Summary
ALS gene expression was studied in the hyposalivatory rat model of oral candidiasis and in clinical specimens collected from HIV-positive patients to assess similarities in expression patterns between the model system and clinical isolates. Two C. albicans strains, SC5314 and OY-2-76, were used in the rat model system and infection progressed for 3 or 5 days. The strains produced similar oral lesions at 3 days. At 5 days, strain OY-2-76 produced more superficial lesions containing relatively more yeast forms compared to invasive hyphal forms observed for strain SC5314. For all infections, the most severe lesions were observed on the tongue and gingiva overlying the mandible. ALS transcripts were easier to detect by RT-PCR later in infection and under other conditions where more fungal cells were present. Expression of ALS1, ALS2, ALS3 and ALS4 was observed in rats infected for 3 days with ALS5 and ALS9 transcripts detected after 5 days of infection. Expression of ALS6 was observed in a single specimen from a 5-day infection while ALS7 transcript was never found. Expression of all ALS genes was observed in oral clinical material collected from HIV-positive patients although ALS6 and ALS7 transcripts required an extra PCR amplification step to be detected. Overall, the patterns of ALS gene expression were similar between the rat model and human clinical specimens, suggesting that the model would be useful for studying the phenotype of als/als mutant strains.
Introduction
Candida albicans is a commensal organism of the human oral cavity, genitalia, and gastrointestinal tract. Certain pathological conditions, however, permit expansion of C. albicans numbers and development of mucocutaneous disease [1]. In general, suppression of cellular immunity, disruption of dominant bacterial flora, or states of chronic irritation predispose patients to oral disease [1]. Infants with congenital defects in cellular immunity frequently manifest symptoms of oral candidiasis shortly after birth [1]. AIDS patients may also present with oroesophageal candidiasis [1]. Candida stomatitis, on the other hand, is a common condition of denture wearers with presumably an intact immune system. Two important predictors of Candida denture stomatitis include chronic irritation and poor oral hygiene [2].
The hyposalivatory rat model of oral candidiasis combines reduction of salivary flow with a high sugar diet and treatment with broad-spectrum antibiotics [3]. Reduction of salivary flow favors oral colonization by C. albicans [4]. High carbohydate diets and broad-spectrum antibiotics also favor colonization and invasion of superficial epithelial layers of the rat oral mucosa [5]. This model has been used to study both the phenotypic effects of virulence genes and gene expression during oral candidiasis [6-8].
One major goal of research in our laboratory is to understand the function of proteins encoded by the C. albicans ALS gene family [9]. Several of the large, cell-surface Als glycoproteins function in adhesion to host surfaces [10-12]. Because interaction of C. albicans with different host sites might result in differential expression of the various ALS genes, studying gene expression would be useful in grouping the various Als proteins into potentially different functions or establishing a hierarchy of importance for Als proteins at different host sites or across different disease states. In order to understand these relationships, we evaluated ALS gene expression in various human clinical specimens and model systems including buccal and vaginal reconstituted human epithelium (RHE), model denture and catheter biofilms, vaginal clinical specimens, a murine model of vaginitis, and a murine model of disseminated candidiasis [13-15].
The studies presented here used an RT-PCR assay [13] to compare ALS gene expression in the hyposalivatory rat model of oral candidiasis with ALS gene expression detected in specimens collected from HIV-positive patients with oral candidiasis. Two different C. albicans strains were used in the rat model and two different time points following inoculation of the animals were studied. In this work, the oral cavity of each experimental animal was carefully dissected so that gene expression and histopathology could be compared at each site. Results from these analyses demonstrate the similarity in ALS gene expression patterns between human clinical specimens and the disease model.
Materials and methods
Rats
Female Sprague-Dawley rats (23 days old) were purchased from Charles River Laboratories. Rats were fed NIH Cariogenic Diet 2000, which consists of 56% sucrose (w/v) and 46% essential nutrients. Water containing 10% (w/v) sucrose and 50 μg ml-1 streptomycin was administered ad libitum [3]. Animal experiments followed the guidelines of the University of Illinois Institutional Animal Care and Use Committee.
