Abstract
Candida albicans ALS3 encodes a large cell-surface glycoprotein that has adhesive properties. Immunostaining of cultured C. albicans germ tubes showed that Als3p is distributed diffusely across the germ tube surface. Two-photon laser scanning microscopy of model catheter biofilms grown using a PALS3-green fluorescent protein (GFP) reporter strain showed GFP production in hyphae throughout the biofilm structure while biofilms grown using a PTPI1-GFP reporter strain showed GFP in both hyphae and yeast-form cells. Model catheter biofilms formed by an als3Δ/als3Δ strain were weakened structurally and had approximately half the biomass of a wild-type biofilm. Reintegration of a wild-type ALS3 allele restored biofilm mass and wild-type biofilm structure. Production of an Als3p-Agα1p fusion protein under control of the ALS3 promoter in the als3Δ/als3Δ strain restored some of the wild-type biofilm structural features, but not the wild-type biofilm mass. Despite its inability to restore wild-type biofilm mass, the Als3p-Agα1p fusion protein mediated adhesion of the als3Δ/als3Δ C. albicans strain to human buccal epithelial cells (BECs). The adhesive role of the Als3p N-terminal domain was further demonstrated by blocking adhesion of C. albicans to BECs with immunoglobulin reactive against the Als3p N-terminal sequences. Together, these data suggest that portions of Als3p that are important for biofilm formation may be different from those that are important in BEC adhesion, and that Als3p may have multiple functions in biofilm formation. Overexpression of ALS3 in an efg1Δ/efg1Δ strain that was deficient for filamentous growth and biofilm formation resulted in growth of elongated C. albicans cells, even under culture conditions that do not favour filamentation. In the catheter biofilm model, the ALS3 overexpression strain formed biofilm with a mass similar to that of a wild-type control. However, C. albicans cells in the biofilm had yeast-like morphology. This result uncouples the effect of cellular morphology from biofilm formation and underscores the importance of Als3p in biofilm development on silicone elastomer surfaces.
INTRODUCTION
Many different micro-organisms can form biofilms. Biofilms are a functional association of surface-attached microbial cells that become encased in extracellular matrix material (Costerton et al., 1987; Donlan & Costerton, 2002). Once committed to biofilm growth, microbial cells exhibit different phenotypic properties compared to free-floating, planktonic cells (Costerton et al., 1995). One clinically relevant consequence of biofilm growth is the development of resistance to antimicrobial compounds (Schwank et al., 1998; Wilson, 1996; Stewart et al. , 2004; Rupp, 2005). Among the fungi, Candida albicans is the most common organism associated with biofilm formation on bioprosthetic materials (reviewed by Douglas, 2003; Ghannoum & O’Toole, 2004).
In the search to identify C. albicans genes that are important for biofilm formation, investigators have assessed the effect of deleting single genes (Kelly et al., 2004; Krueger et al., 2004; Granger et al., 2005; Kumamoto, 2005), conducted systematic searches using collections of mutants (Richard et al., 2005), or studied transcriptional profiles using microarrays (Garcia-Sanchez et al., 2004; Cao et al., 2005; Murillo et al., 2005; K. M. Yeater and others, unpublished). The importance of transcription factors in biofilm formation has been analysed individually for factors such as Efg1p (Ramage et al., 2002b) or by assaying sets of mutant strains (Nobile & Mitchell, 2005). Some of these studies suggested that hyphae are important in biofilm formation and indicated that hypha surface proteins may be important inbiofilm formation (Ramage et al., 2002b; Nobile & Mitchell, 2005; Richard et al., 2005).
During the course of our studies of the C. albicans ALS (agglutinin-like sequence) gene family, we noted that strains in which ALS3 was deleted showed an obvious defect in biofilm formation on silicone elastomer surfaces. ALS3 expression was first associated with germ tube/hypha formation using Northern blot analysis (Hoyer et al., 1998a). Since then, the idea that high-level ALS3 expression is associated with germ tubes and hyphae has been supported by other methods including flow cytometry analysis of a PALS3-GFP (green fluorescent protein) reporter strain 2185 (Zhao et al., 2004; Green et al., 2005b), and real-time RT-PCR quantification of transcript copy number (Green et al., 2005b). Flow cytometry analysis of strain 2185 also showed that ALS3 transcriptional activity increases markedly when germ tubes become visible microscopically, rather than being induced immediately upon inoculation into fresh growth medium as is ALS1 (Zhao et al., 2004; Green et al., 2005b). ALS3 expression in model biofilms was demonstrated by RT-PCR (Green et al., 2004).
The focus of this paper is the role of Als3p in biofilm formation using the catheter biofilm model described by Kuhn et al. (2002). Here, we demonstrate the importance of Als3p in catheter biofilm formation and demonstrate how the Als3p functional domains important for biofilm formation may differ from those important for adhesion to human epithelial cells. We also address the relationship between biofilm formation and filamentous growth by demonstrating that overexpression of ALS3 in the biofilm- and filamentation-defective efg1Δ/efg1Δ strain confers wild-type biofilm mass without the production of hyphae. These results separate C. albicans morphology from biofilm formation and further substantiate the importance of Als3p in biofilm formation.
METHODS
Biofilm growth and measurement
Model catheter biofilms were grown on silicone elastomer discs according to the method of Kuhn et al. (2002). Biofilm mass was measured by dry weight analysis. Prior to incubation in fetal bovine serum, the autoclaved silicone elastomer discs were baked overnight in an 80 °C oven to remove residual moisture. Each disc was preweighed using sterile technique. After 48 h growth of the model biofilm, the medium in each well was removed, and the catheter discs were transferred onto Whatman chromatography paper, dried overnight at 80 °C and weighed immediately after removal from the oven. Biofilm dry weight was calculated by subtracting the preweight of each disc from the disc weight following biofilm growth and baking. Biofilms were grown in triplicate in three separate experiments. Means and SEM were calculated using the LSMEANS option in the MIXED procedure in the SAS/STAT software package (version 8; SAS Institute).
Two-photon laser scanning microscopy
Biofilms were fixed with 4 % paraformaldehyde in Dulbecco’s phosphate-buffered saline without calcium or magnesium (DPBS; Cambrex catalogue no. 17-512Q) for 1 h at room temperature. Yeast-enhanced GFP fluorescence was detected using a Bio-Rad Radiance 2100MP multiphoton laser scanning system (Bio-Rad Microscience) attached to a TE2000E microscope (Nikon USA) with a 60×1·2 NA plan-apochromat water immersion lens (Nikon). GFP was excited at 920 nm by a Mai-Tai titanium sapphire tunable laser system (Spectra-Physics). Biofilms grown from strains that did not encode GFP were stained with Calcofluor White M2R (Molecular Probes). Calcofluor was excited at 818 nm. Images were acquired using a non-descanned external direct detector PMT (Bio-Rad) and image stacks were captured with LaserSharp software (Bio-Rad). All images were processed using Adobe Photoshop software.
Creating serum immunoglobulin (Ig) preparations enriched for Als3p specificity
The Als5p N-terminal domain (Als5p18-329) was produced in Pichia pastoris and purified as described by Hoyer & Hecht (2001). Five hundred micrograms of Als5p18-329 was suspended in TiterMax adjuvant and injected subcutaneously into a New Zealand White Rabbit (Myrtle’s Rabbitry, Thompson Station, TN, USA). Subsequent inoculations consisted of 250 μg of the Als5p18-329 protein in Freund’s incomplete adjuvant and were administered 3 weeks after the previous injection. The rabbit was bled 7 days after every even-numbered inoculation and the anti-serum titre assessed by Western blotting against the Als5p18-329 protein. A total of eight protein inoculations were administered. A final inoculation was administered and the rabbit exsanguinated 7 days afterwards. Serum was collected and stored at -80 °C. The resulting antiserum titre was greater than 1: 60 000 as measured by Western blotting. Lipoproteins were removed from the antiserum by adding sodium dextran sulfate to 0·25 % (v/v) and then calcium chloride to 0·40 % (v/v). Following overnight incubation on ice and centrifugation, the supernatant was recovered and the gammaglobulin fraction precipitated by addition of an equal volume of saturated ammonium sulfate solution. Following overnight on ice and centrifugation, the pellet was resuspended in and dialysed exhaustively against DPBS. Total protein was measured by the Bradford dye-binding procedure (Bio-Rad). Aliquots of the gammaglobulin fraction were stored at -80 °C.
