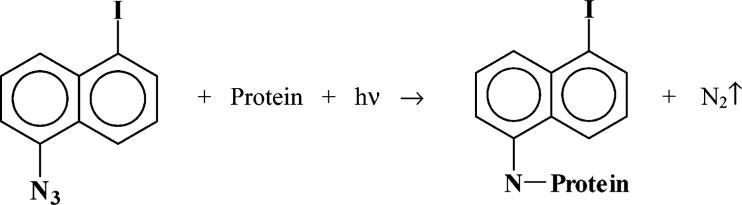Abstract
The interactions of HIV-1 Env (gp120-gp41) with CD4 and coreceptors trigger a barrage of conformational changes in Env that drive the membrane fusion process. Various regions of gp41 have profound effects on HIV entry and budding. However, the precise interactions between gp41 and the membrane have not been elucidated. To examine portions of membrane proteins that are embedded in membrane lipids, we have studied photoinduced chemical reactions in membranes using the lipid bilayer specific probe iodonaphthyl azide (INA). Here we show that in addition to the transmembrane anchor, amphipatic sequences in the cytoplasmic tail (CT) of HIV-1 gp41 are labeled by INA. INA labeling of the HIV-1 gp41 CT was similar whether wild-type or a mutant HIV-1 was used with uncleaved p55 Gag, which does not allow entry. These results shed light on the disposition of the HIV-1 gp41 CT with respect to the membrane. Moreover, our data have general implications for topology of membrane proteins and their in situ interactions with the lipid bilayer.
The human immunodeficiency virus (HIV1) gains entry into its target cell through a fusion process driven by its membrane protein gp41 (1). HIV-1 Env (gp120-gp41) is present on the surface of the virus in a metastable form generated by cleavage with furin of the gp160 Env precursor (2). Upon binding to its target cell, the interactions of HIV-1 Env with CD4 and coreceptors trigger a barrage of conformational changes in Env that drive the membrane fusion process (3). The HIV-1 envelope glycoproteins’ disposition and reorganization as a result of conformational changes is critical for entry and neutralization (4). Although different regions of HIV-1 gp41 have profound effects on its fusogenic potency, the precise interactions of gp41 with membranes have not been elucidated. To examine portions of membrane proteins that are embedded in membrane lipids, we have studied photoinduced chemical reactions in membranes using the lipid bilayer specific probe iodonaphthyl azide (INA).
INA is composed of three moieties. The core of the compound is a naphthalene ring that confers a very high hydrophobicity. Its partition coefficient in biological membranes of ∼105 (5) ensures its selective and exclusive targeting to the lipidic bilayer. Its azido moiety allows its photoactivation. Upon irradiation with near UV light, the azido group undergoes transformation into a highly reactive nitrene that will covalently bind the proteins and lipids in its vicinity (6). Finally, the iodine on this compound provides a very sensitive detection through its radioactive form. [125I]-INA has been extensively used to exclusively label the portions of membrane proteins that are embedded within the lipid domain (6-13). We applied hydrophobic labeling to the HIV-1 Env expressed on the virus or on the surface of cells to assess the topology of the Env glycoprotein.
MATERIALS AND METHODS
Reagents and Cells
Dulbecco's modified Eagle medium (DMEM), RPMI, fetal bovine serum (FBS), G418, hygromycin, and antibiotics were obtained from Invitrogen (Carlsbad, CA). 293T and HeLa cells were cultured in DMEM supplemented with 10% FBS and antibiotics (DMEM-10). Different HIV-1 Env were transiently expressed on the surface of HeLa cells using the recombinant vaccinia vectors vSC60 (IIIB/Lai BH8 Env) or vPE17 (14) (IIIB/Lai BH8 Env truncated at AA733). GHOST (3) X4/R5 (G345), a HOS-derived cell line that stably expresses CD4, CXCR4, and CCR5 was obtained from the AIDS repository. G345 were propagated in DMEM-10 supplemented with 500 μg/ mL G418, 100 μg/mL hygromycin, and 1 μg/mL puromycin. Anti HIV-1 p17 and p24 antibodies were purchased from Advanced Biotechnologies Inc (Columbia, MD). Anti gp41 Mab Chessie 8 (epitope PDRPEG) was originally developed by George Lewis (15) and obtained from the NIH AIDS Research and Reference Reagent Program. Unconjugated mouse IgG True Blot was purchased from eBioscience (San Diego, CA) and was labeled using Alexa fluor 680 succinimidyl ester (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. All other chemicals were obtained from Sigma (St. Louis, MO) and from the highest purity available.
