Abstract
Over the past few years, tissue microarray (TMA) technology has been established as a standard method for assessing the expression of proteins or genes across large sets of tissue specimens. It is being adopted increasingly among leading research institutions around the world and utilized in cancer research in parallel with the cDNA microarray technology. This article summarizes various aspects of cancer understanding and diagnostics in which TMA has had great impact. Although tremendous advances continue to be made to facilitate imaging and archiving of TMA specimens, automatic evaluation and quantitative analysis of TMA still remains an important challenge for modern investigators.
Keywords: Tissue microarray, Molecular profiling, Molecular marker quantification, Automatic tissue analysis
1. Introduction of tissue microarray technology
Tissue microarray (TMA) is a powerful new technology designed to assess the expression of proteins or genes across large sets of tissue specimens assembled on a single glass microscope slide, efficiently and economically [1–3].
Kononen's original TMA creation method extracts tissue cylinders originating from either formalin-fixed paraffin-embedded tumor tissue blocks or freshly frozen tumors fixed in cold ethanol and embedded in paraffin (to reserve RNA or DNA for detection), and precisely arrays them into a “recipient” paraffin block [1], please see Fig. 1. About 200 consecutive sections 4–8 μm in thickness can be cut from each tissue microarray block, with each stained differently to establish a large-scale gene expression profile of cancers. It has been demonstrated that TMA sections can be used to detect DNA (fluorescent in situ hybridization, FISH), RNA (mRNA in situ hybridization, RNA-ISH), or protein (immunohistochemistry, IHC). This new technique should not be confused with DNA microarrays, in which each tiny spot on the grid represents a unique cloned complementary DNA (cDNA) or oligonucleotide.
Fig. 1.
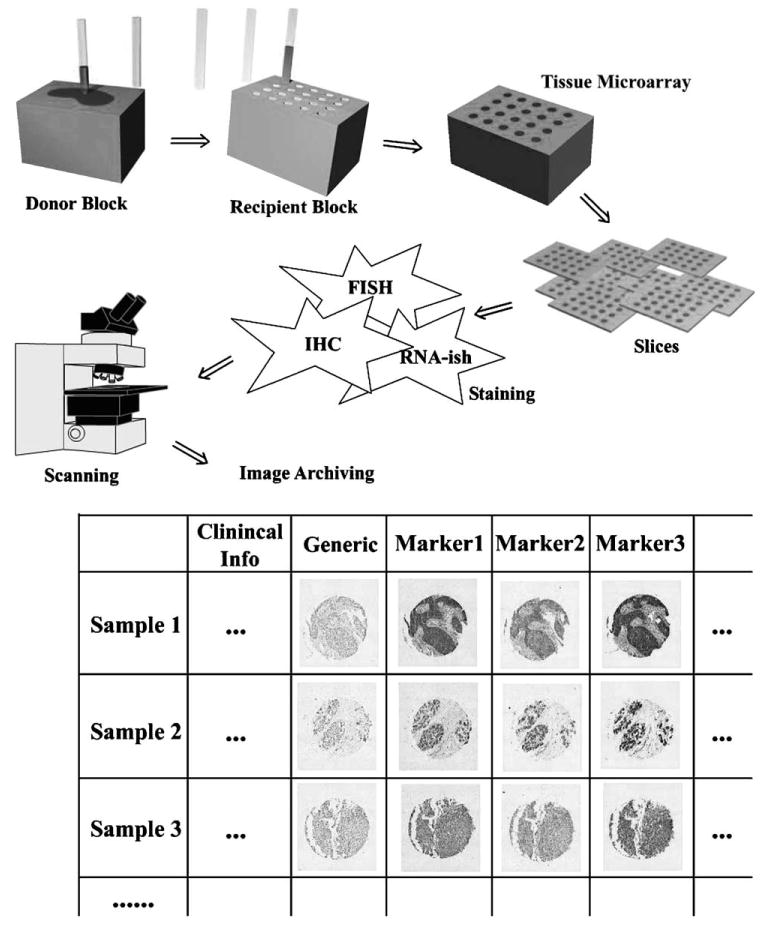
Tissue microarray creation, processing, imaging, and archiving.
Two of the most apparent advantages of TMA technology are that it allows amplification of limited tissue resources by providing the means for producing a large number of small core biopsies, rather than generating a single section, and that it saves time and antibody to analyze multiple specimens at once. Using this technology, a carefully planned array can be constructed with cases from pathology tissue block archives, such that a 20-year survival analysis can be performed on a cohort of 600 or more patients using only a few microliters of antibody. Another major advantage of the TMA technique is the fact that each specimen is treated in an identical manner. Consequently, reagent concentrations are consistent across discs within each TMA specimen, as are the incubation times, temperatures, and washing conditions. The staining results of these discs are hence comparable.
TMA technology continues to evolve and improve in order to simplify the process of constructing arrays [4–7], while accommodating a wider variety of specimens. Although the original TMA technique focused on profiling paraffin-embedded tissue specimens, Abbott et al. established a cell line microarray composed of cell pellets, which had been obtained from human tumor [8], and several groups reported success in creating TMAs from frozen needle biopsies or brain tissue [6,9–11]. Fergenbaum et al. reported their concern that the extended storage time for paraffin-embedded tissue might affect tissue's immunoreactivity and thereby giving rise to an increase in false-negative results in the corresponding TMAs [12]. Combined paraffin coating and nitrogen storage was reported by DiVito et al. [13] to be the best storage condition for preserving TMA antigenicity.