Enrichment of the Ig preparation for reactivity against Als3p was accomplished by absorption against the als3Δ/als3Δ strain 1843 (Table 1; Zhao et al., 2004). Strain 1843 was grown under conditions known to cause high-level transcription of other ALS genes, such as ALS1, which shares considerable sequence identity with ALS3 (Hoyer et al., 1998a; Green et al., 2005b). Conditions included overnight growth in YPD (yeast forms) and 1 h growth in YPD (yeast forms) or RPMI 1640 (RPMI; germ tubes). All incubations were at 37 °C and 200 r.p.m. shaking. All cells from each culture were combined, collected by centrifugation and washed in DPBS. Cells were resuspended in Complete, Mini Protease Inhibitor Cocktail (Roche) to which 1 μM pepstatin (Roche) was added. Three separate rounds of absorption were completed, each with approximately one-third of the cell preparation and end-over-end mixing. Two absorptions were completed at 4 °C for 1 h and the last at room temperature for 1 h. Cells were removed by centrifugation and the protein concentration measured. Aliquots of the anti-Als3p Ig preparation were frozen at -80 °C.
Table 1.
C. albicans strains used in this study
| Strain | Parent | Genotype* | Source |
|---|---|---|---|
| SC5314 | Wild-type | Gillum et al. (1984) | |
| CAI4 | SC5314 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 | Fonzi & Irwin (1993) |
| CAI12 | CAI4 | iro1-ura3Δ : : λimm434/IRO1 URA3 | Porta et al. (1999) |
| 1843 | 1780 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 als3laΔ/als3saΔ-URA3 | Zhao et al. (2004) |
| 2311 | 1843 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 als3laΔ/als3saΔ-ura3 | This study |
| 2322 | 2311 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 als3laΔ/als3saΔ-ALS3LA-URA3 | This study |
| 1926 | 1892 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 ALS3LA/als3saΔ-ura3 | This study |
| 2327 | 1926 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 ALS3-AGa1-URA3/als3saΔ-ura3 | This study |
| HLC52 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 efg1Δ : : hisG/efg1Δ : : hisG-URA3-hisG | Lo et al. (1997) | |
| HLC67 | HLC52 | iro1-ura3Δ : : λimm434/iro1-ura3Δ : : λimm434 efg1Δ : : hisG/efg1Δ : : hisG | Lo et al. (1997) |
| 2296 | HLC67 | HLC67 ALS3SA/ALS3LA : : 1105-PTPI1-ALS3LA-URA3 | This study |
| 2185 | CAI4 | CAI4 ALS3LA/als3saΔ-PALS3SA-GFP-URA3) | Zhao et al. (2004) |
| 1143 | CAI4 | CAI4 (RP10 : : 1105-PTPI1-GFP-URA3) | Green et al. (2005a) |
Indirect immunofluorescence
Indirect immunofluorescent detection of Als proteins on RPMI-grown germ tubes was described previously (Hoyer et al., 1998b). The anti-Als3p (1843-adsorbed) Ig preparation (described above) and normal rabbit IgG (ICN catalogue no. 641471; negative control) were used at 150 μg ml-1 concentration. Fluorescence was detected using a Nikon Eclipse E600 microscope fitted with a Spot camera (Diagnostic Instruments). Images of representative cells were collected using Metamorph software (Universal Imaging Corporation) and processed with Adobe Photoshop. The staining procedure was conducted on three different days with similar results on each day.
Flow cytometry analysis followed a published method (Zhao et al., 2004). Strains CAI12, 1843 and 2327 (see below) were combined with antibody following 30 min incubation in RPMI medium, which was sufficient to form small germ tubes. The anti-Als3p (1843-adsorbed) Ig preparation or normal rabbit IgG, diluted to 60 μg ml-1, was used. Incubations were conducted at either room temperature (normal goat serum block) or 4 °C (primary antibody and FITC-labelled secondary antibody) on an end-over-end mixer. Washing steps used DPBS. Fluorescence was detected by flow cytometry, using a Beckman Coulter EPICS XL, equipped with an argon laser with an excitation wavelength at 488 nm. Flow cytometry analysis of strains HLC52 and 2296 used yeast cells that were grown for 16 h in YPD at 37 °C and 200 r.p.m. shaking.
Reintegration of a wild-type ALS3 allele into an als3Δ/als3Δ strain
Construction and phenotypic evaluation of the als3Δ/als3Δ mutant strain 1843 (Table 1) were described previously (Zhao et al., 2004). In previous work, reintegration of a wild-type ALS3 allele into strain 1843 was complicated because removing the disruption cassette from the ALS3 large allele (ALS3LA) locus also removed several kb of sequence downstream of ALS3. Subsequent efforts finally yielded a strain (named 2311) with the correct construction (Table 1). The ALS3 reintegration cassette was constructed in plasmid pUL, which can be used to make constructs for reintegration of any of the ALS genes (Zhao et al., 2004). ALS3 downstream sequence was amplified from SC5314 genomic DNA using primers ALS3dnF and ALS3dnR (Table 2) and Pfu Turbo polymerase (Stratagene). The amplified fragment was cloned into the SstII/Ngo MIV sites of pUL, generating plasmid 2303. Full-length ALS3 from strain SC5314 [large allele, 12 tandem repeat copies, ALS3LA or ALS3(12), GenBank accession no. AY223552; Zhao et al., 2004; Oh et al., 2005] with 400 bp of upstream sequence was amplified from genomic DNA using the primer pair ALS3upF and ALS3R. This fragment was digested with AvrIIB and XhoI and cloned into AvrII-XhoI-cut plasmid 2303. The AvrII-Ngo MIV fragment, consisting of ALS3 upstream sequence-ALS3LA coding region-URA3-ALS3 downstream sequence, was transformed into strain 2311 and transformants selected as described previously (Zhao et al., 2004). The correct transformant was identified by Southern blotting and named 2322 (Table 1; Fig. 1).
Table 2.
Oligonucleotide primers used in this work
| Primer | Sequence (5′-3′) |
|---|---|
| ALS3dnF | CCC CCG CGG AAG AGC CTG CGA CTA TGA ATT G |
| ALS3dnR | CCC GCC GGC GTT TGG TAA TTA ACA CAT ATT GC |
| ALS3upF | CCC CCT AGG CAG TGA ATT GCA AAT CCT TAT GG |
| ALS3R | CCC CTC GAG CAA CAT TTT CCT TGG ACC TAC TAC |
| ALS3Sph | CCC CGC ATG CAT GCT ACA ACA ATA TAC ATT GTT ACT C |
| ALS3nt1290 | TTG GCA GTG GTA CCT TGT ACA ATG ACA GTG |
| AGα1Xba | CCC TCT AGA TTA GAA TAG CAG GTA CGA CAA AAG CAG |
| AGα1nt980 | CCT CGG TAC CGC TAG CGC CAA AAG CTC |
| 3cdHindIIIF | CCC AAG CTT ATG CTA CAA CAA TAT ACA TTG TTA CTC |
| AGα1XhoR | CCC CTC GAG TTA GAA TAG CAG GTA CGA CAA AAG CAG |
| RTAGα1F | CGT ATC GTC CCT CTC CGT A |
| RTAGα1R | TCC CTG AGA TGA GAG TGC TGT |
Fig. 1.
