Abstract
The 2′-fluoro/2′-O-methyl modified RNA aptamer Macugen is a potent inhibitor of the angiogenic regulatory protein, VEGF165. Macugen binds with high affinity to the heparin-binding domain (HBD) of VEGF165. Hydrogen exchange rates of the imino protons were measured for free Macugen and Macugen bound to the HBD or full-length VEGF to better understand the mechanism for high affinity binding. The results here show that the internal loop and hairpin loop of Macugen are highly dynamic in the free state and are greatly stabilized and/or protected from solvent upon protein binding.
Keywords: NMR, Hydrogen exchange rate, RNA aptamer, VEGF, RNA-protein complex
Introduction
Macugen is the first aptamer to be employed as a human therapeutic and was derived from an in vitro selection against the key angiogenic regulator protein, vascular endothelial growth factor, VEGF165 [1]. VEGFs are a set of essential growth factors that are required for both development of new blood vessels and maintenance of normal vasculature. Binding of VEGFs to various cell surface tyrosine kinase receptors is a critical step in initiation of angiogenesis [2]. VEGFs are also involved in various pathological processes including age-related macular degeneration, diabetic retinopathy and cancer [2]. VEGF165, the most abundant isoform, has been identified as the pathological isoform and therefore represents a drug target for various disease states [2]. The in vitro selection for VEGF165 employed 2′-fluoro pyrimidine modifications to enhance stability against ribonucleases [1]. The clinically used therapeutic, Macugen, is further modified with a 3′ cap, 2′-O-methyl ribose sugars on all but two of the purine residues and a 40 kD PEG on the 5′ end (see Fig. 1A) [1]. The anti-VEGF aptamer (referred to here as Macugen) binds to VEGF165 (referred to here as VEGF) with high affinity (Kd ~ 50 pM), requires Ca2+ ions for binding and has been shown to efficiently inhibit blood vessel growth [1,3]. VEGF consists of two independent domains, a receptor-binding domain and a heparin-binding domain (HBD) [2]. There are NMR and/or X-ray crystal structures for each of the individual domains, and the 55-amino acid HBD has no hydrophobic core but instead four disulfides are critical for stabilizing the protein fold [4,5]. A photo-crosslinking study indicated that Macugen specifically recognizes VEGF by targeting the HBD [3]. Previous studies also showed that Macugen binds to the HBD in vitro with 12 nM affinity and that the isolated HBD efficiently competes with full-length VEGF for Macugen binding in cell culture [6]. NMR studies showed very similar secondary structure and chemical environment of the phosphate backbone for Macugen, whether it is bound to the HBD or to VEGF [6]. These studies also showed new imino proton resonances for Macugen upon binding to either the HBD or VEGF indicating the aptamer is stabilized by complex formation [6]. To more directly address the extent of stabilization of Macugen upon binding protein, the hydrogen exchange rates of imino protons were measured here for free Macugen and Macugen bound to the HBD and to full-length VEGF. The results show that the aptamer employs an induced-fit mechanism to achieve high affinity binding.
Figure 1.
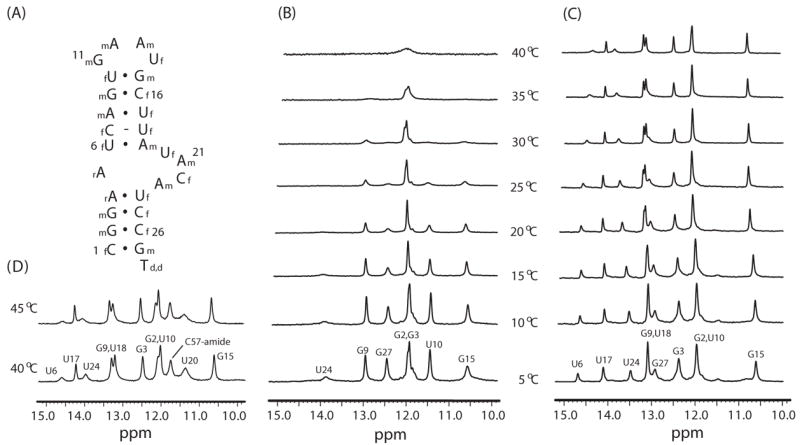
(A) The sequence and secondary structure of Macugen. 2f and 2m indicate 2′-fluoro and 2′-O-methyl modifications. Temperature-dependent imino proton spectra of (B) free Macugen, (C) the HBD-Macugen complex and (D) the VEGF-Macugen complex.