In response to questions posed by many investigators as to whether utilizing 0.6 mm diameter cores of tissue was sufficient in representing the heterogeneous morphology of any given tissue specimen, the TMA technology underwent tremendous scrutiny and was tested and validated repeatedly for use in cancer research over the period from 1998 to 2003. A number of research groups participated in this process by comparing TMA analysis with large tissue sections diagnosis and/or by validating its results with cDNA microarray findings in various cancer types: breast cancer [12,14–19]; prostate cancer [20–23]; gastric cancer [24]; colorectal cancer [25–27]; lymphomas [28–32]; multiple myeloma [33]; soft tissue sarcoma [34]; renal cell carcinoma [35]; bladder tumor [36]; glioma [37,38]; melanoma [39]; lung tumor [40,41]. It is now generally accepted that two to four samples taken from different regions of each donor tissue block provides a sufficient amount of morphologic information for reliably evaluating specimens [18,22,24,31,34,42]. In the case that the protein marker is expressed focally as opposed to homogeneously, the use of TMA is validated by Mucci et al., by increasing number of samples from each specimen [23]. TMA technology has also proven to be an excellent quality assurance tool that can be used to conduct inter-observer, intra-laboratory, and inter-laboratory studies [43–47].
2. TMA in cancer understanding and diagnosis
Although TMA is not currently used to render a primary diagnosis, it does provide unparalleled insight into cancer development, prognosis, and treatment.
TMAs have recently been accepted as a standard method for identifying or validating specific molecular markers associated with cancer [48–52]. Alpha-methylacyl coenzyme A racemase (AMACR) was first identified as a tissue biomarker for prostate cancer through cDNA microarray technology [53,54]. Confirmation of the discovery using TMA has helped to support its use in clinical practice [55–57].
Scientist used TMA to explore various proteins over a large set of specimens for their roles in tumorigenesis, cancer cell survival, and proliferation, metastasis, and tumor invasion [58–65]. Genetic variances have been examined against each other for their contribution in tumor development and progression [66,67]. Members of known signaling pathways were tested using TMA technology to confirm the contribution of these pathways in development of cancer [42,68–70]. For example, over a cohort of over 170 head and neck squamous cell carcinoma (HNSCC) specimens, inactivation of TGF-beta/Smad signaling was found in approximately 15–20% of the cases, and these Smad signaling defects were associated with a greater tendency for metastatic spread and regional or distant recurrence of HNSCC [70]. The loss of certain protein expression has also been successfully utilized to identify high risk patients [71,72].
TMA has also been used to identify possible therapeutic targets for a variety of malignancies [73–78]. Researchers have linked expression of certain markers to specific therapeutic outcomes [79,80].
Molecular profiling has been demonstrated to be a useful approach for providing new insight into cancer development and differentiation although the underlying mechanisms is not yet fully understood. Multivariate statistical analysis, including principal component analysis, hierarchical clustering analysis and other machine learning algorithms, were used in several of these efforts to elucidate the possible relationships between the expression profiles and biological or clinical parameters of the cases [81–88]. Using 13 protein markers, Callagy et al. revealed distinct tumor clusters which divided into two main groups correlating with tumor grade and nodal status, whereas none of the protein biomarkers that were tested could identify these groups individually [82]. Hierarchical clustering analysis applied to immunostaining results for 18 different markers showed that synovial sarcomas, leiomyosarcomas, hemangiopericytomas, and solitary fibrous tumors clustered distinctly [84]. Using principle component analysis to characterize salivary gland tumors resulted in three components using a panel of 15 markers [89].
It has also recently been discovered that, in addition to expression intensity, subcellular location of certain protein markers [90–94] is related to clinical outcome. Berger et al. conducted survival study that included 555 tumors collected from the Yale University Department of Pathology archives [95]. The results of the study indicated that subcellular localization of activating transcription factor 2 (ATF2) could be used as an independent prognostic indicator for melanoma. Subcellular localization of 14-3-3sigma and Ski-related novel protein N (SnoN) are also identified as prognostic factor for breast cancer [92,93].
3. TMA imaging, archiving, and information management
Currently, the primary methods used to evaluate the arrays involve manual, interactive review of TMA samples while they are subjectively evaluated and scored. An alternative, but less utilized approach for evaluation is to digitize each specimen, sequentially, for subsequent semi-quantitative assessment [96]. Both procedures ultimately involve the interactive localization of TMA discs under the microscope, which is a slow, tedious process that is prone to error. For many researchers and technicians, simply navigating among the regularly arranged tissue cores while viewing them under a microscope makes it difficult to keep track the current disc position within the array. This is especially problematic at high magnifications. Research groups and manufacturers have implemented several automatic approaches to image TMA discs for storage and evaluation.
Beyond the algorithmic and software development that is required for analyzing tissue microarrays, reliable tools are also needed to facilitate large-scale, multi-site collaboration for a broad spectrum of research and clinical activities including tissue banking, proteomics, and outcome studies. Future progress in several key areas will rely upon the capacity of individuals to acquire, share, and assess microarrays and correlated data, dynamically.
Although some DNA microarray readers are capable of reading TMA samples, automatic imaging, and evaluation of TMA present a host of unique technical challenges. First, TMA samples often exhibit a higher level of morphological irregularities. For example, aside from the overall rotation of the grid on the slide, tissue discs are sometimes shifted from regular grid positions as a result of mechanical deformations, which can arise during construction of the physical TMA. Such artifacts are often introduced during the sectioning stage of specimen preparation. In addition, discs occasionally become detached and fall out during preparation. To address these issues our group developed a web-based prototype which features automated imaging, registration, and distributed archiving of tissue microarrays in multi-user network environments, please see Fig. 1. The imaging system features a robust alignment algorithm that could reliably recover sample grids and compensate for detached discs [97].
Several other groups of investigators have also developed their own relational databases to manage TMA related information [97–102]. To facilitate exchange of TMA data by researchers across all these different database implementations, the Technical Standards Committee of the Association for Pathology Informatics organized a TMA workshops in order to establish a TMA data exchange specification. The proposed XML-based TMA data exchange specification [103,104] is free and non-proprietary.