Virus Production
Virus particles were obtained through transfection of the pNL43 (AF324493) (PR+) or pNL43/PR− plasmids (16) into 293T cells. The PR+ virus has a normal functioning protease whereas the PR− virus is protease defective. The cells were maintained in culture in DMEM-10 and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Transfections were performed in 10 cm dishes plated at 106 cells/dish. Virus-containing super-natants were harvested 2 days post-transfection, and virions were concentrated (10 to 20×) by centrifugation (20 000×g for 2h). Following centrifugation, pelleted virons were resuspended in 0.5 mL of RPMI supplemented with 10% FBS.
INA Labeling
[125I]-INA was prepared as previously described (7). [125I]-INA or INA was added to the cells/virus in suspension under subdued light. The cells/virus were then incubated at 4 °C for 10 min. Reduced glutathione was added to a final concentration of 20 mM in order to quench any activated INA molecule that would partition out of the lipidic bilayer. The cells were irradiated with UV light using a 100-W ozone-free mercury arc lamp placed in a lamphouse with a collector lens (Olympus, Center Valley, PA). Samples were irradiated through a 310-nm cutoff filter placed in front of the lens (to allow transmission of the 313-, 334-, and 365-nm mercury emission bands) and through a water filter (to prevent sample heating) at a distance of 5 cm from the light source. At that point, the light dose was 10 mW/cm2·s. Irradiation times were 2 min.
Quantification of Protein Labeling
Upon INA activation, the virus/cells were lysed for 2 h at 25 °C. The lysis buffer consisted of 2% Triton X-100 in Tris-buffered saline (TBS; 50 mM Tris, 138 mM NaCl, 2.7 mM KCl, pH 8) supplemented with protease inhibitors. The insoluble material was spun down at 20 000g for 15 min. The supernatant was then diluted twice in TBS, and the samples were subjected to immunoprecipitation using Chessie 8, anti p17, or anti p24 antibodies. Upon overnight incubation with the respective antibody, protein G-agarose was added, and the sample was then mixed by rotation for 2 h. Each sample was washed five times with TBS containing 1% Triton X-100. Proteins were separated by 14% SDS–PAGE and transferred to nitrocellulose membranes. Blots were incubated for 1 h in Odyssey blocking buffer (LICOR, Lincoln, NE). The blots were incubated with the appropriate primary antibody for 2 h at room temperature in Odyssey blocking buffer containing 0.2% Tween-20, washed four times for 10 min each with 0.1% Tween-20 in PBS (PBST), incubated with Alexa Fluor 680 labeled True Blot antibody in Odyssey blocking buffer with 0.2% Tween-20, and washed four times for 10 min with PBST. Immunoreactivity was detected by using an Odyssey infrared imaging system (LI-COR, Lincoln, NE). The blots were then exposed to PhosphorImager screens and quantitated using a Typhoon system (GE Healthcare, Chalfont St. Giles, United Kingdom).