Material and Methods
Non-pegylated Macugen (Figure 1A) was obtained from Transgenomics (Omaha, NE). Uniformly 15N-labeled HBD (VEGF111–165) and 15N, 2H-labeled mouse VEGF164 were expressed in Pichia pastoris and purified as previously described [6]. The 0.5 mM HBD-Macugen and 0.1 mM VEGF-Macugen 1:1 complexes were prepared and exchanged into NMR buffer (10 mM Tris-d11, pH 7.0, 100 mM NaCl, 0.55 mM CaCl2, 0.05 mM EDTA in 90% H2O/10% D2O). NMR experiments were carried out on Varian Inova 500 or 600 MHz spectrometers equipped with room temperature or cyrogenic z-axis pulsed-field gradient probes, respectively. The exchange rates of the imino protons were determined as previously described [7,8]. Briefly, the apparent longitudinal relaxation rate constants, R1a, for the imino protons were measured by semi-selective inversion recovery 1D NMR experiments, using delay times ranging from 0.001 to 5 s. A water-selective inversion recovery experiment was used to determine the longitudinal relaxation rate constants for water, R1w, which were 0.34 and 0.27 s−1 at 15 °C and 35 °C, respectively. Water magnetization transfer experiments were performed using delay times ranging from 5 to 100 ms (Macugen and HBD-Macugen) or from 5 to 50 ms (VEGF-Macugen). The imino hydrogen exchange rate constants (kex) were determined by fitting to Eqn. 1:
| (Eqn. 1) |
where I0 and I(t) are the peak intensities of the imino proton in the water magnetization transfer experiments at times zero and t, respectively [9].
Results and Discussion
Hydrogen exchange properties of the imino protons were measured here to probe changes in base pair dynamics of Macugen upon formation of the Macugen-HBD or Macugen-VEGF complexes. The resonance assignments of the imino proton spectra of free Macugen and Macugen complexed with HBD or VEGF were previously reported [6]. Figs. 1B and 1C show the temperature dependencies of the imino proton spectra of Macugen free and complexed with HBD. Some imino protons show chemical shift changes upon binding to the HBD (U10 and G3) and new resonances are observed (U6, U17, U18 and U24) [6]. These new resonances are in slow exchange with water in the Macugen-HBD complex but exchange too fast to be observed in the free Macugen. All the base-paired imino proton resonances except the terminal G27 are still observed up to 40 °C in the HBD-Macugen complex whereas most of the imino proton resonances are extensively broadened or not observed at 40 °C in free Macugen (Fig. 1). The imino proton spectrum for Macugen complexed with VEGF at 40 °C is very similar to the spectrum for the HBD-Macugen complex except, as previously noted, the U20 resonance is observed in the VEGF but not the HBD complex [6]. These results clearly demonstrate that individual base pairs and the overall secondary structure of Macugen are greatly stabilized upon binding to the isolated HBD or full-length VEGF.