While it is clear that much more research and development is needed to achieve reliable analysis of TMA, several commercial products that have already emerged for imaging and visualization of TMA including BLISS system by Bacus Laboratory (Lombard, IL); MedScan by Trestle (Irvine, CA); ScanScope by Aperio Technologies (Vista, CA); Ariol by Applied Imaging (San Jose, CA).
4. Quantitative analysis of TMA
Gene expression studies using cDNA microarrays have created an unprecedented need for the development and introduction of new analytical tools. Similarly, there now exists an urgent need for tools to analyze TMA specimens efficiently and effectively. However, despite the many commonalities between cDNA microarrays and TMA's, there is one important distinction which makes the analysis of tissue microarrays especially challenging. Unlike cDNA microarrays which can generally be considered homogeneous across a given spot, the discs comprising an array typically consist of a set of heterogeneous, stained tissues which essentially renders simple image analysis strategies completely inadequate in performing any meaningful assessment. The analysis is further complicated by the fact that, depending on the type of tumor or tissue section analyzed, the area of interest may represent nearly the entire disc or only a small percentage thereof (please see examples in Figs. 1 and 2). For example, a pancreatic carcinoma or lobular carcinoma of the breast with substantial dysplastic response may show stromal tissue representing a large percentage of the total area of the disc. If the goal of the assay is to determine epithelial cell expression of a given marker, a protocol must be used that evaluates only that component of the disc. The protocol must not only be able to identify the region of interest but it must also perform normalization operations so that the expression level for any given disc can be reliably compared with the level reported for others.
Fig. 2.
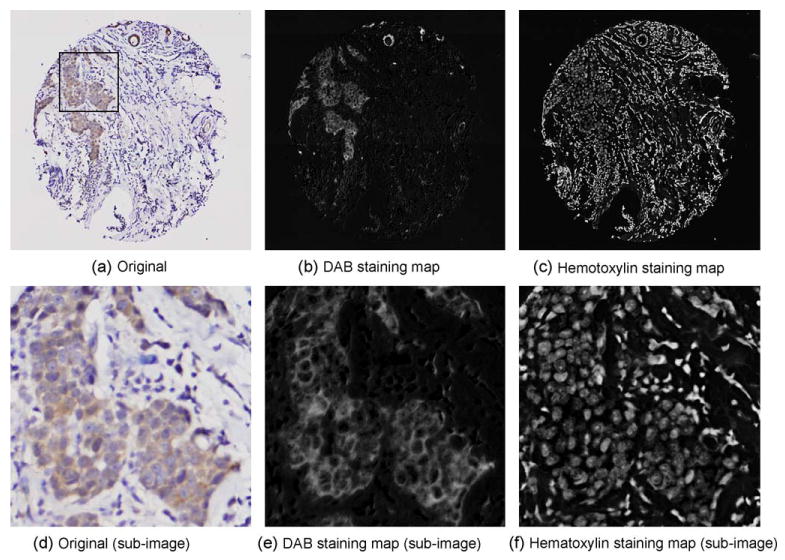
Example of color decomposition results. (a) The original tissue disc image stained by DAB and hematoxylin, showing heterogeneity of tissue. (b) The DAB staining map derived from (a). (c) The hematoxylin staining map derived from (a). (d) A sub-image of (a), as shown, enlarged. (e) The DAB staining map of (d). (f) The hematoxylin staining map of (d).
For these reasons, although a few groups made attempts to read TMA specimens using commercial cDNA microarray readers [105,106], developing reliable and effective methods and protocols specifically for quantitative analysis of tissue microarrays has become an extremely active research area since 2001 [94,107,108]. Since fluorescent TMA's and classic immunohistochemistry (IHC) TMA's each require specific hardware, most of the methods proposed to evaluate these specimens are only suitable for one type of processing.
4.1. Quantitative analysis of immunohistochemistry specimens
IHC is widely used for a broad number of applications throughout both the clinical and biomedical research communities. Research efforts focusing on automated quantification of IHC began with traditional biopsy specimens, but are now also being investigated for potential use in analyzing tissue microarrays. Similar to the TMA quantification, one of the most important tasks in quantification of IHC specimens is to generate tissue-specific or tumor-specific measurements. Another challenge of IHC quantification is how best to normalize the color differences that exist among specimens that have been acquired, stored, and processed under different conditions. Since TMA tissue cores are processed and stained on one single slide, tissue discs within a given slide can generally be considered as having already being normalized, making IHC TMA an ideal platform for developing and validating IHC quantification methods.
Several successful studies based on computerized IHC analysis have been reported including applications involving cell counting [109,110], microvessel counting [111], and cell morphology analysis [109]. There now also exist multiple commercial products which are aimed at quantitative IHC analysis such as the ChromaVision Automated Cellular Imaging System (ChromaVision Medical Systems Inc., San Juan Capistrano, CA) [112], the Image Tiler (Tripath Imaging, Burlington, NC) [113], and QPRODIT (Leica Imaging Systems Ltd., Cambridge, UK) [109], TMAscore (Bacus Instruments, Lombard, IL), and TMAx (Breecher Instruments, Sun Prairie, WI). Most of these approaches rely upon manual delineation of the region of interest (ROI) before analysis can begin [109,114].
4.2. Quantification of fluorescent TMA
Rao et al. developed a TMA-based quantitative fluorescence image analysis (QFIA) method and validated its use by analyzing distinctive BRCA1 expression patterns in high-grade and low-grade sporadic epithelial ovarian cancers and their adjacent dysplastic fields [106]. However, his system could not automatically distinguish epithelial tissue from surroundings, instead it depended on manual segmentation to locate regions of interest.
Haedicke et al. [105] used a cDNA microarray two-color laser scanner to evaluate fluorescent TMA specimens. In his unique approach using double-label indirect immunofluorescence, the target protein expression signal was normalized by a second fluorescent label, cytokeratin, on the same TMA specimen. Since a cDNA microarray scanner only measures total signal strength from each core, his method was based on a debatable underlying assumption, which is uniform expression of cytokeratin in epithelial tissue.