In Gel Digestion and MS Analysis of gp41
Full-length glycosylated gp41 was purified as described earlier (17) and submitted to digestion with V8 protease (Sigma, St. Louis, MO) as follows. The V8 protease was resuspended in 125 mM Tris HCl, pH 6.8. Two to four micrograms of V8 was then added to the purified gp41 (about 30 μg), and the digestion was left to proceed for 1 h at 37 °C. The sample was then submitted to SDS-PAGE and the gel stained with sypro ruby (BioRad, Hercules, CA). The gel bands corresponding to gp41 were destained with 1% hydrogen peroxide. Gel slices were washed with water and 1% formic acid, dried in a Speedvac, rehydrated in 25 mM NH4HCO3 (pH 8.2) containing 10 mM DTT, and incubated at 56 °C for 1 h. The rehydration solution was replaced with 25 mM NH4HCO3, pH 8.2 containing 55 mM iodoacetamide and incubated for 45 min at ambient temperature in the dark. The gel slices were washed with 25 mM NH4HCO3, pH 8.2, dehydrated in a SpeedVac, and rehydrated with 25 mM NH4HCO3, pH 8.2, to which 20 ng/μL porcine trypsin had been added. After overnight digestion at 37 °C, peptides were extracted three times with 70% ACN/ 5% trifluoroacetic acid. The combined supernatants were ZipTipped, lyophilized, and stored at −20 °C until analysis. The sample (1 μL) was cocrystalized with an equal volume of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% ACN/1% trifluoroacetic acid and spotted directly on a stainless steel matrix-assisted laser desorption ionization (MALDI) plate. Mass spectra were acquired using a 4700 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA) operating at a laser frequency of 200 Hz. The mass spectra were internally calibrated (<20 ppm) using trypsin autolysis products. Postacquisition baseline correction and smoothing was carried out using software provided as part of the mass spectrometer. Alternatively, mass spectra were acquired using a TofSpec 2E (Micromass, Manchester, UK) operated using an acceleration voltage of 15 kV. In that case, bradykin and ACTH clip were used as internal calibrants, and postacquisition baseline correction was achieved using the MassLynx software (Waters, Milford, MA).
In Silico Analysis of INA-Labeled Peptides
The monoisotopic masses of the peptides were recovered and analyzed in silico. We first computed the anticipated peptide size pattern resulting from the digestion of gp41 with trypsin and V8 protease. Upon irradiation, INA loses a nitrogen molecule to generate the nitrene radical according to Scheme 1. Since little is known on the exact chemical reaction that nitrene undergoes with the different amino acids, we modified the predicted pattern with the addition of the monoisotopic MW of the moiety that binds to the protein, 266.954 Da, and the possible loss of hydrogen or hydroxyl moiety that can result from the addition. This gave us the in silico peptide size pattern. In order to identify any site of modification in the protein, we studied the discrepancies between this in silico pattern and the actual pattern experimentally obtained in the INA labeled sample. Any difference lower than 25 ppm was considered as a possible identification for INA modification.
Scheme 1.
RESULTS
Identification of gp41 Peptides by μLC/MS2 Analysis
The analysis of gp41 membrane inserted regions by photosensitized labeling necessitated the development of new technologies for membrane protein purification and fragmentation. HIV-1IIIB Env was expressed on the surface of HeLa cells using the recombinant vaccinia construct vSC60 and labeled with INA. Full-length gp41 was purified to homogeneity by continuous elution electrophoresis with simultaneous detergent exchange followed by affinity chromatography (17). Following in-gel tryptic digestion, sequence analysis was performed by microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry (μLC/MS2) using a Finnigan LCQ DECA XP quadrupole ion trap mass spectrometer at the Harvard Microchemistry Facility. Optimization of the in-gel tryptic digestion protocol resulted in 49% coverage of the protein by amino acid count (data not shown). However, the coverage did not identify peptides corresponding to the N-terminal region of HIV-1 gp41 (fusion peptide). Moreover, using this method, we could not ascertain the presence of INA in any of the peptides of gp41. This artifact could result from the INA group interfering with the peptide's ability to undergo ionization, the running of the peptide on the chromathography column or the peptide's fragmentation pattern.