The hydrogen exchange rate constants of the imino protons, kex, were determined as described in Methods. The R1a measured here (Fig. 5S in Supporting Material) represents the sum of the spin lattice relaxation rate constant R1 and kex. The R1a values were determined for free Macugen at 15 °C and for the HBD-Macugen complex at 15 and 35 °C and the VEGF-Macugen complex at 35 °C (Table 2S Supporting Material). Fig. 2A shows 1D difference spectra of the magnetization transfer experiment as a function of delay time between the selective water inversion and detection pulse for free Macugen at 15 °C. The non-exchangeable H2′ and H2″ resonances on dT28 are used as controls to identify interference arising from NOEs with water and show no peaks in these difference spectra below 100 ms. In contrast, there are large changes in the intensity of many imino proton resonances indicating exchange with water. The relative peak intensities of the difference spectra, [I0−I(t)]/I0, for the imino proton resonances of free Macugen at 15 °C are plotted as a function of delay time in Fig. 2B and these data were used to determine kex by fitting to Eqn. 1. Analogous difference spectra for the Macugen-HBD complex at 15 and 35 °C are shown in Figs. 3A and 3B respectively. Plots of [I0−I(t)]/I0 for several imino protons in the Macugen-HBD are shown in Fig. 4. The kex values for individual imino protons in free Macugen and Macugen in the HBD and VEGF complexes are given in Table 1. In free Macugen at 15 °C, residues G2 and G3 in the lower stem and G9 in the upper stem all form Watson-Crick base pairs and the imino protons for G2 and G3 exchange over 4-fold slower than for G9. As expected, relatively rapid exchange is observed for the imino proton on the terminal C1·G27 base pair (kex = 76 s−1 for G27) as well as for the imino protons in the G15·U10 wobble in the upper stem (kex = 32 and 25 s−1 for U10 and G15). The imino protons in the internal and hairpin loops, the A4·U24 and U6·A19 base pairs neighboring the internal loop and the A8·U17 base pairs in the upper stem all exchanged too fast with solvent to be observed in the NMR spectra (kex > 400 s−1 Table 1). These results show that a large part of Macugen, including the internal loop, the hairpin loop and parts of the upper and lower stem are quite dynamic in the free state.
Figure 2.
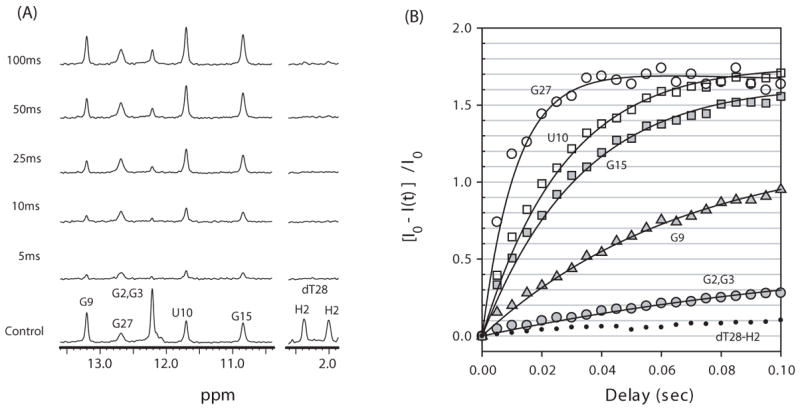
(A) Imino proton difference spectra between a control spectrum (shown at bottom) and spectra acquired by varying the delay time after water inversion for free Macugen at 15 °C. The control spectrum is the same watergate solvent suppression experiment except that no water inversion was performed. The H2′ and H2″ resonances of dT28 are shown on the right to compare exchange and NOE effects. (B) The relative peak intensities in the difference spectra, [I0−I(t)]/I0, for various imino protons in free Macugen as a function of delay time after water inversion. Solid lines indicate the best fit to Eqn. 1. The peak intensities of dT28-H2″ resonances, which arise from an NOE effect, are represented as small closed circles.
Figure 3.