Surgical procedures
Rats were weighed and anesthetized. Anesthesia was begun in an induction chamber, using a gas mixture of oxygen at 1 L min-1 flow with 3% isofluorane to sedate the rats. Sedated rats were injected intraperitoneally with 75 mg kg-1 ketamine and 10 mg kg-1 xylazine to maintain anesthesia. Immediately prior to surgery, the rat oral cavity was swabbed with a sterile cotton applicator and streaked to YPD and then to Sabouraud dextrose agar with chloramphenicol (50 μg ml-1). Agar plates were grown for 48 h; none of the animals had any detectable C. albicans in its oral cavity.
The ventral mandibular and cervical region of each rat was shaved and the area scrubbed with 7.5% betadine solution followed by 70% ethanol. A midline incision was made from the mandibular symphysis to the manubrium. The skin edges were retracted laterally to expose the mandibular salivary glands. The blood vessels at the caudolateral aspect of the right mandibular salivary gland were ligated and divided using 6-0 polydioxanone suture (PDS). The duct at the cranial aspect of the gland was ligated, divided and the gland was excised. The procedure was repeated on the left side. The major parotid duct was then exposed by lateral retraction of the skin and blunt separation of subcutaneous tissues from the masseter muscle. The major parotid duct was ligated with 6-0 PDS. Ligation of the major parotid duct was repeated on the left side. Simple, interrupted sutures were used to close the subcutaneous tissues and dead space associated with the removal of the mandibular salivary glands, and the skin was closed in a simple interrupted pattern with 6-0 PDS. Sterile saline was used to clean the perioperative site following completion of the surgery. The surgical site on each animal was evaluated twice daily for any signs of infection, including swelling, pain, and discharge.
Infection of rats with C. albicans
C. albicans strains SC5314 [16] and OY-2-76 were used in this study. Strain OY-2-76 was isolated from an AIDS patient with oropharyngeal candidiasis and was provided by Mahmoud Ghannoum (Case Western Reserve University). Strains were streaked to YPD agar plates (per liter: 10 g yeast extract, 20 g peptone, 20 g dextrose, 20 g Bacto agar) and incubated at 37°C for 24 h. An isolated colony was resuspended in 1 ml YPD medium and 5 μl used to inoculate a 10 ml YPD culture. The 10 ml culture was incubated for 24 h in a 37°C water bath with shaking at 200 rpm.
Seven days following surgery, rats were weighed and sedated with isofluorane as described above. The oral cavity of each rat was swabbed with a sterile, cotton-tipped applicator that had been saturated in the overnight culture of the appropriate C. albicans strain. Aliquots of the inoculum culture were flash-frozen in a dry ice/ethanol bath and stored at -80°C. As predicted from previous experiments [13], growth of C. albicans under these conditions resulted in RT-PCR detection of expression of all ALS genes in each strain (data not shown).
Rat infections progressed for either 3 or 5 days. At these endpoints, rats were sedated with isofluorane and euthanized with an intraperitoneal injection of Lethal Plus® (Vortech). Animals were monitored for respiratory arrest, then dissected.
Rat dissection and tissue processing
The rat oral cavity was dissected to remove the right and left buccal mucosa, the mandible, tongue, and palatal and pharyngeal mucosa. Half of each sample of oral tissue was fixed in 10% neutral-buffered formalin for histopathology. The other half of each sample was flash frozen in liquid nitrogen and stored at -80°C until used for RNA analysis. Mandibles were decalcified in DeltaFORM® (Delta Products Group Aurora, IL) prior to trimming and sectioning. Tissues were embedded in paraffin, sectioned and stained with hematoxylin and eosin or using the Gomori methenamine silver method (GMS) [17].