Analysis of INA-Labeled Peptides by MALDI-MS
Since MS2 sequencing analysis did not reveal INA-labeled portions of HIV-1 gp41, an alternative strategy was employed. Full-length INA labeled gp41 was purified to homogeneity according to our published method (17). Upon digestion, the samples were submitted to MALDI-MS analysis to avoid the possible artifacts inherent to the MS2 analysis mentioned above. We yielded an average coverage of about 35%. Our goal is to identify any site of modification in the protein. The attachment of INA to the protein produced discrepancies between the anticipated peptide sizes predicted from an in silico digest of the unmodified protein and the actual MS peptide map obtained for the INA labeled sample. When the experimental masses were matched against an in silico digest of the protein along with the predicted mass of the modification, gp41 peptides labeled by 0−4 INA molecules were identified (Figure 1A,B). Two peptides from the C-terminal tail (LLP-1 and LLP-2) that interact with lipid membranes were labeled with several INA molecules each. The peptide that includes the transmembrane domain was labeled with one INA molecule. The labeled peptide was at the C-terminal end of the transmembrane domain. In order to examine portions of HIV-1 gp41 labeled as a result of fusion, we performed the same analysis following the coculture of env-expressing and target cells. We used as effector cells Hela infected with vSC60 and as targets G345. These cells were incubated together for 3 h at 37 °C, at which time fusion is complete (18, 19). The pattern of gp41 labeling with INA after fusion yielded a similar result, i.e., the labeling with INA of the transmembrane domain and the two amphipathic peptides LLP-1 and LLP-2 (Figure 1B).
Figure 1.
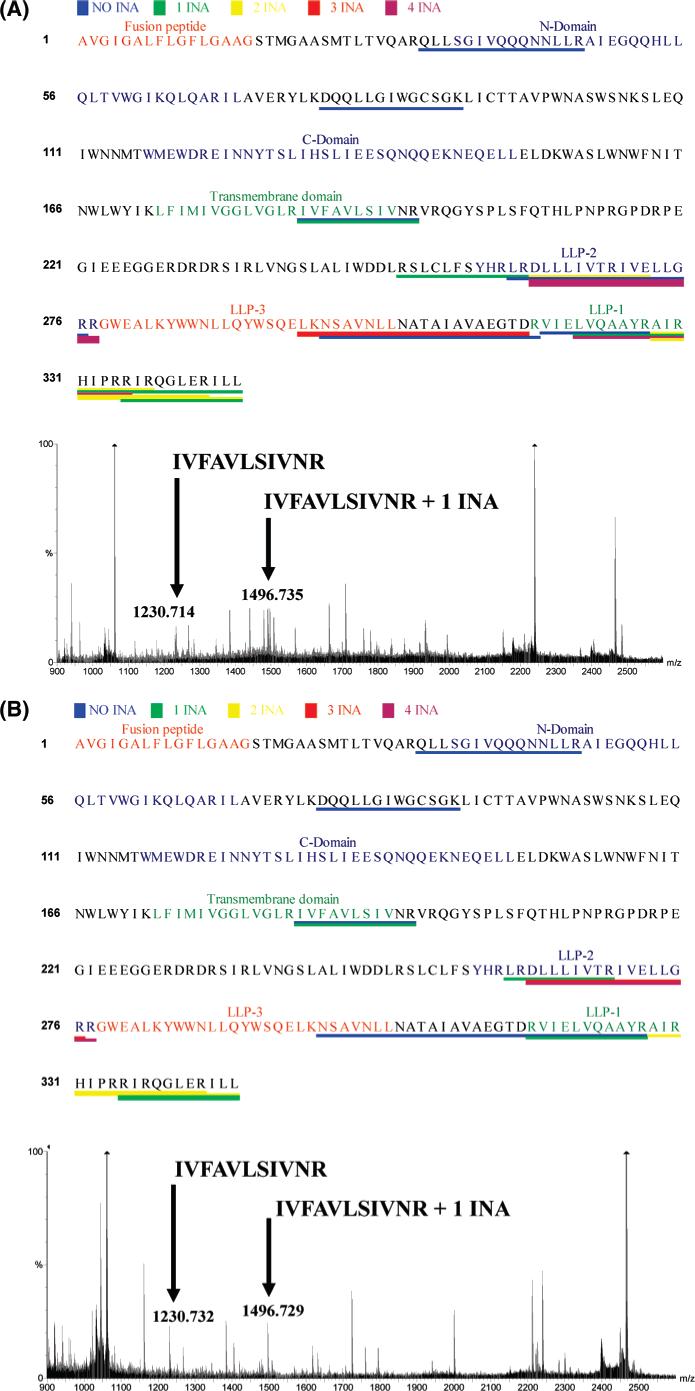
In silico mapping of INA incorporation into gp41. gp41 was labeled with INA and purified to homogeneity. Upon digestion with V8 and trypsin, the peptides generated were analyzed by MALDI analysis (representative spectra shown). The observed peptides were compared to the theoretical computed ones. Differences corresponding to the possible incorporation of INA molecules within 25 ppm were conserved. (A) gp41 recovered before fusion. (B) gp41 recovered after fusion.