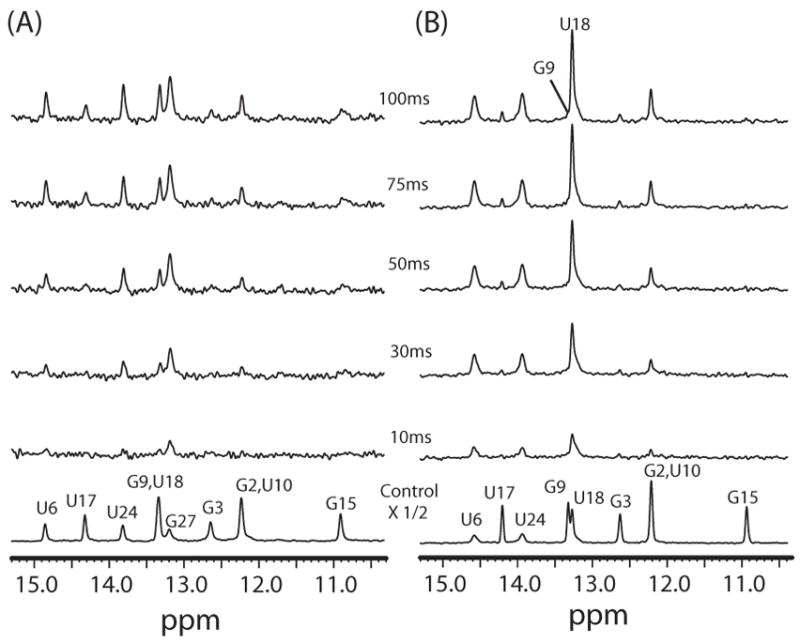
Imino proton difference spectra between control spectra (shown on bottom) and spectra acquired by varying the delay time after water inversion for the HBD-Macugen complex at (A) 15 °C and (B) 35 °C. The control spectra are the watergate solvent suppression experiment except that no water inversion was performed.
Figure 4.
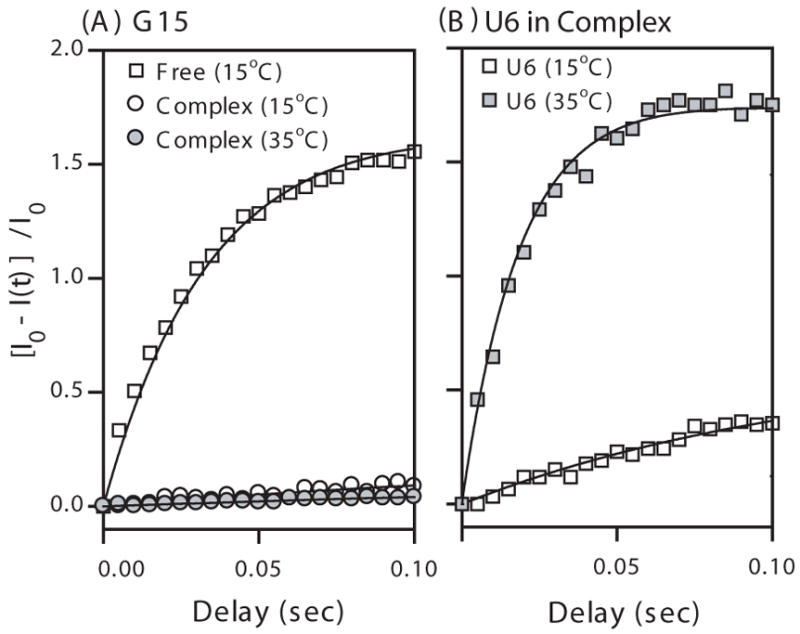
The relative peak intensities in the difference spectra, [I0−I(t)]/I0, as a function of delay time (A) for the G15 imino protons in free Macugen (15 °C) and in the HBD-Macugen complex (15 °C and 35 °C) and (B) for the U6 imino proton of the HBD-Macugen complex (15 °C and 35 °C). Solid lines indicate the best fitting of these data using Eqn. 1. The U6 imino proton exchanges too fast to be observed in free Macugen.
Table 1.