Collection of human oral candidiasis specimens
Specimens from HIV-positive patients with clinical signs of oral candidiasis were collected at Case Western Reserve University. Informed consent was obtained from patients and collection of clinical specimens followed the guidelines of the Institutional Review Boards of Case Western Reserve University and of the University of Illinois at Urbana-Champaign. The patient group included five men and one woman. Patient ages ranged from 34 to 42 years (mean and standard deviation = 38.3 ± 2.9 years). Disease severity was assessed by examination of the oropharynx, hard and soft palate, tongue, and cheeks. In five of the patients, two of the areas were affected and, in one patient, three areas had clinical lesions. The thick curd-like material characteristic of oral thrush was scraped with a tongue blade, placed into a plastic vial and flash-frozen in liquid nitrogen. Samples were shipped on dry ice and stored at -80°C until RNA was extracted using methods described below. A portion of each sample was plated onto Sabouraud agar with 50 μg ml-1 chloramphenicol and the fungal strain recovered into pure culture. Isolated colonies from the pure culture were used for yeast identification similar to the methods described by Naglik et al. [18]. These included API 20C AUX strips (BioMerieux), growth on CHROMagar Candida plates (CHROMagar Microbiology), germ tube tests in brain heart infusion broth (Difco) containing 10% horse serum (GibcoBRL), and growth at 42°C. Yeasts were also grown on corn meal/Tween agar plates (Remel) to observe microscopic morphology. All samples used for gene expression analysis yielded colonies that were identified as C. albicans. Since all C. albicans strains do not encode every ALS gene [9], genomic DNA was extracted from each clinical strain and tested by PCR using primers specific for each ALS gene [13]. This analysis confirmed that the clinical isolates in this work encoded all eight ALS genes (data not shown).
RNA isolation and RT-PCR analysis
RNA was extracted from both rat oral tissues and human clinical specimens using an acid-phenol extraction [19]. Glass beads (425-600 μm, Sigma) were added to the rat tissues and vortexed to aid disruption of tissue structure and fungal cell integrity. This treatment did not homogenize the tissue, but was effective for disrupting superficial structures where the majority of fungal cells were located. For rat specimens, Poly(A) RNA was isolated from each sample using the Poly(A) Purist kit (Ambion). One hundred fifty to 300 μg of total RNA were used for Poly(A) enrichment with a predicted 1-2% recovery. After enrichment, Poly(A) RNA was treated overnight at 37°C with 6 units of RNAse-free DNAse (Ambion). The resulting sample was tested for the presence of C. albicans genomic DNA with an ALS9-specific primer pair as previously described [13]. RNA was precipitated with ethanol, glycogen, and ammonium acetate. The final mRNA pellet was resuspended in 10 μl of nuclease-free water. The entire aliquot of mRNA was used as template in the reverse transcription reaction. The RT-PCR protocol and sensitivity of the assay were described previously [13]. Controls to test whether the pairs of ALS-specific primers amplified DNA with equivalent efficiency were also described [13]. In these analyses, all primer pairs displayed similar amplification efficiency except the ALS6 pair, which was more efficient, presumably because it amplified the smallest product [13]. For human clinical specimens, RT-PCR analysis was conducted twice using total RNA as the starting material. In the first experiment, RNA was pelleted from a fixed volume of the ethanol-RNA suspension resulting from the acid-phenol extraction protocol, the RNA purified with RNeasy (Qiagen), and its quantity determined spectrophotometrically. The entire quantity was DNase treated, checked for genomic DNA and analyzed by RT-PCR as described above. In the second experiment, the remaining volume from each RNA sample was subjected to the same procedure. PCR using a set of nested ALS primers were performed as described by Cheng et al. [14]. Prior to PCR with the nested primers, product from the initial RT-PCR PCR Preps DNA Purification System (Promega).