[125I]-INA Labeling of the Cytoplasmic Tail (CT) of gp41
In order to confirm that massive INA labeling of WT HIV-1 gp41 occurred at the CT, we examined INA labeling of HIV-1 gp41 whose C-terminal tail was truncated (104 amino acids from the C-terminal). The truncated gp41 was expressed in HeLa cells through the vaccinia vPE17 vector, and the wild type gp41 was expressed in the same cells using vaccinia vSC60 vector. Following photoreaction with [125I]-INA, both forms of gp41 were isolated by immunoprecipitation, and the extent of [125I]-INA labeling was measured by autoradiography of the isolated proteins using a “Typhoon” phosphorimager. The truncated form of gp41 loses about 80% of its [125I]-INA labeling, as shown in Figure 2. Since this truncation corresponds only to the loss of one-third of the protein, this experiment shows that there is a highly specific labeling of the truncated domain and that a preponderant fraction of the labeling observed in the wild type protein can be attributed to the interaction of the C-terminal domain with the lipid phase of the membrane. To rule out the possibility that INA traverses the viral membrane and reacts with the gp41 CT outside of the lipidic environment, we examined Gag labeling by INA. p17 is presumably associated with gp41 (20) and only interacts with the membrane through its myristoyl moiety (21) whereas p24 is a soluble protein that has no known direct interaction with the lipidic bilayer. Figure 3 shows only background labeling of matrix proteins as compared with gp41, indicating that the gp41 CT regions are indeed embedded in the viral membrane.
Figure 2.
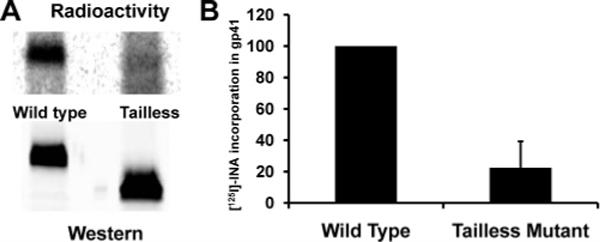
Labeling with [125I]-INA of wild type and tailless mutant of gp41 expressed in Hela cells with the vSC60 and vPE17 vaccinia constructs respectively. (A) Western blot detection of gp41 with Chessie 8 and reading of the same blot by phosphorimager. (B) Quantitation of the radioactivity in gp41 normalized by the amount of protein recovered as detected by western analysis.
Figure 3.
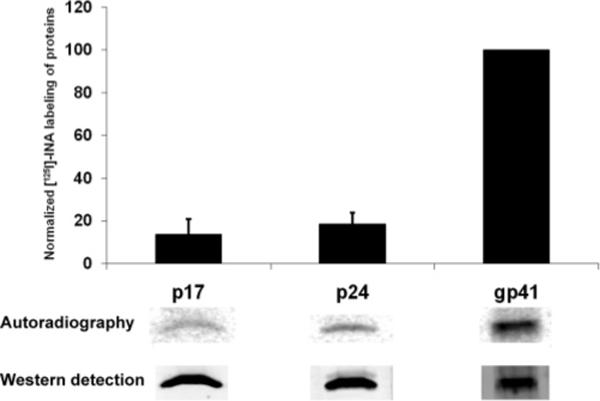
Incorporation of [125I]-INA into viral proteins. PR+ (wild type NL43) virus was labeled with [125I]-INA and p17, p24, and gp41 were recovered by immunoprecipitation. Upon running on a gel and blotting, each protein was detected by western analysis and their incorporation of [125I]-INA was measured on a phosphoimager. The resulting quantitation shown represents the average of three experiments.