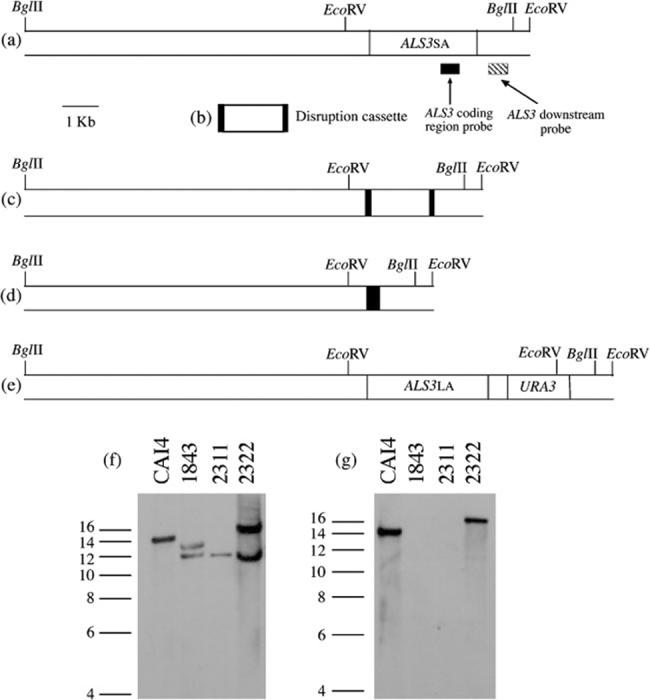
Reintegration of a wild-type ALS3 allele into a C. albicans als3Δ/als3Δ strain. The large allele of ALS3 [12 tandem repeat copies, called ALS3LA or ALS3(12), Gen-Bank accession no. AY223552; Zhao et al., 2004; Oh et al., 2005] was reintegrated into the C. albicans als3Δ/als3Δ mutant strain 1843 (Table 1). (a) Diagram of the ALS3 small allele [9 tandem repeat copies, called ALS3SA or ALS3(9), GenBank accession no. AY223551; Zhao et al., 2004; Oh et al., 2005] locus in strains derived from SC5314. Probes to detect the ALS3 coding region (Hoyer et al., 1998a) and the ALS3 downstream region (Zhao et al., 2004) are shown. (b) The PCR-amplified disruption cassette used to delete ALS3 alleles in strain 1843 (Zhao et al., 2004). (c) ALS3SA locus with the disruption cassette integrated as in strain 1843. (d) ALS3 small allele locus after selection of strain 1843 on 5-FOA to identify strain 2311. (e) ALS3SA locus with reintegrated copy of ALS3LA as in strain 2322. (f, g) Southern blots of BglII-digested genomic DNA from the strains indicated, probed with the ALS3 downstream fragment (f) or the ALS3 coding-region fragment (g). Molecular size (in kb) is indicated at the left of each image.
Construction of a C. albicans strain that produces an Als3p-Agα1p fusion protein on its germ tube surface
An inframe fusion between the 5′ end of ALS3 and the 3′ end of AGα1 (Lipke et al., 1989; Hauser & Tanner, 1989) was constructed in the Saccharomyces cerevisiae expression vector pYES2 (Invitrogen). Primers ALS3Sph and ALS3nt1290 (Table 2) were used to amplify a fragment encoding the N-terminal 430 aa of Als3p. The fragment encoding the C-terminal 323 aa of Agα1p was amplified from S. cerevisiae genomic DNA using primers AGα1Xba and AGα1nt980 (Table 2). The C-terminal half of Agα1p does not contain the adhesive domain (Cappellaro et al., 1994; Chen et al., 1995; de Nobel et al., 1996; Zhao et al., 2001) and shares similar cellular localization signals and amino acid composition with the C-terminal domains of Als proteins. The C-terminal half of Agα1p, therefore, provides a cell-surface-localized, highly glycosylated protein stalk that can display the Als3p N-terminal domain on the C. albicans cell surface. The ALS3 PCR product was digested with SphI/KpnI and the AGα1 PCR product was digested with KpnI/XbaI. These fragments were ligated into SphI/XbaI-cut pYES2 DNA and transformed into Escherichia coli TOP10F′ (Invitrogen) to form plasmid 881. Restriction mapping and DNA sequencing verified the construct. The ALS3-AGα1 fusion was amplified from plasmid 881 using primers 3cdHind IIIF and AGα1XhoR (Table 2). The PCR product was digested with Hind III/XhoI and ligated into similarly digested plas-mid 2303, generating plasmid 2326, which contained (5′-3′) the ALS3-AGα1 fusion-URA3-ALS3 downstream sequence between the HindIII and SstI sites. Plasmid 2326 was digested with HindIII/SstI and the fragment transformed into strain 1926 (iro1-ura3Δ:: λimm434/iro1-ura3Δ:: λimm434 ALS3LA/als3saΔ-ura3), from which the ALS3 small allele (ALS3SA) had been deleted previously. Integration of the fusion construct into the ALS3 large allele (ALS3LA) locus replaced the ALS3LA coding sequence, creating C. albicans strain 2327 (Table 1), which did not produce Als3p, but instead produced the Als3p-Agα1p fusion under control of the ALS3 promoter (Fig. 2). Southern blotting verified construction of clone 2327 (Fig. 2). The AGα1 probe was amplified by PCR using primers RTAGα1F and RTAGα1R (Table 2). The ALS3 downstream sequence probe was synthesized using primers ALS3dnF and ALS3dnR (Table 2). Growth rate of strain 2327 in YPD medium and germ tube formation in RPMI 1640 were measured according to published methods (Zhao et al., 2004) and were not significantly different from the wild-type strain CAI12. The presence of the Als3p N-terminal domain on the surface of strain 2327 germ tubes was verified by indirect immunolabelling using the anti-Als3p Ig preparation (see below) and flow cytometry (Green et al., 2005b). Flow cytometry readings for strain 2327 showed that production of the fusion protein increased surface fluorescence of the als3Δ/als3Δ strain to the level observed for the CAI12 wild-type control.
Fig. 2.
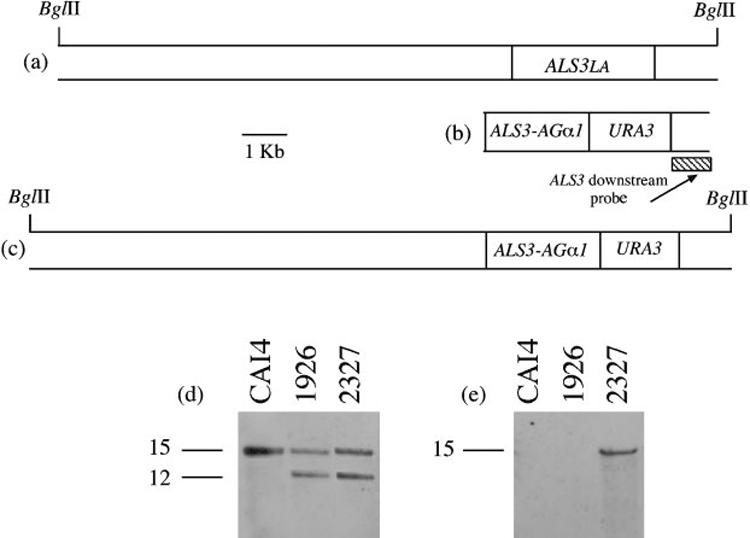
Construction and verification of a C. albicans als3Δ/als3Δ strain that produces an Als3p-Agα1p fusion protein under control of the ALS3 promoter. (a) ALS3LA locus in strains derived from SC5314. (b) Transformation fragment that includes the fused ALS3 and AGα1 coding regions and the URA3 selectable marker. The ALS3 down-stream probe fragment is shown. (c) Integration of the fusion construct into the ALS3LA locus. (d, e) Southern blots of BglII-digested genomic DNA hybridized with the ALS3 downstream probe (d) or an AGα1-specific fragment (e). Molecular size (in kb) is indicated at the left of each image.
Epithelial cell adhesion assays
The method for assaying C. albicans adhesion to pooled, fresh human buccal epithelial cells (BECs) was described previously (Zhao et al., 2004). Briefly, C. albicans cells were incubated in RPMI medium at 37 °C for 1 h to form germ tubes, buccal epithelial cells were added to the incubation flask and the incubation continued for an additional 30 min. Non-adherent fungal cells were separated from the BECs by filtration and washing over a 12 μm pore-size filter. Results were evaluated by counting the number of fungal cells adhered to each of 50 randomly selected BECs. Results were reported as the mean±SEM for each treatment. To assess the effects of the anti-Als3p Ig preparation on C. albicans adhesion to the BECs, 150 μg Ig (37·5 μgml-1 final concentration in the flask) were added to the RPMI medium 30 min after C. albicans germ tubes of wild-type strain CAI12 started to form. After an additional 30 min, the BECs were added to the culture flask and the assay completed as described above. Normal rabbit IgG was used as a negative control. The assay was conducted on three separate days.