Results
Histopathology examination of oral tissues from C. albicans-infected hyposalivatory rats
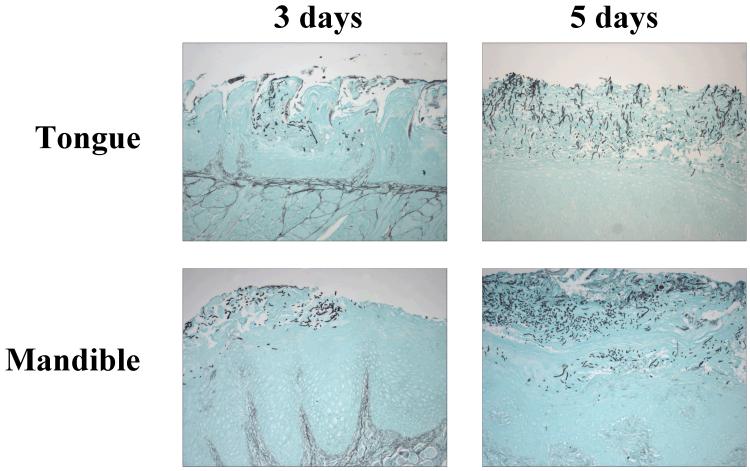
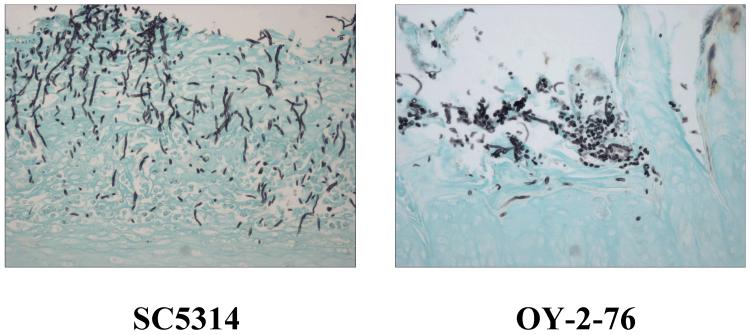
Two separate experiments were conducted with the rat model of oral candidiasis. In the first, three rats were inoculated with C. albicans SC5314 and three with OY-2-76. Two different C. albicans strains were used to determine if there were strain effects in gene expression, and disease severity and progression. Following 3 days of infection, rats were euthanized and the oral cavity dissected. After examining the resulting lesions, a second experiment was conducted. Infection progressed for 5 days in the second experiment in order to develop more severe lesions. Representative images of tongue and mandible sections from each time point are shown in Fig. 1. Histopathology analysis of the rat tissues indicated that lesions produced by strains SC5314 and OY-2-76 were similar in character at 3 days of infection. Although examination of some tissue sections suggested that SC5314 may have produced more severe lesions than strain OY-2-76, overall conclusions showed there was overlap in severity of lesions produced by the two strains. The perception of variation between the strains could be attributed to animal-to-animal variation in lesion severity or to bias due to sample selection for histopathology analysis. When noted, increased severity of the SC5314 lesions was characterized by larger individual lesions, deeper ulceration, and a tendency to invade deeper into tissue. Although most lesions produced by SC5314 were limited to the stratum corneum at 3 days, occasionally a few hyphae could be observed invading into the superficial layers of the stratum granulosum. SC5314 also seemed to have more inflammatory crusting and intraepithelial pustules in comparison to strain OY-2-76. These overall tendencies were also true for tissues following 5 days of infection. In general, a longer time of disease progression produced more severe lesions for each C. albicans strain (Fig. 1). At the 5-day time point, strain OY-2-76 produced more superficial yeast forms compared to SC5314, which had predominantly more hyphae that invaded deeper into the tissue (Fig. 2). From analysis of the tissue sections, a hierarchy of lesion severity emerged with the most severe lesions in the tongue, followed in severity by mandibular mucosal lesions. The buccal mucosa lesions were next most severe and the least damage was observed in the palatal and pharyngeal mucosa.
Fig. 1.
Light micrographs of GMS-stained tissue sections from SC5314-infected rats to show increased severity of lesions at the 5-day time point, compared to the 3-day time point. Magnification = 100x.
Fig. 2.
Light micrographs of GMS-stained tongue sections from SC5314- and OY-2-76-infected rats to show the relative presence of hyphal and yeast forms for the two strains. Magnification = 200x.
RT-PCR detection of ALS gene expression in C. albicans-infected hyposalivatory rats
RT-PCR analysis with a set of ALS-specific primers [13] was used to detect ALS gene expression in the different oral tissues from each rat. Results for rats infected for 3 days are shown in Table 1 and results from the 5-day infections are shown in Table 2. In general, a greater number of different ALS transcripts were detected in animals infected for 5 days. However, it is important to note that an extended period of infection was also characterized by an increased abundance of fungal cells. At the 3-day time point, ALS1-specific transcript was the most commonly detected, even in specimens where fungal cells were relatively rare (Table 1). Specific transcripts for ALS2, ALS3 and ALS4 were also documented. At the 5-day time point, ALS1, ALS2, ALS3 and ALS4 were commonly observed, with the addition of ALS5 and ALS9 (Table 2). ALS6-specific transcript was observed in only one specimen and ALS7 transcript was not found. At the 5-day time point, relatively more ALS transcripts were detected in rats inoculated with strain OY-2-76.
Table 1.