Effect of p55 Cleavage on gp41 [125I]-INA Labeling
It has been shown that gp41 is more stably associated with immature rather than mature viral particles (22, 23), and cleavage of the p55 Gag precursor protein by the viral protease is required to generate Envs with maximal fusogenicity. In order to examine the gp41 topology in HIV-1 virions with uncleaved p55 Gag, we generated such particles from protease-defective HIV-1 (PR−). As shown in Figure 4 the amount of INA incorporation into HIV-1 gp41 was similar for PR− and PR+ particles. These results are consistent with the hypothesis that the gp41-Gag associations that prevail with uncleaved Gag do not affect the association of the gp41 CT with the viral membrane.
Figure 4.
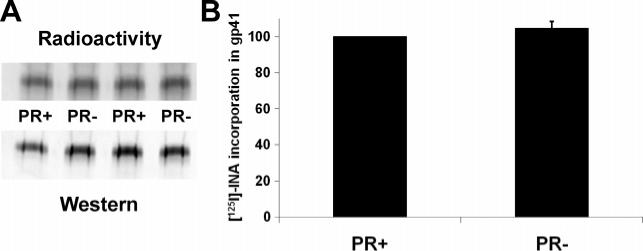
Incorporation of [125I]-INA into gp41 of PR+ and PR− viruses. PR+ and PR− viruses were labeled with INA and gp41 was recovered by immunoprecipitation. (A) Western blot detection of gp41 with Chessie 8 and reading of the same blot by a phosphorimager. (B) Quantitation of the [125I]-INA incorporation in gp41 normalized by the amount of protein recovered as detected by western analysis. The resulting quantitation shown represents the average of three experiments.
DISCUSSION
Membrane fusion mediated by HIV-1 envelope glycoproteins (gp120/gp41) is a critical step in the entry of the virus into susceptible cells. The fusion reaction involves the binding of the trimeric viral envelope glycoprotein gp120/gp41 to cell surface receptor CD4 and chemokine coreceptor CXCR4 or CCR5. These interactions trigger conformational changes in the envelope proteins (3) that ultimately lead to the formation of a six-helix bundle core structure, comprising the N- (N-HR) and C- (C-HR) terminal heptad repeat regions of the gp41 ectodomain (24-26), and membrane fusion (1, 27). In previous studies, we have dissected these steps kinetically and analyzed the molecular features of the intermediates (28, 29). Although the resolution of the gp41 core structure has guided our thinking about the way gp41 mediates membrane fusion, some crucial pieces of the puzzle are still missing. The high-resolution gp4l structures do not include the fusion peptide (FP), the membrane proximal external region (MPER), the transmembrane anchor (TM), or the cytoplasmic tail (CT). Generally, information regarding the interactions of these regions with the bilayer membrane has been gleaned from studies of interactions between peptides representing sequences of these regions and lipid membranes (30-32). However, a direct comparison of membrane-interactive properties of these peptides with liposomes and their function in the intact envelope glycoprotein is fraught with pitfalls. For instance, the ability of HIV-1 gp41 to promote membrane fusion can be completely abolished by a single amino acid G10V mutation within the fusion peptide (33). However, that same mutation did not affect the ability of the peptide to mediate lipid mixing in liposomes (Viard et al., unpublished). Nieva and Suarez (34) have performed a theoretical analysis to detect amino acid sequences of viral envelope glycoproteins putatively engaged in interactions with the target membranes using the classical Kyte–Doolittle hydropathicity scales as well as the hydrophobicity-at-interface scale, as proposed by Wimley and White (35). Of interest in this context is the fact that peptide sequences that score high on the Kyte–Doolittle scale but low on the Wimley–White scale (such as TM, LLP-1, and LLP-2) show significant INA labeling in this study. Conversely, a peptide sequence, like MPER, that scores low on the Kyte–Doolittle scale but high on the Wimley–White scale and is membrane-active in liposome assays does not seem to be labeled by INA. This observation is consistent with the hypothesis that INA labeling requires some measure of insertion of the peptide into the hydrophobic portion of the lipid bilayer, and that in the context of the full length protein, interactions with the membrane will be altered.