Fluorescent multi-labeling technique was also used in the Automated Quantitative Analysis (AQUA) system developed by Camp et al. to detect target protein subcellular-localization in TMA specimens [55,94,108]. This method automatically localized tumor cells and subcellular compartments using three specific tags: anti-cytokeratin for tumor cells, DAPI for nuclei, and α-catenin for membrane. The resulting co-localization masks were subsequently used to measure compartment-specific expression of estrogen receptor (ER) and β-catenin, which were visualized using DAB.
The capacity to co-localize multiple targets is the most important advantage of using multi-fluorescent-labeling in immunohistochemistry analysis. However, this approach also has its limitations. Developing appropriate multi-labeling staining protocols is complicated, time-consuming, and expensive. Fluorochromes used in these protocols must be carefully selected according to their excitation and emission wavelength in order to be effective.
4.3. Quantification of light microscopy IHC TMA
Light microscopy IHC assays still remain the most utilized techniques in diagnostic pathology when establishing protein expression status at the tissue level. Such assays can be conducted relatively inexpensively during routine processing without the need for specialized equipment. Theses approaches are also widely used in TMA research.
Although some commercial IHC quantification systems were quickly adapted to support TMA analysis, the majority cannot distinguish between tumor cells and non-malignant tissue components such as stroma or inflammation, in their analysis.
Elie et al. attempted automatic tissue specific nuclei quantification [115]. Their method for delineation of epithelial from stromal compartments was based on intensity difference in the green color channel, which is not necessarily applicable to other IHC specimens. Lacking the technology to un-mix multiple stains on a given IHC specimen, they generated their measurements by manipulating RGB color channels while approximately isolating DAB stained colors.
We have developed the principal color decomposition method to detect and characterize the staining signatures for each dye within the specimen based upon vector decomposition in L*u*v* color space [97]. The method explored the fact that color vectors that are generated for a representative TMA specimen stained with DAB and hematoxylin distributes along a hyperplane in L*u*v* color space. By performing a polar transformation of the L*u*v* color space plot and identifying the two principal peak colors in this representation, the principal color vectors are determined. Therefore, the staining characteristics of each tissue disc appear as a continuous representation of staining intensity for each of the dyes. It is interesting to note that the protocol that we have reported is able to unveil and quantify the underlying staining characteristics of even those cells that suffer from visual masking due to the counterstaining, please see Fig. 2. We also explored the use of the color decomposition algorithms for a broader set of immunohistochemical staining agents. Recent feasibility studies showed that this approach was also effective in analyzing histological specimens stained with Nova Red and Vector Red against hematoxylin.
5. Conclusion
The power and efficiency of TMA technology rests in its capacity to provide investigators with a means for evaluating gene expression in tissue or cell lines while processing large cohorts of specimens simultaneously. Due to the heterogeneous and complex nature of histological samples, the staining characteristics of TMA specimens can be described and quantified at multiple levels of detail. The most common approaches for assessment have focused primarily on determining the overall staining intensity of the individual tissue discs comprising the arrays. At the tissue level, however, this overall staining intensity can be normalized according to the amount of tissue which is present each disc. The next two levels of detail are at the cellular and subcellular resolution where the assessment can provide even greater insight related to the marker distribution in vivo. A tremendous amount of research efforts are targeting these levels of detail in order to promote better understanding of the underlying mechanisms of disease progression. This level of detailed analyses accentuates the need for higher standards for TMA imaging resolution and data management solutions. It is anticipated that the advances in TMA image analysis will also serve to impact the manner in which routine diagnostic IHC specimens are imaged, archived, and analyzed.
Although it is not anticipated that TMA's will have direct clinical application in the near future, incremental steps are being taken by the clinical and research communities to design, develop, and evaluate the computational tools which are necessary for performing reliable, reproducible comparative analysis of expression patterns in arrayed tissues. As these tools become increasingly reliable they will have tremendous potential in the areas of drug discovery, clinical outcomes, and therapy planning.
Acknowledgments
The research of the authors mentioned in this review is supported, in part, by a grant from The Cancer Institute of New Jersey and NIH contract 5R01LM007455-03 from the National Library of Medicine and 1R01EB003587-01A2 from the National Institute on Biomedical Imaging and Bioengineering.