ALS gene expression detected by RT-PCR of poly(A) RNA isolated from oral tissues from hyposalivatory rats at 3 days after C. albicans inoculation.*
| Left | Right | ||||||
|---|---|---|---|---|---|---|---|
| Rat | Buccal | Buccal | Palatal | Pharyngeal | |||
| Strain | Number | Tongue | Mandible | Mucosa | Mucosa | Mucosa | Mucosa |
| OY-2-76 | 1 | - | 1 | 1,2 | 1,2 | 1,4 | - |
| 2 | 1 | 1 | 1,3,4 | - | - | - | |
| 3 | 1,2,3,4 | 1 | - | - | - | - | |
| SC5314 | 1 | 1 | 1 | 1,2 | - | - | 1 |
| 2 | 1,3 | 3 | 1,2,3 | 2 | 1 | - | |
| 3 | 1,2,3,4 | 1,2,3 | 2,3,4 | 2 | - | - | |
| Uninoculated | - | - | - | - | - | - | |
Oral infections with C. albicans strains SC5314 or OY-2-76 progressed for 3 days. RT-PCR for ALS transcripts was performed on each region of the dissected oral cavity. Numbers shown denote the identity of specific ALS transcripts that were detected in a given sample. “-” indicates no ALS transcripts were detected in that specimen.
Table 2.
ALS gene expression detected by RT-PCR of poly(A) RNA isolated from oral tissues from hyposalivatory rats at 5 days after C. albicans inoculation.*
| Left | Right | ||||||
|---|---|---|---|---|---|---|---|
| Rat | Buccal | Buccal | Palatal | Pharyngeal | |||
| Strain | Number | Tongue | Mandible | Mucosa | Mucosa | Mucosa | Mucosa |
| OY-2-76 | 1 | 1,2,3,4,5,9 | 1,2,3,4,5,9 | 1,2,9 | 1 | 1,2 | 1 |
| 2 | 1,2,3,4,5,9 | 1,2,3,4,5,9 | 1,3 | 1,2,3,4 | 1,2,3 | 1 | |
| 3 | - | 1,2,3,4,5,9 | 1,2,3,4 | 1,2,3 | - | - | |
| SC5314 | 1 | 1,2,3 | 1,2,3,9 | 1,2,3 | 1,2,3 | 1,2,3 | 1,3 |
| 2 | - | 1,3 | - | 1,3 | - | 6 | |
| 3 | 1,2,3,4,5 | 1,3 | 1,3 | 1,2,3 | 1 | 1 | |
| Uninoculated | - | - | - | - | - | - | |
Oral infections with C. albicans strains SC5314 or OY-2-76 progressed for 5 days. RT-PCR for ALS transcripts was performed on each region of the dissected oral cavity. Numbers shown denote the identity of specific ALS transcripts that were detected in a given sample. “-” indicates no ALS transcripts were detected in that specimen.
Detection of ALS gene expression in human clinical specimens
The major goal of this work was to understand how the rat disease model relates to human clinical specimens. To assess ALS gene expression during human disease, oral scrapings were collected from HIV-positive patients with signs of oral candidiasis. RT-PCR analysis was conducted using the same method as for the rat tissues. Since the oral microbiota of the human patients may include organisms that cross-react with the ALS RT-PCR primers and there is a lack of negative control for these specimens, nested PCR was also run. The nested PCR protocol has been used previously to verify that the initial RT-PCR products are derived from transcription of C. albicans ALS genes [14]. Because of the additional amplification step associated with it, the nested PCR protocol also serves to increase the assay sensitivity, providing evidence of transcription of more ALS genes. RT-PCR analysis was conducted twice as described in the Materials and Methods section. Both quantities of total RNA assayed are shown in Table 3 along with results indicating the positive reactions from the assays. In the initial RT-PCR analysis, ALS2 transcript was detected in all specimens, followed in frequency by detection of ALS1 and ALS3 (Table 3). Transcription of ALS4 and ALS5 was detected in two of the specimens, transcription of ALS9 in one specimen, and evidence of ALS6 and ALS7 transcription was absent. Reamplification of each PCR product with the nested primers specific for that ALS gene increased the ability to detect specific PCR products for each gene and confirmed the specificity of the ALS2 products. The aggregate of observations from this analysis indicates that transcription of each ALS gene was evident in human clinical material, with increased assay sensitivity required to document expression of ALS6 and ALS7.