According to the “sticky finger” models (36), it is generally assumed that the fusion peptide inserts into the target cell following the triggering of gp120-gp41 complexes by host cell CD4 and coreceptors. However, in the case of HIV-1 gp41 fusion peptide insertion has yet to be demonstrated. This has been done for influenza hemagglutinin (37) and VSV (38) by the group of Joseph Brunner, the pioneer of hydrophobic membrane protein photolabeling techniques, using carbene-based hydrophobic markers incorporated in liposomes. Liposomes are, however, not suitable targets for HIV, and our labeling approach is based on the use of aryl azides that can be added to viruses or cells and activated in situ. This technique was proved efficient in following the incorporation of viral proteins into target cells as a result of fusion of influenza virus (11), VSV (12), and HIV/SIV (18). We could not, however, detect differential labeling of portions of gp41 resulting from the fusion process. If for VSV and influenza all the spikes can be simultaneously activated and undergo conformational change upon low pH triggering, HIV relies on the sequential interactions with its receptor and coreceptor. The percentage of total Env molecules that are actually engaged in the fusion process is thought to be low (39, 40), and their study might require a specific isolation.
Nevertheless, this study has been able to reveal some important aspects of the topology of HIV-1 gp41 in the membrane, specifically located in the CT. HIV and SIV CTs are remarkably long, and alteration in their length or mutations in conserved sequences have revealed their participation in different steps of the virus life cycle. During the circulation of the virus, the length of the tail has been correlated to a differential exposure of the virus to neutralizing antibodies (41). At the stage of entry, the cytoplasmic tail has also been implicated in regulating the kinetics of fusion and in the ability of the Env to promote syncytia (42, 43). Mutational analysis has shown that the tail interacts with p55 Gag protein at the stage of budding, allowing the incorporation of the envelope protein into the virion (44-46). Interestingly, this interaction has been related to the ability of the virus to fuse with its target. Inhibition of the protease activity encoded in the virus prevents the cleavage of p55 and has been shown to regulate fusion (22, 23). Studies with peptides derived from two α helical “lentivirus lytic peptide” domains (LLP-1 and LLP-2) that are highly conserved in HIV-1 have suggested that these domains strongly interact with the cytoplasmic leaflet of plasma membrane (47-50). The INA-labeling data presented in this study are consistent with this model (Figures 1-3). We find that INA labeling of the HIV-1 gp41 CT was the same in a wild type virus as in a mutant HIV-1 virus with uncleaved p55 Gag, which does not allow entry (Figure 4). Although these data do not reveal differences in Gag-gp41 CT interactions between PR+ and PR− virions, they do set limits on models that consider effects of p55 Gag on HIV-1 Env glycoprotein-mediated fusion. In very general terms, our data show how topology of membrane proteins can be determined and how membrane proteins interact with the lipid bilayers in living cells and viruses.
ACKNOWLEDGMENT
We are grateful to NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, for providing reagents whose provenance is listed in Materials and Methods. We thank the Keck Facility at Yale and in particular Kathryn Stone and Tom Abbot for their help in carrying some of the analysis of digested gp41 by MALDI-TOF. We thank members of the Blumenthal Lab for their help with the studies and insightful comments. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Further funding was provided by a grant from the NIH Intramural AIDS Targeted Antiviral Program (IATAP) and by a grant from the NIAID Intramural Biodefense Research Program. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the United States Government.
Footnotes
This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by a grant from the NIH Intramural AIDS Targeted Antiviral Program (IATAP), and by a grant from the NIAID Intramural Biodefense Research Program. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400.