Construction of C. albicans strains that overexpress ALS3
ALS3 overexpression used plasmid 1105 (Green et al., 2005a), which is a modified version of CIp10 (Murad et al., 2000). Plasmid 1105 encodes the C. albicans TPI1 promoter and terminator sequences, separated by a polylinker that includes restriction sites (5′-3′) XhoI-SmaI-NotI-BglII. The XhoI-BglII sites allow cloning for overexpression of any full-length ALS gene due to lack of these restriction sites in any of the ALS gene coding regions. These sites are used in various vectors in the laboratory and allow interchangeable cloning of ALS genes between the different constructs. ALS3LA (GenBank accession no. AY223552) from strain SC5314 was excised from previously built constructs and ligated into XhoI-BglII-cut plasmid 1105. The PTPI1-ALS3 overexpression construct was linearized with HindIII, which cuts once within the 3′ end of ALS3 to direct integration of the plasmid to the ALS3 locus in the efg1Δ/efg1Δ strain HLC52. The resulting strain, 2296, was verified by Southern blotting and showed integration of the plasmid at the ALS3LA locus (Fig. 3). The growth rate of strain 2296 was the same as the wild-type control, CAI12. Real-time RT-PCR analysis of ALS3 expression (Green et al., 2005b) in C. albicans yeast cells grown for 16 h in YPD at 37 °C showed that strain 2296 produced 15-foldmore ALS3 transcript than strain HLC52. Flow cytometry analysis of immunolabelled yeast cells of strains HLC52 and 2296 indicated a shifted fluorescence peak consistent with greater surface expression of Als3p epitopes (Fig. 3).
Fig. 3.
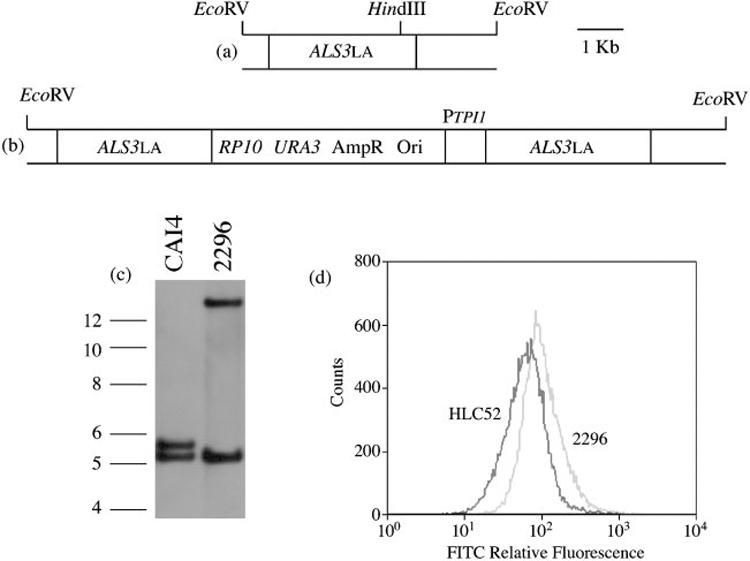
Construction and verification of a C. albicans efg1Δ/efg1Δ strain that overproduces Als3p under control of the constitutive TPI1 promoter. (a) The ALS3LA locus in strains derived from SC5314, such as the efg1Δ/efg1Δ strain HLC52 (Lo et al., 1997). (b) The ALS3LA locus with the overexpression cassette integrated. (c) Southern blot of EcoRV-digested genomic DNA, hybridized with the ALS3 downstream probe. Molecular size (in kb) is indicated at the left of the image. (d) Relative fluorescence (FITC) intensities of immunostained strains HLC52 and 2296. Overexpression of ALS3 in strain HLC52 increased the surface fluorescence, consistent with increased production and correct localization of Als3p on the C. albicans cell surface.
Evaluation of C. albicans cellular morphology
C. albicans yeast forms were grown to stationary phase in YPD, washed in DPBS and inoculated into YPD+10 % fetal calf serum or RPMI at a density of 5×106 cells ml-1. After 2 h at 37 °C, aliquots of each culture were fixed with 1 % (v/v) glutaraldehyde and photographed as described above.
RESULTS
ALS3 is expressed in model catheter biofilms
Construction and characterization of the C. albicans PALS3-GFP reporter strain, 2185, was reported previously (Zhao et al., 2004). In strain 2185, GFP is transcribed under control of the ALS3 promoter and translated into soluble protein that is localized within the cytoplasm. Growth of strain 2185 in a model catheter biofilm showed fluorescent hyphae when visualized by two-photon laser scanning microscopy, suggesting that GFP is produced following transcription from the ALS3 promoter, which is active under these growth conditions (Fig. 4a). Fluorescence of hyphae from strain 2185 was evident throughout the biofilm, suggesting that ALS3 expression is not limited to a particular area of the biofilm. A control strain, 1143 (Green et al., 2005a), in which GFP is transcribed under control of the constitutive TPI1 promoter, was also tested (Fig. 4b). As expected, biofilm growth of strain 1143 showed fluorescence of both hyphae and yeast throughout the biofilm. Dry weight analysis of biofilms formed from strain 2185 showed a similar mass to those formed by wild-type control strain CAI12 (1·50±0·12 vs 1·52±0·12 mg; P=0·92).
Fig. 4.
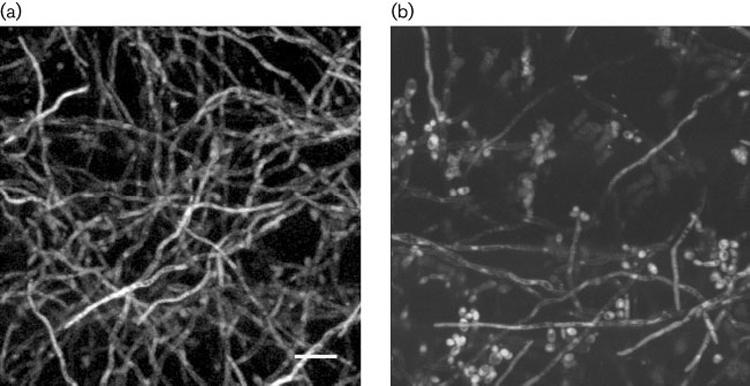
Two-photon laser scanning micrograph of the model catheter biofilms grown using the PALS3-GFP strain 2185 (a) or PTPI1-GFP strain 1143 (b). Fluorescence is detectable only in hyphae for strain 2185, but in hyphae and yeast forms for strain 1143. Bar, 5 μm.
Als3p is localized diffusely on the germ tube surface
Although Als3p is cross-linked to β-1, 6-glucan in the C. albicans cell wall (Kapteyn et al., 2000), its distribution on the cell surface has not been reported. To determine the cell-surface distribution of Als3p, serum Ig enriched for Als3p specificity was prepared by absorption of a polyclonal anti-Als serum with various morphological forms of the als3Δ/als3Δ strain 1843 (Zhao et al., 2004; see Methods). Immunostaining using the anti-Als3p Ig preparation showed diffuse fluorescence that covered nearly the entire length of the germ tube (Fig. 5a). Immunostaining with normal rabbit IgG (negative control) showed very faint fluorescence of the mother yeast and germ tube (Fig. 5c). Immunofluorescence was not detectable when the als3Δ/als3Δ strain was stained with the anti-Als3p Ig preparation (Fig. 5e).
Fig. 5.
