuynder M, Bougueleret L, Cherif D, Medhioub M, Pasturaud P, Alvaro V, et al. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linares-Cruz G, Bruzzoni-Giovanelli H, Alvaro V, Roperch J-P, Tuynder M, Schoevaert D, Nemani M, Prieur S, Lethrosne F, Piouffre L, et al. Proc Natl Acad Sci USA. 1998;95:1131–1135. doi: 10.1073/pnas.95.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vito P, Lacana E, D'Adiamo L. Science. 1996;271:521–524. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 20.Wolozin B, Iwasaki K, Vito P, Ganjei J K, Lacanà E, Sunderland T, Zhao B, Kusiak J W, Wasco W, D'Adamio L. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 21.Czech C, Lesort M, Tremp G, Terro F, Blanchard V, Schombert B, Carpentier N, Dreisler S, Bonici B, Takashima A, et al. Neuroscience. 1998;87:325–336. doi: 10.1016/s0306-4522(98)00162-6. [DOI] [PubMed] [Google Scholar]
- 22.Roperch J-P, Lethrosne F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron M-C, Dausset J, et al. Proc Natl Acad Sci USA. 1999;96:8087–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 24.Sah V P, Attardi L D, Mulligan G J, Williams B O, Bronson R T, Jacks T. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong J F, Kaufman M H, Harrison D J, Clarke A R. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 26.Morris R G M. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 27.Chapillon P, Roull P. Dev Psychobiol. 1996;29:529–545. doi: 10.1002/(SICI)1098-2302(199609)29:6<529::AID-DEV5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 29.Archer J. Anim Behav. 1973;21:205–235. doi: 10.1016/s0003-3472(73)80065-x. [DOI] [PubMed] [Google Scholar]
- 30.Gershenfeld H K, Paul S M. Genomics. 1997;46:1–8. doi: 10.1006/geno.1997.5002. [DOI] [PubMed] [Google Scholar]
- 31.Duyckaerts C, Bennecib M, Grignon Y, Uchihara T, He Y, Piette F, Hauw J-J. Neurobiol Aging. 1997;18:267–273. doi: 10.1016/s0197-4580(97)80306-5. [DOI] [PubMed] [Google Scholar]