Table 3.
ALS gene expression detected by RT-PCR of total RNA isolated from oral scrapings from HIV-positive patients with symptoms of oral candidiasis.*
| Total RNA used | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | for cDNA | ||||||||
| Number | synthesis (μg) | ALS1 | ALS2 | ALS3 | ALS4 | ALS5 | ALS6 | ALS7 | ALS9 |
| 1 | 0.5, 1.1 | × | × | × | × | × | × | ||
| Nested | × | × | × | × | × | × | × | × | |
| 2 | 1.6, 2.0 | × | × | × | × | ||||
| Nested | × | × | × | × | × | × | × | × | |
| 3 | 1.3, 2.4 | × | × | ||||||
| Nested | × | × | × | × | |||||
| 4 | 0.1, 0.5 | × | |||||||
| Nested | × | × | × | × | × | × | |||
| 5 | 1.2, 1.6 | × | × | ||||||
| Nested | × | × | × | × | × | × | × | × | |
| 6 | 0.4, 0.8 | × | × | ||||||
| Nested | × | × | × | × | × | × | × | ||
| % positive | |||||||||
| Initial RT-PCR | 50 | 100 | 50 | 33 | 33 | 0 | 0 | 17 | |
| Nested PCR | 100 | 100 | 100 | 66 | 83 | 50 | 83 | 100 | |
For each patient sample, results for the initial PCR analysis of reverse-transcribed RNA are shown on the top line. Each assay was run twice using the quantities of total RNA indicated. Positive reactions from the assays are denoted by an “×” under the appropriate gene name heading. Products amplified with the nested primer pair are indicated on the lower line.
Discussion
The goal of the studies presented here was to evaluate ALS gene expression in the hyposalivatory rat model of oral candidiasis and to compare these results to ALS gene expression patterns from human clinical specimens. ALS3 expression had already been detected in the rat model by differential display analysis of RNA taken from C. albicans cells collected from the rat oral cavity with a sterile swab [8]. In the work presented here, the oral cavity was dissected to study lesion severity and gene expression at specific sites. The data defined a hierarchy of lesion severity (tongue > mandible > buccal mucosa > palatal and pharyngeal mucosa) and also associated increased diversity of ALS transcript detection with increased length of infection. From these observations, it is tempting to conclude that additional ALS genes are transcribed as infection progresses, suggesting an ordered pattern of gene expression and conclusions regarding the temporal role of various proteins in disease. However, data from this study more likely suggest that transcript detection is a function of transcriptional abundance and are consistent with the idea that some ALS genes are strongly expressed, while others are expressed more weakly during oral disease. This conclusion is supported by data from other studies. Previous RT-PCR analysis of ALS gene expression focused on the buccal reconstituted human epithelium (RHE) in vitro disease model and on model C. albicans biofilms [13]. All ALS transcripts could be detected in both systems, although ALS6- and ALS7-specific transcripts more readily fell below the detection limit of the assay, suggesting that they were relatively less abundant than transcripts from the other ALS genes. Controls for the amplification efficiency of the various ALS-specific primer pairs showed that all were similar except ALS6, which amplified the smallest product and appeared more efficient than the others [13]. C. albicans cell number was not limiting in these analyses, providing abundant fungal RNA in the context of little contaminating RNA from other sources. In the rat model presented here, more ALS transcripts were detected when relatively more fungal RNA was present such as for lesions from the 5-day infections and lesions that were superficial rather than invasive. This relationship was apparent when comparing data from 3 and 5 days of infection (Fig. 1). More ALS transcripts were also detected for strain OY-2-76 compared to SC5314 at 5 days of infection (Table 2). The OY-2-76 cells tended to be yeast forms rather than the invasive hyphae formed by SC5314. The yeast forms are likely to be more readily dissociated from the rat tissue surface and broken during the RNA preparation procedure yielding an increased abundance of fungal RNA for RT-PCR. However, alternate explanations such as yeast forms requiring additional adhesins to maintain association with a host surface cannot be discounted.