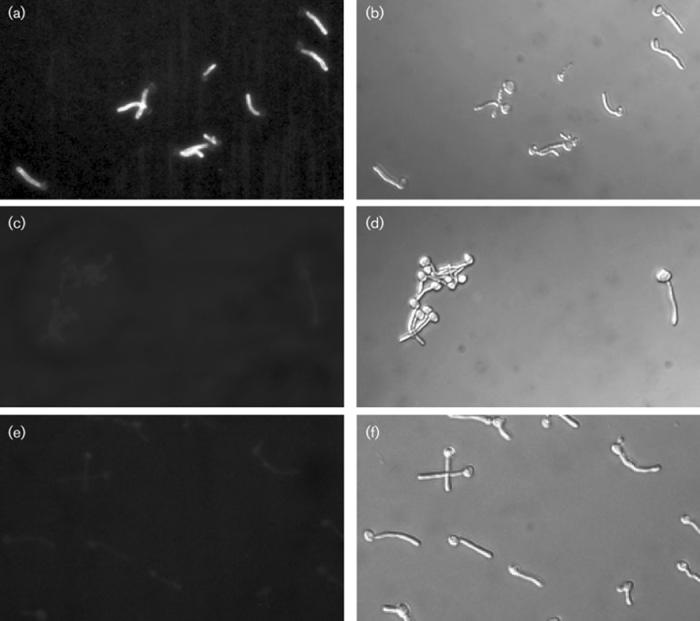
Fluorescence micrographs of C. albicans CAI12 germ tubes that were immunostained with either the anti-Als3p Ig preparation (a) or normal rabbit control IgG (c) and a secondary anti-rabbit FITC-labelled antibody. (e) Immunostaining of the als3Δ/als3Δ strain 1843 with the anti-Als3p Ig preparation and the FITC-labelled secondary antibody. Light micrographs (b, d, f) are matched with each fluorescence image. Anti-Als3p Ig produced a diffuse staining pattern on nearly the entire germ tube length. The control rabbit IgG produced a very faint background signal on both the mother yeast and germ tube. Because the antiserum used for preparation of the anti-Als3p Ig was raised against the N-terminal domain of Als5p, the anti-Als3p Ig cross-reacts with Als5p. This cross-reactivity is undetectable in the experiments presented here, presumably because of the low ALS5 transcriptional activity under these growth conditions (Green et al., 2005b).
ALS3 deletion results in a disorganized and weakened biofilm structure
Growth of the als3Δ/als3Δ strain 1843 in the catheter biofilm model resulted in a weakened structure that was obvious immediately when viewing the biofilms in the 12-well plate in which they were grown. The mature biofilm was dislodged easily from the catheter disc and readily crumbled into many pieces when the 12-well plate was moved from the incubator. The dry weight of the mutant 48 h biofilm was significantly less than that for the wild-type control (1·15±0·12 mg vs 1·52±0·12 mg; P=0·04). Microscopic imaging of calcofluor-stained biofilms showed parallel hyphae in the mutant biofilm (Fig. 6a) that contrasted in appearance with the intertwining hyphae present in the wild-type biofilm structure (Fig. 6b). This difference in biofilm architecture is unlikely to be due to differences in filamentation or growth rate between the two strains since their phenotypes were indistinguishable when tested in various growth media (Zhao et al., 2004). Construction of a strain in which a copy of ALS3LA from strain SC5314 was reintegrated into strain 1843 was described in Methods (Fig. 1). The resulting strain (2322) restored wild-type dry weight of the model catheter biofilm (1·55±0·12 mg vs 1·52±0·12 mg; P=0·84) and restored wild-type biofilm structure. These observations suggest that ALS3 deletion caused the reduced biofilm mass and structural instability.
Fig. 6.
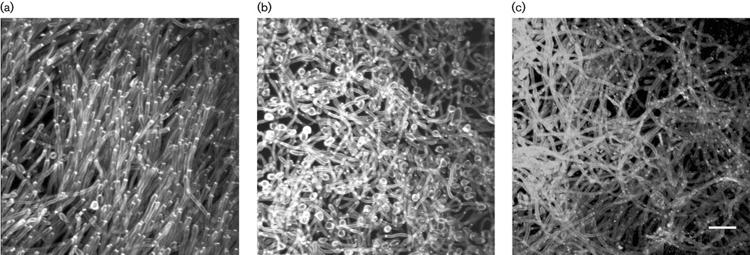
Two-photon laser scanning micrograph of calcofluor-stained model catheter biofilms grown using the als3Δ/als3Δ strain 1843 (a) the wild-type control CAI12 (b), or strain 2327, which lacks ALS3 coding sequences, but produces a cell-surface Als3p-Agα1p fusion protein under control of the ALS3 promoter. Compared to hyphae in the wild-type biofilm (b), hyphae in the als3Δ/als3Δ biofilm (a) are parallel and less densely packed. Production of the Als3p-Agα1p fusion protein (c) restores intertwined hyphae that are characteristic of the wild-type biofilm (b), but does not restore wild-type biofilm dry weight. Bar=5 μm.
Evaluation of the Als3p N-terminal domain function in biofilm formation
Als proteins are composed of three domains (Hoyer, 2001). The N-terminal domain contains approximately 433 aa and shares sequence similarities with the N-terminal half of S. cerevisiae alpha-agglutinin (Agα1p; Lipke et al., 1989; Hauser & Tanner, 1989), which contains residues critical for adhesive function (de Nobel et al., 1996). Therefore, it is reasonable to expect that adhesive function resides within the Als protein N-terminal domain. To investigate the role of the Als3p N-terminal domain in biofilm formation, and to probe the relationship between adhesion and biofilm formation, C. albicans strain 2327 was created (Fig. 2). Strain 2327, from which both ALS3 coding regions were deleted, produced a cell-surface fusion of the N-terminal 430 aa of Als3p to the C-terminal 323 aa of S. cerevisiae alpha-agglutinin under control of the native ALS3 promoter. The fusion protein displayed the Als3p N-terminal domain on a stalk-like portion of Agα1p that is targeted to the cell wall and heavily glycosylated with N- and O-linked carbohydrate, but does not contain any sequences involved in the Agα1p adhesive interactions (Cappellaro et al., 1994; Chen et al., 1995; de Nobel et al., 1996; Zhao et al., 2001). Growth of strain 2327 in the catheter biofilm model showed intertwined hyphae that were more characteristic of a wild-type biofilm (Fig. 6c), but production of the fusion protein did not restore wild-type biofilm mass (1·05±0·12 vs 1·52±0·12 mg; P=0·01).
This result prompted additional experimentation to characterize the phenotype of the Als3p-Agα1p-producing strain. Most work to define Als protein function has been conducted from the standpoint of adhesion. Previouslypublished data showed that the als3Δ/als3Δ mutant strain 1843 has a significantly decreased ability to adhere to human BECs (Zhao et al., 2004). Strain 2327 was tested for BEC adhesion to place it into the context of this knowledge. Consistent with the previous data (Zhao et al., 2004), strain 1843 was significantly less adherent to BECs than was wild-type CAI12 (Fig. 7a). Production of the Als3p-Agα1p fusion protein on the surface of strain 2327 significantly increased C. albicans adhesion to BECs compared to the als3Δ/als3Δ mutant, but did not fully restore wild-type adhesion levels. This intermediate adhesion level may be due to the length of the Als3p-Agα1p fusion protein. Previous studies showed that longer ALS3 alleles (more tandem repeat copies) produce proteins with greater adhesive function than shorter ALS3 alleles (Oh et al., 2005). It is possible that the greater number of tandem repeat copies in the longer alleles displays the N-terminal domain at a greater distance from the cell surface, making it more accessible to binding interactions. However, a role for the tandem repeat sequences in binding interactions has not been ruled out. The Als3p-Agα1p fusion protein in the current study was only 753 aa, which was considerably shorter than the smaller Als3p (1047 aa) and larger Als3p (1155 aa) tested by Oh et al. (2005).
Fig. 7.