Hydrogen exchange rate constants of the imino protons, kex, for Macugen free and in the HBD-Macugen and VEGF-Macugen complexes (s−1)
| Base Pair | Imino proton | Free Macugen
|
HBD-Macugen
|
VEGF-Macugen
|
|
|---|---|---|---|---|---|
| 15 °C | 15 °C | 35 °C | 35 °C | ||
| G1·C27 | G27 | 76 ± 5 | 16 ± 5 | >400a | >400a |
| G2·C26 | G2 | ≤2.1b | <1.5c,d | <1.5c,d | <1.5c,d |
| G3·C25 | G3 | ≤2.1b | <1.5c | <1.5c | <1.5c |
| A4·U24 | U24 | >400a | 4.2 ± 0.5 | 33 ± 2 | 20 ± 4 |
| n. d. f | U20 | >400a | >400a | >400a | 44 ± 5 |
| U6·A19 | U6 | >400a | 2.7 ± 0.5 | 48 ± 3 | 29 ± 8 |
| C7·U18 | U18 | >400a | <1.5c,e | 16 ± 1 | 8 ± 3 |
| A8·U17 | U17 | >400a | <1.5c | <1.5c | <1.5c |
| G9·C16 | G9 | 9.0 ± 0.6 | <1.5c,e | <1.5c | <1.5c |
| U10·G15 | U10 | 32 ± 2 | <1.5c,d | <1.5c,d | <1.5c,d |
| G15 | 25 ± 1 | <1.5c | <1.5c | <1.5c | |
No resonances were observed for these imino protons under these conditions. Simulated two-site chemical exchange of the imino protons with water yielded a lower limit of 400 s−1 for these exchange rates (data not shown).
The G2 and G3 resonances overlap and only an upper limit for each proton is reported.
Only an upper limit of kex < 1.5 s−1 is reported due to a potential NOE with water interfering with measurement of the exchange rate (see text).
The G2 and U10 resonances overlap and only an upper limit on the kex for each proton is reported.
The G9 and U18 resonances overlap and only an upper limit on the kex for each proton is reported.
The base pair for U20 was not determined.
A high degree of solvent protection was observed for the Macugen-HBD complex at 15 °C, where many of the imino protons exchanged too slowly to be accurately determined from this NMR hydrogen exchange method (kex < 1.5 s−1). As seen in the 1D difference spectrum at 100 ms in Figs. 2A and 2B there are small but observable intensities for the nonexchangeable H2′ and H2″ resonances of dT28. These peaks cannot be due to solvent exchange and thus arise from direct or spin-diffused NOE interactions with water [10]. Therefore, it was only possible to place an upper limit of < 1.5 s−1 for the slowest exchanging imino protons. In addition, resonance kex overlap for G2/U10 and G9/U18 prevented the kex values for these protons from being uniquely determined so only an upper limit for each pair is reported (Table 1). The U14 and U20 imino proton resonances were not observed which means the kex > 400 s−1 in the HBD-Macugen complex. Interestingly, the kex for G15 of the G·U wobble pair was too slow to accurately measure in the complex (Fig. 4B) and therefore is reduced by at least a factor of 25 upon binding the HBD (Table 1). The U6 and U24 imino protons are in base pairs neighboring the internal loop and have kex of 2.7 and 4.2 s−1 respectively, in the complex as compared with their kex of > 400 s−1 in free Macugen (Table 1). These results demonstrate that the secondary structure of Macugen and the internal loop and hairpin loop are greatly stabilized by formation of the HBD-Macugen complex.