Regardless of the reason for increased detection of transcripts, the analyses presented here and in earlier work [13] suggest that all C. albicans ALS genes are detectable in the rat oral cavity and in the RHE model, as well as in human clinical specimens. Most importantly, the hierarchy of transcript abundance in the clinical specimens and model systems is the same [13]. ALS6 and ALS7 transcripts were difficult to detect in these studies; methods for preparing RT-PCR tubes for the analyses ensure these results are not in error. In this study and others where ALS gene expression was analyzed, large batches of RT-PCR tubes containing all ingredients except the template cDNA were prepared and frozen at -20°C until used. Each batch was tested with genomic DNA as a positive control for each primer pair, and tested without template to ensure a lack of contamination. This method of RT-PCR tube preparation also lent consistency to the overall analysis since tubes in each batch were essentially identical. The scarcity of ALS6 and ALS7 transcript suggests that either the conditions to induce high-level expression of these genes have not been identified or that a very low level of ALS6 and ALS7 transcript is sufficient for their encoded proteins to function. The latter possibility becomes more favored as the low level of ALS6 and ALS7 transcription is documented in an increasing number of experimental systems. Recent development of a quantitative real-time RT-PCR assay for the ALS family has substantiated the wide divergence in transcript copy number for highly expressed genes such as ALS1 and ALS3 and the relatively quiet ALS6 and ALS7 [15]. This apparent divergence in transcriptional activity suggests the potential for different functions for the various Als proteins.
Considering the obvious differences between the RHE and rat models of oral candidiasis, it was unexpected to find that patterns of ALS expression matched so closely. Results from these analyses validate the use of either system for phenotypic evaluation of alsΔ/alsΔ mutant strains. Interestingly, patterns of ALS gene expression in oral models differed from those observed in clinical vaginal specimens and vaginal disease models where transcription of ALS4 and ALS5 was most difficult to detect [14]. Taken together, these data suggest that expression of the ALS genes is influenced by host site.
After our work with the rat model had begun, a murine model of oropharyngeal candidiasis (OPC) was described that relies on immunosuppression with cortisone acetate and addition of tetracycline to drinking water to promote disease [20]. The authors reported that white patches, comparable to the pseudomembrane observed in human thrush, became visible on the dorsum of the tongue 2 or 3 days after inoculation and remained through day 9. While gross lesions were observed on the rat tongues in our experiments, they were never as extensive as those described for the murine model. Because the rats in this model have an intact immune system, the observed differences in pathology highlight a fundamental difference in the two model systems. Additionally, the basis of the rat model is the profound change in ecology of the oral cavity, while immunosuppression is the basis for disease in the mouse model. Perhaps the rat model represents more of a picture of what would be expected in individuals with chronically irritated mouths like Sjogren’s syndrome patients, while the murine model reflects the oral ecology of an AIDS patient. However, gene expression results from analysis of clinical material from HIV-positive patients closely match results from the rat model system suggesting similarities in oral ALS gene expression regardless of the underlying host factors that contribute to disease.
Despite the differences between the rat and mouse models, ALS1 potentially serves as a unifying link between them. Kamai et al. [21] demonstrated that fungal burden within the tongue and associated soft tissue was diminished at day 1 in mice infected with an als1/als1 mutant compared to the wild-type organism and that tongue lesions were smaller in mice infected with the mutant strain. The authors reasoned that adhesion in the early stages of infection could account for the apparently diminished virulence, so that strains were assayed for adherence to mouse tongues ex vivo, where the als1/als1 mutant was the least adherent strain. These studies provided functional data to support the importance of Als1p in the early stages of development of oropharyngeal candidiasis and are consistent with our observations that the ALS1 transcript is easily detected in the oral cavity. Additional analysis of alsΔ/alsΔ mutant strains in oral disease models will provide greater insight into the role of other Als proteins in development of oral candidiasis.
Acknowledgements
We thank Jane Chladny and the histopathology staff of the Veterinary Diagnostic Laboratory for preparation and staining of slides. We also thank Kimberly Bunag for her work with the rat model. This work was supported by Public Health Service grant DE14158 from the National Institutes of Health.