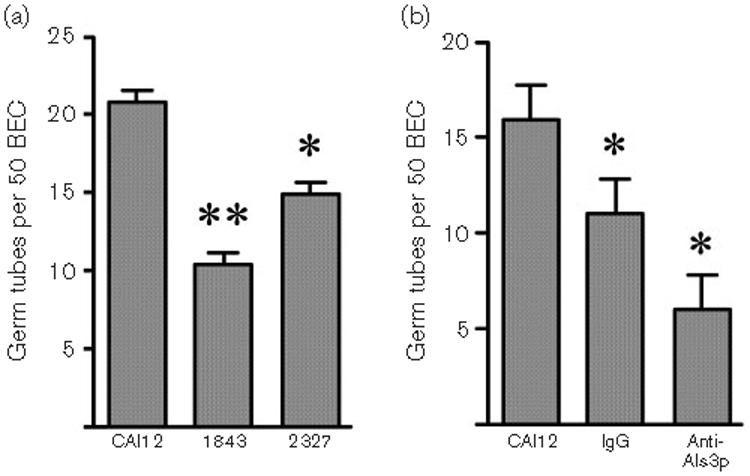
BEC adhesion assay data. (a) Wild-type CAI12, als3Δ/als3Δ mutant 1843, and the Als3p-Agα1p fusion protein-producing strain 2327 were tested for adhesion to BECs. Loss of ALS3 significantly reduced adhesion of C. albicans to BECs (**P<0·0001). Production of the Als3p-Agα1p fusion protein on the surface of strain 2327 significantly increased C. albicans adhesion to BECs compared to the als3Δ/als3Δ mutant (*P=0·01), but did not fully restore wild-type adhesion levels. (b) CAI12 germ tubes were pre-incubated without antibody (CAI12), with normal rabbit IgG (IgG) or with the anti-Als3p Ig preparation prior to their addition to a BEC adhesion assay. Pre-incubation with normal rabbit IgG significantly decreased adherence of C. albicans to BECs (*P=0·01). Pre-incubation with the anti-Als3p Ig preparation significantly reduced adhesion compared to the rabbit IgG control (*P=0·01). Collectively, these results suggest that Als3p is involved in C. albicans adhesion to human BECs and that the N-terminal domain of the protein mediates this interaction.
These data are consistent with the conclusion that Als3p adhesive function is found predominantly within the N-terminal domain. To further evaluate this idea, we blocked adhesion of C. albicans to human BECs using the anti-Als3p Ig preparation. Wild-type CAI12 cells were grown in RPMI 1640 medium to form germ tubes, which were then pre-incubated with the anti-Als3p Ig preparation, or with a normal rabbit IgG control, before incubating them with BECs. Compared to cells to which no antibody was added, pre-incubation with normal rabbit IgG significantly decreased adherence of wild-type C. albicans to BECs (Fig. 7b). Pre-incubation with the anti-Als3p Ig preparation significantly reduced adhesion compared to the rabbit IgG control (Fig. 7b). These results further support the role of Als3p in C. albicans adhesion to human BECs and the role of the N-terminal domain in this interaction.
Wild-type biofilm mass is restored to a C. albicans efg1Δ/efg1Δ strain by overexpression of ALS3
Leng et al. (2001) showed that EFG1 is required for activation of ALS8, and more recent work demonstrated that ALS3 and ALS8 are the same locus (Zhao et al., 2004). Deletion of EFG1 destroyed the ability of C. albicans to form a biofilm; the efg1Δ/efg1Δ strain grew as a sparse monolayer of cells (Ramage et al., 2002b). Integration of a wild-type EFG1 copy restored biofilm formation to the mutant strain (Ramage et al., 2002b). These results suggest the importance of hyphae in C. albicans biofilm formation. In the context of its role in hypha formation, Efg1p regulates expression of many C. albicans genes (Liu, 2002). Therefore, many different proteins could be responsible for deficient biofilm formation by the mutant strain. The importance of hypha-associated cell-surface proteins in biofilm formation has also been suggested by subsequent studies (Nobile & Mitchell, 2005; Richard et al., 2005). The significance of ALS3 and EFG1 in wild-type biofilm formation, as well as the established regulatory connection between the two genes, suggested that ALS3 might reverse the biofilm-deficient phenotype of an efg1Δ/efg1Δ strain. To test this idea, ALS3 was overexpressed in strain HLC52 under control of the constitutive TPI1 promoter (Fig. 3). In a previous study, constitutive ALS1 overexpression in the efg1Δ/efg1Δ strain HLC52 resulted in formation of elongated morphologies under growth conditions that produce hyphae in wild-type cells (Fu et al., 2002). Because of these observations, we examined the cellular morphology of our efg1Δ/efg1Δ ALS3 overexpression strain, 2296. Wild-type strain CAI12 and the efg1Δ/efg1Δ strain were included as controls (Fig. 8). When grown in conditions that did not promote filamentation of the wild-type strain (YPD medium), strain 2296 formed elongated cells. Incubation in YPD+serum or in RPMI 1640 medium resulted in germ-tube-like structures for strain 2296, although these cells exhibited an obvious altered morphology compared to the wild-type control (Fig. 8). Growth of strain 2296 in the catheter model produced a biofilm with wild-type dry weight although the biofilm lacked hyphae and, instead, was composed mainly of yeast forms or short, elongated cells (Fig. 9). Therefore, although wild-type hypha formation was not restored in strain 2296, overproduction of Als3p was able to produce a biofilm of wild-type mass. These results dissect the effects of biofilm formation from those of cellular morphology (specifically hypha growth) and demonstrate a specific role for a hypha-associated surface protein in biofilm formation.
Fig. 8.
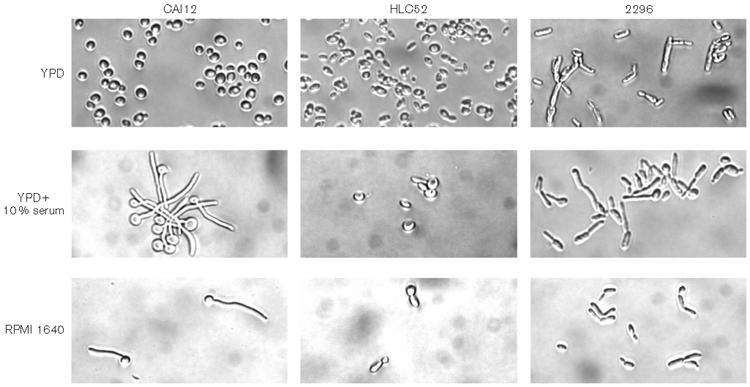
Light micrographs of the cellular morphology of the ALS3 overexpression and control strains. Strains CAI12, HLC52 (efg1Δ/efg1Δ) and 2296 (efg1Δ/efg1Δ PTPI1-ALS3) were grown in YPD overnight at 37 6C, washed, counted and transferred to YPD, YPD+serum or RPMI 1640 for 2 h at 37 6C. Images shown are representative of the morphologies present for each culture.
Fig. 9.
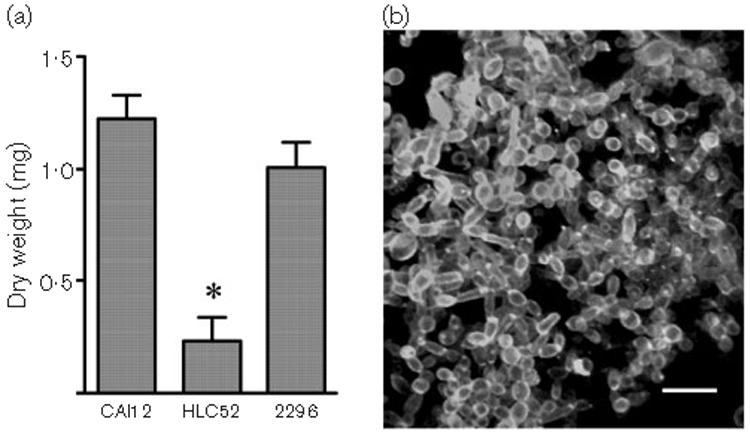
(a) Histogram comparing dry weights for biofilms formed from strains CAI12, HLC52 or 2296. Mass of the biofilm formed by the efg1Δ/efg1Δ mutant strain (HLC52) was significantly reduced compared to a CAI12 biofilm (*P=0·0002). The inability of HLC52 to form biofilm was demonstrated previously (Ramage et al., 2002b). Under biofilm-forming conditions, HLC52 grows as a sparse cellular monolayer. (b) Two-photon laser scanning micrograph of a calcofluor-stained model catheter biofilm grown using strain 2296, in which ALS3 is overexpressed in the HLC52 background. Overexpression of ALS3 in strain HLC52 restores wild-type biofilm mass (a). In contrast to the morphology of a wild-type biofilm (Fig. 6b), the ALS3 overexpression strain does not form hyphae (b). Bar, 5 μm.