References
- 1.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Nat Med. 1998;4:844. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 2.Simon R, Mirlacher M, Sauter G. Biotechniques. 2004;36:98. doi: 10.2144/04361RV01. [DOI] [PubMed] [Google Scholar]
- 3.Rimm DL, Camp RL, Charette LA, Costa J, Olsen DA, Reiss M. Cancer J. 2001;7:24. [PubMed] [Google Scholar]
- 4.Dan HL, Zhang YL, Zhang Y, Wang YD, Lai ZS, Yang YJ, Cui HH, Jian YT, Geng J, Ding YQ, Guo CH, Zhou DY. World J Gastroenterol. 2004;10:579. doi: 10.3748/wjg.v10.i4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matysiak BE, Brodzeller T, Buck S, French A, Counts C, Boorsma B, Datta MW, Kajdacsy-Balla AA. Appl Immunohistochem Mol Morphol. 2003;11:269. doi: 10.1097/00129039-200309000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Datta MW, Kahler A, Macias V, Brodzeller T, Kajdacsy-Balla A. Appl Immunohistochem Mol Morphol. 2005;13:96. doi: 10.1097/00129039-200503000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Hidalgo A, Pina P, Guerrero G, Lazos M, Salcedo M. J Clin Pathol. 2003;56:144. doi: 10.1136/jcp.56.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott RT, Tripp S, Perkins SL, Elenitoba-Johnson KS, Lim MS. Mod Pathol. 2003;16:607. doi: 10.1097/01.MP.0000067423.83712.74. [DOI] [PubMed] [Google Scholar]
- 9.Fejzo MS, Slamon DJ. Am J Pathol. 2001;159:1645. doi: 10.1016/S0002-9440(10)63011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoos A, Cordon-Cardo C. Lab Invest. 2001;81:1331. doi: 10.1038/labinvest.3780347. [DOI] [PubMed] [Google Scholar]
- 11.Kylaniemi M, Koskinen M, Karhunen P, Rantala I, Peltola J, Haapasalo H. Neuropathol Appl Neurobiol. 2004;30:39. doi: 10.1046/j.0305-1846.2003.00502.x. [DOI] [PubMed] [Google Scholar]
- 12.Fergenbaum JH, Garcia-Closas M, Hewitt SM, Lissowska J, Sakoda LC, Sherman ME. Cancer Epidemiol Biomarkers Prev. 2004;13:667. [PubMed] [Google Scholar]
- 13.DiVito KA, Charette LA, Rimm DL, Camp RL. Lab Invest. 2004;84:1071. doi: 10.1038/labinvest.3700131. [DOI] [PubMed] [Google Scholar]
- 14.Ginestier C, Charafe-Jauffret E, Bertucci F, Eisinger F, Geneix J, Bechlian D, Conte N, Adelaide J, Toiron Y, Nguyen C, Viens P, Mozziconacci MJ, Houlgatte R, Birnbaum D, Jacquemier J. Am J Pathol. 2002;161:1223. doi: 10.1016/S0002-9440(10)64399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi OP, Sauter G. Am J Pathol. 2001;159:2249. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van den Eynden GG, Van der Auwera I, Van Laere S, Colpaert CG, van Dam P, Merajver S, Kleer CG, Harris AL, Van Marck EA, Dirix LY, Vermeulen PB. Breast Cancer Res Treat. 2004;85:13. doi: 10.1023/B:BREA.0000021028.33926.a8. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Salto-Tellez M, Putti TC, Do E, Koay ES. Mod Pathol. 2003;16:79. doi: 10.1097/01.MP.0000047307.96344.93. [DOI] [PubMed] [Google Scholar]
- 18.Camp RL, Charette LA, Rimm DL. Lab Invest. 2000;80:1943. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 19.Gancberg D, Di Leo A, Rouas G, Jarvinen T, Verhest A, Isola J, Piccart MJ, Larsimont D. J Clin Pathol. 2002;55:315. doi: 10.1136/jcp.55.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merseburger AS, Kuczyk MA, Serth J, Bokemeyer C, Young DY, Sun L, Connelly RR, McLeod DG, Mostofi FK, Srivastava SK, Stenzl A, Moul JW, Sesterhenn IA. Oncol Rep. 2003;10:223. [PubMed] [Google Scholar]
- 21.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. Nature. 2002;419:624. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 22.Rubin MA, Dunn R, Strawderman M, Pienta KJ. Am J Surg Pathol. 2002;26:312. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 23.Mucci NR, Akdas G, Manely S, Rubin MA. Hum Pathol. 2000;31:406. doi: 10.1053/hp.2000.7295. [DOI] [PubMed] [Google Scholar]
- 24.Gulmann C, Butler D, Kay E, Grace A, Leader M. Histopathology. 2003;42:70. doi: 10.1046/j.1365-2559.2003.01556.x. [DOI] [PubMed] [Google Scholar]
- 25.Hendriks Y, Franken P, Dierssen JW, De Leeuw W, Wijnen J, Dreef E, Tops C, Breuning M, Brocker-Vriends A, Vasen H, Fodde R, Morreau H. Am J Pathol. 2003;162:469. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jourdan F, Sebbagh N, Comperat E, Mourra N, Flahault A, Olschwang S, Duval A, Hamelin R, Flejou JF. Virchows Arch. 2003;443:115. doi: 10.1007/s00428-003-0833-z. [DOI] [PubMed] [Google Scholar]
- 27.Fernebro E, Dictor M, Bendahl PO, Ferno M, Nilbert M. Arch Pathol Lab Med. 2002;126:702. doi: 10.5858/2002-126-0702-EOTTMT. [DOI] [PubMed] [Google Scholar]
- 28.Zettl A, Meister S, Katzenberger T, Kalla J, Ott MM, Muller-Hermelink HK, Ott G. Histopathology. 2003;43:209. doi: 10.1046/j.1365-2559.2003.01702.x. [DOI] [PubMed] [Google Scholar]