References
- 1.Odds FC. Candida and Candidosis. 2nd edn. Balliere Tindall; London: 1988. [Google Scholar]
- 2.Budtz-Jorgensen E, Mojon P, Rentsch A, Deslauriers N. Effects of an oral health program on the occurrence of oral candidosis in a long-term care facility. Community Dent Oral Epidemiology. 2000;28:141–149. doi: 10.1034/j.1600-0528.2000.028002141.x. [DOI] [PubMed] [Google Scholar]
- 3.Meitner SW, Bowen WH, Haidaris CG. Oral and esophageal Candida albicans infection in hyposalivatory rats. Infect Immun. 1990;58:2228–2236. doi: 10.1128/iai.58.7.2228-2236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Totti MAG, Jorge AOC, dos Santos EB, de Almeida OP, Scully C. Implantation of Candida albicans and other Candida species in the oral cavity of rats. J Oral Pathol Med. 1996;25:308–310. doi: 10.1111/j.1600-0714.1996.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 5.Samaranayake YH, Samaranayake LP. Experimental oral candidiasis in animal models. Clin Microbiol Rev. 2001;14:398–429. doi: 10.1128/CMR.14.2.398-429.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole MF, Bowen WH, Zhao XJ, Cihlar RL. Avirulence of Candida albicans auxotrophic mutants in a rat model of oropharyngeal candidiasis. FEMS Microbiol Lett. 1995;126:177–180. doi: 10.1111/j.1574-6968.1995.tb07413.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhao XJ, McElhaney-Feser GE, Bowen WH, Cole MF, Broedel SE, Jr., Cihlar RL. Requirement for the Candida albicans FAS2 gene for infection in a rat model of oropharyngeal candidiasis. Microbiology. 1996;142:2509–2514. doi: 10.1099/00221287-142-9-2509. [DOI] [PubMed] [Google Scholar]
- 8.Zhao XJ, Newsome JT, Cihlar RL. Up-regulation of two Candida albicans genes in the rat model of oral candidiasis detected by differential display. Microb Pathog. 1998;25:121–129. doi: 10.1006/mpat.1998.0218. [DOI] [PubMed] [Google Scholar]
- 9.Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 10.Gaur NK, Klotz SA. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun. 1997;65:5289–5294. doi: 10.1128/iai.65.12.5289-5294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol. 2002;44:61–72. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X, Oh S-H, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJP, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparison between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 13.Green CB, Cheng G, Chandra J, Ghannoum MA, Hoyer LL. RT-PCR detection of Candida albicans gene family expression in the reconstituted human epithelium (RHE) model of superficial oroesophageal candidiasis. Microbiology. 2004;150:267–275. doi: 10.1099/mic.0.26699-0. [DOI] [PubMed] [Google Scholar]
- 14.Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr., Trupin SR, Hoyer LL. Comparison between Candida albicans agglutinin-like sequence (ALS) gene expression patterns in human clinical specimens and models of vagainal candidiasis. Infect Immun. 2005 doi: 10.1128/IAI.73.3.1656-1663.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green CB, Zhao X, Yeater KM, Hoyer LL. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology. 2005 doi: 10.1099/mic.0.27696-0. in press. [DOI] [PubMed] [Google Scholar]
- 16.Gillum AM, Tsay EYH, Kirsh DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae URA3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 17.Luna L, editor. Manual of Histologic Staining mMethods of the Armed Forces Institute of Pathology. 3rd ed. McGraw-Hill; New York: 1968. [Google Scholar]
- 18.Naglik JR, Newport G, White TC, Fernandes-Naglik LL, Greenspan JS, Greenspan D, Sweet SP, Challacombe SJ, Agabian N. In vivo analysis of secreted aspartyl proteinase expression in human oral candidiasis. Infect Immun. 1999;67:2482–2490. doi: 10.1128/iai.67.5.2482-2490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collart MA, Oliviero S. Preparation of yeast RNA. In: Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley and Sons; New York: 1993. pp. 13.12.11–13.12.15. [DOI] [PubMed] [Google Scholar]
- 20.Kamai Y, Kubota M, Kamai Y, Hosokawa T, Fukuoka T, Filler SG. New model of oropharyngeal candidiasis in mice. Antimicrob Agents Chemother. 2001;45:3195–3197. doi: 10.1128/AAC.45.11.3195-3197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamai Y, Kubota M, Kamai Y, Hosokawa T, Fukuoka T, Filler SG. Contribution of Candida albicans ALS1 to the pathogenesis of experimental oropharyngeal candidiasis. Infect Immun. 2002;9:5256–5258. doi: 10.1128/IAI.70.9.5256-5258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]