DISCUSSION
Previous analysis suggested that high-level ALS3 expression is associated with germ tubes and hyphae, and that Als3p is a strong adhesin that interacts with both epithelial and endothelial surfaces (Hoyer et al., 1998b; Zhao et al., 2004). ALS3 expression was previously demonstrated within model biofilms (Green et al., 2004) and in this work ALS3 expression was detected in hyphae throughout a model catheter biofilm using two-photon laser scanning microscopy of a PALS3-GFP reporter strain. Indirect immunofluorescence of germ tubes grown in vitro showed that Als3p is localized diffusely across the cell surface. In the absence of Als3p, model catheter biofilms are weakened and disorganized and lack the mass associated with a wild-type biofilm. Although the Als3p N-terminal domain is responsible for adhesion of C. albicans to BECs, display of the Als3p N-terminal domain on the C. albicans surface does not restore biofilm mass, but restores the intertwining of hyphae observed in a wild-type biofilm. Overproduction of Als3p is sufficient to restore wild-type biofilm mass to a C. albicans strain lacking EFG1, even though the strain does not form hyphae. This result uncouples the effects of a hypha-associated protein from cellular morphology and suggests a major role for Als3p in biofilm formation.
Results from this work begin to define Als3p function in biofilm formation. The most straightforward explanation to consider is that Als3p is involved in adhesion of C. albicans to silicone elastomer. However, because the adhesion phase of biofilm growth uses yeast forms that do not appear to express ALS3, this explanation is unlikely to account for the importance of Als3p in biofilm formation. It is more likely that Als3p becomes important at a later stage of biofilm development when germ tubes and hyphae are present. The ability of a C. albicans strain displaying the Als3p N-terminal domain to confer greater association between hyphae, but not wild-type biofilm mass, suggests that Als3p may have multiple roles in biofilm formation. The lack of association between hyphae in the mutant biofilm suggests that, in the absence of Als3p, C. albicans cells are not attracted to, or are repelled by, each other. These observations suggest that Als3p may mediate cellular aggregation by either homotypic or heterotypic interactions, alter the surface charge or hydrophobic properties of hyphae, or alter interactions between the extracellular matrix and cellular components of the biofilm. The requirement for Als3p to achieve wild-type biofilm mass suggests that the entire protein is either needed for cellular proliferation within the biofilm, or is a key component of the extracellular matrix. Previous work suggested that Als proteins are shed from the C. albicans surface at various stages of culture growth (Hoyer et al., 1998b). Current data suggest that the extracellular matrix is composed primarily of molecules that are present in planktonic C. albicans cells, rather than being made from biofilm-specific components (Vediyappan & Chaffin, 2006; D. A. Coleman, M. S. Kuhlenschmidt & L. L. Hoyer, unpublished observations). During biofilm development, C. albicans grows in adherent microcolonies (Kuhn et al., 2002) that could trap Als3p shed from the cell surface. The large, mucin-like Als3p glycoprotein could act as glue that holds together the biofilm structure. Since the Als3p tandem repeat and C-terminal domains are predicted to be the most heavily glycosylated portions of the mature protein, these domains might have greater significance in accumulation of biofilm mass. Perhaps wild-type biofilm mass is not restored for strains producing the Als3p-Agα1p fusion protein because the non-native C-terminal sequences in the fusion construct do not permit shedding from the C. albicans surface. Additional experimentation is required to test these ideas and to further define the role of Als3p in biofilm formation.
Questions regarding Als3p function were placed into the context of C. albicans-host cell adhesion since this direction has dominated Als protein functional analysis. Production of the Als3p-Agα1p fusion protein in an als3Δ/als3Δ C. albicans strain demonstrated function of the Als3p N-terminal domain in BEC adhesion. The idea that the Als N-terminal domain is involved in adhesion came from comparisons to S. cerevisiae alpha-agglutinin, in which adhesive function resides within the N-terminal half of the protein (Cappellaro et al., 1994; Chen et al., 1995; de Nobel et al., 1996; Zhao et al., 2001). Previous studies addressed adhesive function of the Als N-terminal domain. Adhesion of a C. albicans ALS1 overexpression strain to vascular endothelial cells was blocked by a monoclonal antibody that was raised against the Als1p N-terminal domain (Fu et al., 2002). S. cerevisiae, expressing ALS1 with mutations within the N-terminal domain-encoding sequences, showed reduced adhesion compared to a S. cerevisiae strain expressing the wild-type allele (Loza et al., 2004). Another study claimed to demonstrate adhesive specificity within the N-terminal domain of Als5p and Als6p using a domain-swapping approach (Sheppard et al., 2004). However, rather than swapping the N-terminal domain of Als5p onto the tandem repeat and C-terminal domain of Als6p (and vice versa), the investigators included the tandem repeat sequences in the swap. In addition, the sequences of the C-terminal domain of Als5p and Als6p are over 90 % identical with conservative replacements for many of the mismatched amino acids. Therefore, the experiment essentially recreated native Als5p and Als6p, rather than testing the effects of placing the Als N-terminal domain onto a non-native C-terminal sequence. In our Als3p domain-swapping experiment, only the N-terminal domain of Als3p was fused to sequences of alpha-agglutinin that are C-terminal to the adhesive domain. The ability of the fusion protein to restore epithelial adhesion to the als3Δ/als3Δ mutant C. albicans strain supports the conclusion that the Als3p N-terminal domain has adhesive function.
Data presented here also address the relationship between C. albicans cellular morphology and biofilm formation. Several lines of evidence demonstrated the importance of filamentous growth in biofilm formation (reviewed by Lopez-Ribot, 2005). Hyphae are essential for the structural stability of mature biofilms (Baillie & Douglas, 1999). C. albicans strains with impaired hypha formation due to mutation in genes encoding the transcription factors EFG1 (Ramage et al., 2002b) or TEC1 (Nobile & Mitchell, 2005) cannot form wild-type biofilms. Also, blocking of hypha formation with farnesol inhibits biofilm formation (Ramage et al., 2002a). In our work, we restored wild-type biofilm mass by overexpression of ALS3 in a strain lacking Efg1p. Previous work showed that EFG1 is required for ALS3 expression (Leng et al., 2001) and ALS3 is now recognized as part of the ‘hyphal regulon’ (Kumamoto & Vinces, 2005). Although ALS3 overexpression caused the efg1Δ/efg1Δ strain to grow in a somewhat elongated form under hypha-inducing conditions, biofilms formed from this strain showed mainly yeast forms (Fig. 9). These results dissociate biofilm formation from hypha formation and suggest that the need for hyphae in biofilm growth is due, at least in part, to the role of hypha-associated proteins. Nobile & Mitchell (2005) also reached this conclusion by testing C. albicans transcription factor mutants for deficiencies in biofilm development. In their work, disruption of the zinc finger transcription factor Bcr1p did not affect the ability of planktonic cells to form hyphae, but resulted in a strain that was deficient in biofilm formation. Microarray analysis showed that Bcr1p regulates expression of several genes that encode hypha cell-surface proteins, including ALS3.
Nobile & Mitchell (2005) assayed ALS gene expression in the bcr1Δ/bcr1Δ strain in their catheter biofilm model and found that, in addition to the 16-fold reduction in ALS3 expression, ALS1 and ALS9 expression were reduced twofold in the mutant strain. Expression of other ALS genes, including ALS2, was not affected by Bcr1p loss. These results contrast with data from systematic testing of C. albicans strains with ALS gene mutations that showed reduced ALS2 expression caused a significant loss of biofilm mass in the catheter biofilm model (Zhao et al., 2005). In both studies, a silicone elastomer surface was used for biofilm formation, although growth media were different. Using our model, the contribution of Als3p to biofilm formation is clearly superior to that of Als2p. These results suggest the potential for model-specific effects on biofilm formation, but clearly support the importance of ALS3 in both cases. Additional comparisons will reveal the potential role of other Als proteins in biofilm formation.