- 29.Tzankov A, Zimpfer A, Lugli A, Krugmann J, Went P, Schraml P, Maurer R, Ascani S, Pileri S, Geley S, Dirnhofer S. J Pathol. 2003;199:201. doi: 10.1002/path.1279. [DOI] [PubMed] [Google Scholar]
- 30.Garcia JF, Camacho FI, Morente M, Fraga M, Montalban C, Alvaro T, Bellas C, Castano A, Diez A, Flores T, Martin C, Martinez MA, Mazorra F, Menarguez J, Mestre MJ, Mollejo M, Saez AI, Sanchez L, Piris MA. Blood. 2003;101:681. doi: 10.1182/blood-2002-04-1128. [DOI] [PubMed] [Google Scholar]
- 31.Hedvat CV, Hegde A, Chaganti RS, Chen B, Qin J, Filippa DA, Nimer SD, Teruya-Feldstein J. Hum Pathol. 2002;33:968. doi: 10.1053/hupa.2002.127438. [DOI] [PubMed] [Google Scholar]
- 32.Rassidakis GZ, Jones D, Thomaides A, Sen F, Lai R, Cabanillas F, McDonnell TJ, Medeiros LJ. Am J Clin Pathol. 2002;118:328. doi: 10.1309/HKMV-VMPP-0CH8-3DPQ. [DOI] [PubMed] [Google Scholar]
- 33.Natkunam Y, Warnke RA, Montgomery K, Falini B, van De Rijn M. Mod Pathol. 2001;14:686. doi: 10.1038/modpathol.3880373. [DOI] [PubMed] [Google Scholar]
- 34.Engellau J, Akerman M, Anderson H, Domanski HA, Rambech E, Alvegard TA, Nilbert M. Appl Immunohistochem Mol Morphol. 2001;9:358. doi: 10.1097/00129039-200112000-00012. [DOI] [PubMed] [Google Scholar]
- 35.Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, Sauter G. Am J Pathol. 1999;154:981. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nocito A, Bubendorf L, Maria Tinner E, Suess K, Wagner U, Forster T, Kononen J, Fijan A, Bruderer J, Schmid U, Ackermann D, Maurer R, Alund G, Knonagel H, Rist M, Anabitarte M, Hering F, Hardmeier T, Schoenenberger AJ, Flury R, Jager P, Luc Fehr J, Schraml P, Moch H, Mihatsch MJ, Gasser T, Sauter G. J Pathol. 2001;194:349. doi: 10.1002/1096-9896(200107)194:3<349::AID-PATH887>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 37.Fuller CE, Wang H, Zhang W, Fuller GN, Perry A. J Neuropathol Exp Neurol. 2002;61:1078. doi: 10.1093/jnen/61.12.1078. [DOI] [PubMed] [Google Scholar]
- 38.Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J. Cancer Res. 2000;60:6617. [PubMed] [Google Scholar]
- 39.Pacifico MD, Grover R, Richman P, Daley F, Wilson GD. Melanoma Res. 2004;14:39. doi: 10.1097/00008390-200402000-00006. [DOI] [PubMed] [Google Scholar]
- 40.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Nat Med. 2002;8:816. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 41.Leversha MA, Fielding P, Watson S, Gosney JR, Field JK. J Pathol. 2003;200:610. doi: 10.1002/path.1374. [DOI] [PubMed] [Google Scholar]
- 42.Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M. Cancer Res. 2002;62:497. [PubMed] [Google Scholar]
- 43.van de Rijn M, Gilks CB. Histopathology. 2004;44:97. doi: 10.1111/j.1365-2559.2004.01766.x. [DOI] [PubMed] [Google Scholar]
- 44.Parker RL, Huntsman DG, Lesack DW, Cupples JB, Grant DR, Akbari M, Gilks CB. Am J Clin Pathol. 2002;117:723. doi: 10.1309/PEF8-GL6F-YWMC-AG56. [DOI] [PubMed] [Google Scholar]
- 45.Hsu FD, Nielsen TO, Alkushi A, Dupuis B, Huntsman D, Liu CL, van de Rijn M, Gilks CB. Mod Pathol. 2002;15:1374. doi: 10.1097/01.MP.0000039571.02827.CE. [DOI] [PubMed] [Google Scholar]
- 46.Mengel M, Kreipe H, von Wasielewski R. Appl Immunohistochem Mol Morphol. 2003;11:261. doi: 10.1097/00129039-200309000-00010. [DOI] [PubMed] [Google Scholar]
- 47.Diaz LK, Gupta R, Kidwai N, Sneige N, Wiley EL. J Histochem Cytochem. 2004;52:501. doi: 10.1177/002215540405200408. [DOI] [PubMed] [Google Scholar]
- 48.Han JH, Kang Y, Shin HC, Kim HS, Kang YM, Kim YB, Oh SY. Arch Pathol Lab Med. 2003;127:1330. doi: 10.5858/2003-127-1330-MEILNI. [DOI] [PubMed] [Google Scholar]
- 49.Hofer MD, Kuefer R, Varambally S, Li H, Ma J, Shapiro GI, Gschwend JE, Hautmann RE, Sanda MG, Giehl K, Menke A, Chinnaiyan AM, Rubin MA. Cancer Res. 2004;64:825. doi: 10.1158/0008-5472.can-03-2755. [DOI] [PubMed] [Google Scholar]
- 50.Kielhorn E, Provost E, Olsen D, D'Aquila TG, Smith BL, Camp RL, Rimm DL. Int J Cancer. 2003;103:652. doi: 10.1002/ijc.10893. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Ballesteros E, Mollejo M, Rodriguez A, Camacho FI, Algara P, Martinez N, Pollan M, Sanchez-Aguilera A, Menarguez J, Campo E, Martinez P, Mateo M, Piris MA. Blood. 2005;106:1831. doi: 10.1182/blood-2004-10-3898. [DOI] [PubMed] [Google Scholar]
- 52.Rakha EA, Abd El Rehim D, Pinder SE, Lewis SA, Ellis IO. Histopathology. 2005;46:685. doi: 10.1111/j.1365-2559.2005.02156.x. [DOI] [PubMed] [Google Scholar]
- 53.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. JAMA. 2002;287:1662. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 54.Jiang Z, Woda BA, Rock KL, Xu Y, Savas L, Khan A, Pihan G, Cai F, Babcook JS, Rathanaswami P, Reed SG, Xu J, Fanger GR. Am J Surg Pathol. 2001;25:1397. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 55.Rubin MA, Zerkowski MP, Camp RL, Kuefer R, Hofer MD, Chinnaiyan AM, Rimm DL. Am J Pathol. 2004;164:831. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang Z, Woda BA, Wu CL, Yang XJ. Am J Clin Pathol. 2004;122:275. doi: 10.1309/EJUY-UQPE-X1MG-68MK. [DOI] [PubMed] [Google Scholar]