To further identify the most stable regions of Macugen in the complex, kex measurements were performed at 35 °C in the HBD- and VEGF-Macugen complexes. The imino protons for G2 and G3 in the lower stem and G9 and U17 in the upper stem still exchanged too slowly to accurately measure (Table 1). Interestingly, the U10 and G15 in the wobble pair also exchanged too slowly to measure in either complex at 35 °C (Fig. 4A). This compares with the imino protons of the central U·G wobble pair in an 11 base-paired RNA duplex which had kex values of 13 to 16 s−1 under similar conditions [8]. These results indicate that the base pairs in the upper stem, including the U10·G15 wobble pair, are strongly stabilized and/or much more protected from solvent, upon protein binding. This is consistent with previous photo-crosslinking of U14 in the hairpin loop with C137 in the HBD of VEGF [3]. Excluding the terminal G·C base pair, the U6·A19 and A4·U24 base pairs that neighbor the internal loop had the fastest exchange for any Watson-Crick base pairs in the HBD complex at both temperatures (Table 1, Fig. 4B). The VEGF-Macugen complex (Table 1) shows slightly smaller kex for residues U6, U18 and U24 in the internal loop compared to the complex with the HBD. The major difference between these complexes is for the imino proton on U20 in the internal loop, which goes from being in too fast exchange to observe in the HBD complex (kex > 400 s−1) to kex of 44 s−1 in the VEGF complex at 35 °C. Thus, amino acids outside the HBD are directly or indirectly having a large effect on hydrogen exchange of the U20 imino proton in the VEGF complex. These data show that for Macugen in the HBD and VEGF complexes, base pairs in the upper stem and the U·G wobble pair in hairpin loop have a higher degree of stabilization and/or are more protected from solvent than residues in the internal loop.
The hydrogen exchange data obtained here show very rapid exchange for many imino protons in the internal loop and base pairs neighboring the internal loop in the free aptamer. There are very large changes in for many of these imino protons upon binding of the HBD or VEGF kex protein. These results demonstrate that even RNAs that bind their target protein with extremely high affinities (Macugen binds VEGF with Kd = 50 pM) can be very dynamic in their free state. A dynamic free state has been previously observed for the U1A RNA which also forms a high affinity complex with the U1A protein [11]. The hydrogen exchange data here support an induced-fit type mechanism for the aptamer binding its protein target [12,13].
Supplementary Material
Acknowledgments
This work was supported by NIH Grant AI33098 to A.P. and KRF Grant (MOEHRD, Basic Research Promotion Fund) (KRF-2006-331-C00188) and KOSEF Grant (MOST, R01-2007-000-10691-0) to J.-H.L. We thank Dr. Richard Shoemaker for performing the simulations for rapid exchange of the imino protons with water.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 3.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165) J Biol Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 4.Wiesmann C, Christinger HW, Cochran AG, Cunningham BC, Fairbrother WJ, Keenan CJ, Meng G, de Vos AM. Crystal structure of the complex between VEGF and a receptor-blocking peptide. Biochemistry. 1998;37:17765–17772. doi: 10.1021/bi9819327. [DOI] [PubMed] [Google Scholar]
- 5.Stauffer ME, Skelton NJ, Fairbrother WJ. Refinement of the solution structure of the heparin-binding domain of vascular endothelial growth factor using residual dipolar couplings. J Biomol NMR. 2002;23:57–61. doi: 10.1023/a:1015346504499. [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF(165) Proc Natl Acad Sci USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guéron M, Leroy JL. Studies of base pair kinetics by NMR measurement of proton exchange. Meth Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Pardi A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007;35:2965–2974. doi: 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snoussi K, Leroy JL. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry. 2001;40:8898–8904. doi: 10.1021/bi010385d. [DOI] [PubMed] [Google Scholar]
- 10.Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci U S A. 2002;99:12697–12702. doi: 10.1073/pnas.202477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 12.Westhof E, Patel DJ. Nucleic acids. From self-assembly to induced-fit recognition. Curr Opin Struct Biol. 1997;7:305–309. doi: 10.1016/s0959-440x(97)80044-9. [DOI] [PubMed] [Google Scholar]
- 13.Williamson JR. Induced fit in RNA-protein recognition. Nature Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