ACKNOWLEDGEMENTS
We thank Al Brown, Richard Cannon, Brendan Cormack, Gerry Fink and Cheryl Gale for strains, plasmids and reagents used in this work. We thank Liping Wang and the staff of the Carver Biotechnology Center Immunological Resource Center for production of the rabbit polyclonal antiserum, Barbara Pilas and Ben Montez of the Flow Cytometry Laboratory for assistance with flow cytometry experiments, and Mariangela Segre for helpful comments on the manuscript. This work was supported by Public Health Service grant DE14158 (to L. L. H.) and AI2392 (to D. R. S.) from the National Institutes of Health. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR16515-01 from the National Center for Research Resources, National Institutes of Health.
Abbreviations
- ALS
agglutinin-like sequence
- BEC
buccal epithelial cell
- GFP
green fluorescent protein
- Ig
immunoglobulin
REFERENCES
- Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- Cao YY, Cao YB, Xu Z, Ying K, Li Y, Xie Y, Zhu ZY, Chen WS, Jiang YY. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother. 2005;49:584–589. doi: 10.1128/AAC.49.2.584-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro C, Baldermann C, Rachel R, Tanner W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 1994;13:4737–4744. doi: 10.1002/j.1460-2075.1994.tb06799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Shen ZM, Bobin S, Kahn PC, Lipke PN. Structure of Saccharomyces cerevisiae alpha-agglutinin. Evidence for a yeast cell wall protein with multiple immunoglobulin-like domains with atypical disulfides. J Biol Chem. 1995;270:26168–26177. doi: 10.1074/jbc.270.44.26168. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- de Nobel H, Lipke PN, Kurjan J. Identification of a ligand-binding site in an immunoglobulin fold domain of the Saccharomyces cerevisiae adhesion protein α-agglutinin. Mol Biol Cell. 1996;7:143–153. doi: 10.1091/mbc.7.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol. 2002;44:61–72. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo J-M, d’Enfert C. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell. 2004;3:536–545. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, O’Toole GA. Microbial Biofilms. American Society for Microbiology; Washington, DC: 2004. [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans genes for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. Yeast wall protein 1 of Candida albicans. Microbiology. 2005;151:1631–1644. doi: 10.1099/mic.0.27663-0. [DOI] [PubMed] [Google Scholar]
- Green CB, Cheng G, Chandra J, Mukherjee P, Ghannoum MA, Hoyer LL. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology. 2004;150:267–275. doi: 10.1099/mic.0.26699-0. [DOI] [PubMed] [Google Scholar]
- Green CB, Zhao X, Hoyer LL. Use of green fluorescent protein and reverse transcription-PCR to monitor Candida albicans agglutinin-like sequence gene expression in a murine model of disseminated candidiasis. Infect Immun. 2005a;73:1852–1855. doi: 10.1128/IAI.73.3.1852-1855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Zhao X, Yeater KM, Hoyer LL. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology. 2005b;151:1051–1060. doi: 10.1099/mic.0.27696-0. [DOI] [PubMed] [Google Scholar]
- Hauser K, Tanner W. Purification of the inducible α-agglutinin of S. cerevisiae and molecular cloning of the gene. FEBS Lett. 1989;255:290–294. doi: 10.1016/0014-5793(89)81108-1. [DOI] [PubMed] [Google Scholar]
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Hecht JE. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast. 2001;18:49–60. doi: 10.1002/1097-0061(200101)18:1<49::AID-YEA646>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998a;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Hecht JE. Identification of Candida albicans ALS2 and ALS4 and localization of Als proteins to the fungal cell surface. J Bacteriol. 1998b;180:5334–5343. doi: 10.1128/jb.180.20.5334-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn JC, Hoyer LL, Hecht JE, Muller WH, Andel A, Verkleij AJ, Makarow M, Van Den Ende H, Klis FM. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol. 2000;35:601–611. doi: 10.1046/j.1365-2958.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJP, Butler G. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- Krueger KE, Ghosh AK, Krom BP, Cihlar RL. Deletion of the NOT4 gene impairs hyphal development and pathogenicity in Candida albicans. Microbiology. 2004;150:229–240. doi: 10.1099/mic.0.26792-0. [DOI] [PubMed] [Google Scholar]
- Kuhn DM, Chandra J, Mukherjee PK, Ghannoum MA. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun. 2002;70:878–888. doi: 10.1128/iai.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc Natl Acad Sci U S A. 2005;102:5576–5581. doi: 10.1073/pnas.0407097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Leng P, Lee PR, Wu H, Brown AJ. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J Bacteriol. 2001;183:4090–4093. doi: 10.1128/JB.183.13.4090-4093.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke PN, Wojciechowicz D, Kurjan J. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interaction during mating. Mol Cell Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol. 2002;292:299–311. doi: 10.1078/1438-4221-00215. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lopez-Ribot JL. Candida albicans biofilms: more than filamentation. Curr Biol. 2005;15:R453–R455. doi: 10.1016/j.cub.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Loza L, Fu Y, Ibrahim AS, Sheppard DC, Filler SG, Edwards JE., Jr Functional analysis of the Candida albicans ALS1 gene product. Yeast. 2004;21:473–482. doi: 10.1002/yea.1111. [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Murillo LM, Newport G, Lan C-Y, Habelitz S, Dungan J, Agabian NM. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell. 2005;4:1562–1573. doi: 10.1128/EC.4.9.1562-1573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Oh S-H, Cheng G, Nuessen JA, Jajko R, Yeater KM, Zhao X, Pujol C, Soll DR, Hoyer LL. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology. 2005;151:673–681. doi: 10.1099/mic.0.27680-0. [DOI] [PubMed] [Google Scholar]
- Porta A, Ramon AM, Fonzi WA. PRR1, a homolog of Aspergillus nidulans palF, control pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol. 1999;181:7516–7523. doi: 10.1128/jb.181.24.7516-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 2002a;68:5459–5463. doi: 10.1128/AEM.68.11.5459-5463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002b;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Richard ML, Nobile CJ, Bruno VM, Mitchell AP. Candida albicans biofilm-defection mutants. Eukaryot Cell. 2005;4:1493–1502. doi: 10.1128/EC.4.8.1493-1502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME. Microbial biofilms. N Engl J Med. 2005;352:846. [Google Scholar]
- Schwank S, Rajacic Z, Zimmerli W, Blaser J. Impact of bacterial biofilm formation on in vitro and in vivo activities of antibiotics. Antimicrob Agents Chemother. 1998;42:895–898. doi: 10.1128/aac.42.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DC, Yeaman MR, Welch WH. Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem. 2004;279:30480–30489. doi: 10.1074/jbc.M401929200. 7 other authors. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Mukherjee PK, Ghannoum MA. Biofilm antimicrobial resistance. In: Ghannoum MA, O’Toole GA, editors. Microbial Biofilms. American Society for Microbiology; Washington, DC: 2004. pp. 250–268. [Google Scholar]
- Vediyappan G, Chaffin WL. Non-glucan attached proteins of Candida albicans biofilm formed on various surfaces. Mycopathologia. 2006;161:3–10. doi: 10.1007/s11046-005-0167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. Susceptibility of oral bacterial biofilms to anti-microbial agents. J Med Microbiol. 1996;44:79–87. doi: 10.1099/00222615-44-2-79. [DOI] [PubMed] [Google Scholar]
- Zhao H, Shen ZM, Kahn PC, Lipke PN. Interaction of α-agglutinin and a-agglutinin, Saccharomyces cerevisiae sexual cell adhesion molecules. J Bacteriol. 2001;183:2874–2880. doi: 10.1128/JB.183.9.2874-2880.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Oh S-H, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJP, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparison between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Oh S-H, Yeater KM, Hoyer LL. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology. 2005;151:1619–1630. doi: 10.1099/mic.0.27763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