- 57.Harding A. The Scientist. 2005;11:26. [Google Scholar]
- 58.Cao Y, Becker C, Lundwall A, Christensson A, Gadaleanu V, Lilja H, Bjartell A. Prostate. 2003;57:196. doi: 10.1002/pros.10296. [DOI] [PubMed] [Google Scholar]
- 59.Chaib H, Rubin MA, Mucci NR, Li L, Taylor JMG, Day ML, Rhim JS, Macoska JA. Cancer Res. 2001;61:2390. [PubMed] [Google Scholar]
- 60.Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, Huang M, Wang Y, Wu MC, Huang JF, Zeng WF, Sham JS, Yang M, Guan XY. Oncogene. 2004;23:298. doi: 10.1038/sj.onc.1206483. [DOI] [PubMed] [Google Scholar]
- 61.Ivan D, Diwan AH, Esteva FJ, Prieto VG. Mod Pathol. 2004;17:811. doi: 10.1038/modpathol.3800123. [DOI] [PubMed] [Google Scholar]
- 62.Dai DL, Makretsov N, Campos EI, Huang C, Zhou Y, Huntsman D, Martinka M, Li G. Clin Cancer Res. 2003;9:4409. [PubMed] [Google Scholar]
- 63.Fields AC, Cotsonis G, Sexton D, Santoianni R, Cohen C. Mod Pathol. 2004;17:1378. doi: 10.1038/modpathol.3800203. [DOI] [PubMed] [Google Scholar]
- 64.Jacobsen J, Grankvist K, Rasmuson T, Bergh A, Landberg G, Ljungberg B. BJU Int. 2004;93:297. doi: 10.1111/j.1464-410x.2004.04605.x. [DOI] [PubMed] [Google Scholar]
- 65.Toncheva DI, Zaharieva BM. Urol Int. 2003;71:408. doi: 10.1159/000074095. [DOI] [PubMed] [Google Scholar]
- 66.Haven CJ, van Puijenbroek M, Karperien M, Fleuren GJ, Morreau H. J Pathol. 2004;202:86. doi: 10.1002/path.1489. [DOI] [PubMed] [Google Scholar]
- 67.Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H. Am J Hum Genet. 2002;71:432. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al-Aynati MM, Radulovich N, Riddell RH, Tsao MS. Clin Cancer Res. 2004;10:1235. doi: 10.1158/1078-0432.ccr-03-0087. [DOI] [PubMed] [Google Scholar]
- 69.Altomare DA, Wang HQ, Skele KL, Rienzo AD, Klein-Szanto AJ, Godwin AK, Testa JR. Oncogene. 2004;23:5853. doi: 10.1038/sj.onc.1207721. [DOI] [PubMed] [Google Scholar]
- 70.Xie W, Bharathy S, Kim D, Haffty BG, Rimm DL, Reiss M. Oncol Res. 2003;14:61. doi: 10.3727/000000003108748612. [DOI] [PubMed] [Google Scholar]
- 71.Boltze C. Pathol Res Pract. 2005;200:783. doi: 10.1016/j.prp.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 72.Schneider-Stock R, Boltze C, Lasota J, Peters B, Corless CL, Ruemmele P, Terracciano L, Pross M, Insabato L, Di Vizio D, Iesalnieks I, Dirnhofer S, Hartmann A, Heinrich M, Miettinen M, Roessner A, Tornillo L. Clin Cancer Res. 2005;11:638. [PubMed] [Google Scholar]
- 73.Allander SV, Illei PB, Chen Y, Antonescu CR, Bittner M, Ladanyi M, Meltzer PS. Am J Pathol. 2002;161:1587. doi: 10.1016/S0002-9440(10)64437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dupont J, Wang X, Marshall DS, Leitao M, Hedvat CV, Hummer A, Thaler H, O'Reilly RJ, Soslow RA. Gynecol Oncol. 2004;94:449. doi: 10.1016/j.ygyno.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 75.Ferbus D, Bovin C, Validire P, Goubin G. J Pathol. 2003;200:177. doi: 10.1002/path.1337. [DOI] [PubMed] [Google Scholar]
- 76.Freedland SJ, Seligson DB, Liu AY, Pantuck AJ, Paik SH, Horvath S, Wieder JA, Zisman A, Nguyen D, Tso CL, Palotie AV, Belldegrun AS. Prostate. 2003;55:71. doi: 10.1002/pros.10202. [DOI] [PubMed] [Google Scholar]
- 77.Gray PJ, Jr, Bearss DJ, Han H, Nagle R, Tsao MS, Dean N, Von Hoff DD. Mol Cancer Ther. 2004;3:641. [PubMed] [Google Scholar]
- 78.July LV, Beraldi E, So A, Fazli L, Evans K, English JC, Gleave ME. Mol Cancer Ther. 2004;3:223. [PubMed] [Google Scholar]
- 79.Hu YC, Komorowski RA, Graewin S, Hostetter G, Kallioniemi OP, Pitt HA, Ahrendt SA. Clin Cancer Res. 2003;9:4165. [PubMed] [Google Scholar]
- 80.Huang Y, Sadee W. Drug Discov Today. 2003;8:356. doi: 10.1016/s1359-6446(03)02654-0. [DOI] [PubMed] [Google Scholar]
- 81.Alkushi A, Irving J, Hsu F, Dupuis B, Liu CL, Rijn M, Gilks CB. Virchows Arch. 2003;442:271. doi: 10.1007/s00428-002-0752-4. [DOI] [PubMed] [Google Scholar]
- 82.Callagy G, Cattaneo E, Daigo Y, Happerfield L, Bobrow LG, Pharoah PD, Caldas C. Diagn Mol Pathol. 2003;12:27. doi: 10.1097/00019606-200303000-00004. [DOI] [PubMed] [Google Scholar]
- 83.Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola J, van Diest PJ, Brandt B, Boecker W, Buerger H. Lab Invest. 2002;82:1525. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- 84.Nielsen TO, Hsu FD, O'Connell JX, Gilks CB, Sorensen PH, Linn S, West RB, Liu CL, Botstein D, Brown PO, van de Rijn M. Am J Pathol. 2003;163:1449. doi: 10.1016/S0002-9440(10)63502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Nat Med. 2002;8:68. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 86.Shi T, Seligson D, Belldegrun AS, Palotie A, Horvath S. Mod Pathol. 2005;18:547. doi: 10.1038/modpathol.3800322. [DOI] [PubMed] [Google Scholar]
- 87.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. Int J Cancer. 2005;116:340. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 88.Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, Aburatani H. Genomics. 2005;86:127. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 89.Iwafuchi H, Mori N, Takahashi T, Yatabe Y. Mod Pathol. 2004;17:803. doi: 10.1038/modpathol.3800122. [DOI] [PubMed] [Google Scholar]
- 90.Kang YK, Hong SW, Lee H, Kim WH. Hum Pathol. 2004;35:1340. doi: 10.1016/j.humpath.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 91.Berger AJ, Camp RL, Divito KA, Kluger HM, Halaban R, Rimm DL. Cancer Res. 2004;64:8767. doi: 10.1158/0008-5472.CAN-04-1384. [DOI] [PubMed] [Google Scholar]
- 92.Zhang F, Lundin M, Ristimaki A, Heikkila P, Lundin J, Isola J, Joensuu H, Laiho M. Cancer Res. 2003;63:5005. [PubMed] [Google Scholar]
- 93.Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Steele D, Cossu A, Budroni M, Palmieri G, Lakhani SR. J Pathol. 2004;202:274. doi: 10.1002/path.1530. [DOI] [PubMed] [Google Scholar]
- 94.Camp RL, Chung GG, Rimm DL. Nat Med. 2002;8:1323. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 95.Berger AJ, Kluger HM, Li N, Kielhorn E, Halaban R, Ronai Z, Rimm DL. Cancer Res. 2003;63:8103. [PubMed] [Google Scholar]
- 96.Bova GS, Parmigiani G, Epstein JI, Wheeler T, Mucci NR, Rubin MA. Hum Pathol. 2001;32:417. doi: 10.1053/hupa.2001.23517. [DOI] [PubMed] [Google Scholar]
- 97.Chen W, Reiss M, Foran DJ. IEEE Trans Inform Technol Biomed. 2004;8:89. doi: 10.1109/titb.2004.828891. [DOI] [PubMed] [Google Scholar]
- 98.Shaknovich R, Celestine A, Yang L, Cattoretti G. Arch Pathol Lab Med. 2003;127:492. doi: 10.5858/2003-127-0492-NRDFTM. [DOI] [PubMed] [Google Scholar]
- 99.Manley S, Mucci NR, De Marzo AM, Rubin MA. Am J Pathol. 2001;159:837. doi: 10.1016/S0002-9440(10)61759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coombes KR, Zhang L, Bueso-Ramos C, Brisbay S, Logothetis C, Roth J, Keating MJ, McDonnell TJ. Appl Bioinform. 2002;1:155. [PubMed] [Google Scholar]
- 101.Chen W, Foran DJ, Reiss M. Arch Pathol Lab Med. 2002;126:781. [Google Scholar]
- 102.Chen W, Foran DJ, Reiss M. Proc AMIA Symp. 2002;2002:136. [PMC free article] [PubMed] [Google Scholar]
- 103.Berman JJ, Edgerton ME, Friedman BA. BMC Med Inform Decis Mak. 2003;3:5. doi: 10.1186/1472-6947-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berman JJ, Datta M, Kajdacsy-Balla A, Melamed J, Orenstein J, Dobbin K, Patel A, Dhir R, Becich MJ. BMC Bioinformatics. 2004;5:19. doi: 10.1186/1471-2105-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haedicke W, Popper HH, Buck CR, Zatloukal K. Biotechniques. 2003;35:164. doi: 10.2144/03351md04. [DOI] [PubMed] [Google Scholar]
- 106.Rao J, Seligson D, Hemstreet GP. Biotechniques. 2002;32:924. doi: 10.2144/02324pt04. [DOI] [PubMed] [Google Scholar]
- 107.Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, Wheeler TM, Lu KP, Bao L. Cancer Res. 2003;63:6244. [PubMed] [Google Scholar]
- 108.Camp RL, Dolled-Filhart M, King BL, Rimm DL. Cancer Res. 2003;63:1445. [PubMed] [Google Scholar]
- 109.van Sandick JW, Baak JP, van Lanschot JJ, Polkowski W, ten Kate FJ, Obertop H, Offerhaus GJ. J Pathol. 2000;190:177. doi: 10.1002/(SICI)1096-9896(200002)190:2<177::AID-PATH508>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 110.Benali A, Leefken I, Eysel UT, Weiler E. J Neurosci Methods. 2003;125:33. doi: 10.1016/s0165-0270(03)00023-2. [DOI] [PubMed] [Google Scholar]
- 111.Wester K, Ranefall P, Bengtsson E, Busch C, Malmstrom PU. Br J Cancer. 1999;81:1363. doi: 10.1038/sj.bjc.6693399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang Z, Wu CL, Woda BA, Iczkowski KA, Chu PG, Tretiakova MS, Young RH, Weiss LM, Blute RD, Jr, Brendler CB, Krausz T, Xu JC, Rock KL, Amin MB, Yang XJ. Histopathology. 2004;45:218. doi: 10.1111/j.1365-2559.2004.01930.x. [DOI] [PubMed] [Google Scholar]
- 113.Nakabayashi T, Kumagai T, Yamauchi K, Sugano M, Kuramoto A, Fujita K, Hidaka H, Tozuka M. Am J Clin Pathol. 2001;115:424. doi: 10.1309/MATM-BCUL-96KL-FUCJ. [DOI] [PubMed] [Google Scholar]
- 114.Polkowski W, Baak JP, van Lanschot JJ, Meijer GA, Schuurmans LT, Ten Kate FJ, Obertop H, Offerhaus GJ. J Pathol. 1998;184:161. doi: 10.1002/(SICI)1096-9896(199802)184:2<161::AID-PATH971>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 115.Elie N, Plancoulaine B, Signolle JP, Herlin P. Cytometry. 2003;56A:37. doi: 10.1002/cyto.a.10075. [DOI] [PubMed] [Google Scholar]