Abbreviations: AA, amino acid; ACN, acetonitrile; AIDS, acquired immune deficiency syndrome; CD4, cluster of differenciation 4; CT, cytoplasmic tail; DMEM, Dulbecco's modified Eagle medium; DMEM-10; Dulbecco's modified Eagle medium supplemented with 10 % fetal bovine serum and antibiotics; DTT, dithiothreitol; Env, envelope; FBS, fetal bovine serum; FP, fusion peptide; G345, GHOST (3) X4/R5 cells; HIV-1, human immunodeficiency virus type 1; HOS, human osteogenic sarcoma; HPLC, high-performance liquid chromatography; HR, heptad repeat; gp, glycoprotein; IgG, immunoglobulin G; INA, 5-iodonaphthalene-1-azide; [125I]-INA, INA labeled with radioactive iodine 125; LLP, lentivirus lytic peptide; MALDI, matrix-assisted laser desorption/ionization; MPER, membrane proximal region; MS, mass spectrometry; MS2, tandem mass spectrometry; MW, molecular weight; NIH, National Institute of Health; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PBST, phosphate-buffered saline supplemented with 0.1% Tween 20; ppm, parts per million; PR−, protease deficient virus; PR+, virus with an efficient protease; RPMI, Roswell Park memorial institute medium; SDS, sodium dodecyl sulfate; SIV, simian immunodeficiency virus; TBS, Tris-buffered saline; TM, transmembrane anchor; TOF, time-of-flight; UV, ultraviolet; VSV, vesicular stomatitis virus; WT, wild-type.
REFERENCES
- 1.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed EO, Martin MA. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 3.Gallo SA, Finnegan CM, Viard M, Raviv Y, Dimitrov A, Rawat SS, Puri A, Durell S, Blumenthal R. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 5.Bayley H, Knowles JR. Photogenerated reagents for membranes: selective labeling of intrinsic membrane proteins in the human erythrocyte membrane. Biochemistry. 1980;19:3883–3892. doi: 10.1021/bi00558a001. [DOI] [PubMed] [Google Scholar]
- 6.Raviv Y, Salomon Y, Gitler C, Bercovici T. Selective labeling of proteins in biological systems by photosensitization of 5-iodonaphthalene-1-azide. Proc. Natl. Acad. Sci. U.S.A. 1987;84:6103–6107. doi: 10.1073/pnas.84.17.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bercovici T, Gitler C. 5-[125I]Iodonaphthyl azide, a reagent to determine the penetration of proteins into the lipid bilayer of biological membranes. Biochemistry. 1978;17:1484–1489. doi: 10.1021/bi00601a020. [DOI] [PubMed] [Google Scholar]
- 8.Gilk SD, Raviv Y, Hu K, Murray JM, Beckers CJ, Ward GE. Identification of PhIL1, a novel cytoskeletal protein of the Toxoplasma gondii pellicle, through photosensitized labeling with 5-[125I]iodonaphthalene-1-azide. Eukaryot. Cell. 2006;5:1622–1634. doi: 10.1128/EC.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holowka D, Gitler C, Bercovici T, Metzger H. Reaction of 5-iodonaphthyl-1-nitrene with the IgE receptor on normal and tumour mast cells. Nature. 1981;289:806–808. doi: 10.1038/289806a0. [DOI] [PubMed] [Google Scholar]
- 10.Kahane I, Gitler C. Red cell membrane glycophorin labeling from within the lipid bilayer. Science. 1978;201:351–352. doi: 10.1126/science.663661. [DOI] [PubMed] [Google Scholar]
- 11.Pak CC, Krumbiegel M, Blumenthal R, Raviv Y. Detection of influenza hemagglutinin interaction with biological membranes by photosensitized activation of [125I]iodonaphthylazide. J. Biol. Chem. 1994;269:14614–14619. [PubMed] [Google Scholar]
- 12.Pak CC, Puri A, Blumenthal R. Conformational Changes and Fusion Activity of Vesicular Stomatitis Virus Glycoprotein: 125Iodonaphthylazide Photo Labeling Studies in Biological Membranes. Biochemistry. 1997;36:8890–8896. doi: 10.1021/bi9702851. [DOI] [PubMed] [Google Scholar]
- 13.Raviv Y, Bercovici T, Gitler C, Salomon Y. Detection of nearest neighbors to specific fluorescently tagged ligands in rod outer segment and lymphocyte plasma membranes by photosensitization of 5-iodonaphthyl 1-azide. Biochemistry. 1989;28:1313–1319. doi: 10.1021/bi00429a055. [DOI] [PubMed] [Google Scholar]
- 14.Earl PL, Koenig S, Moss B. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J. Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 16.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viard M, Blumenthal R, Raviv Y. Improved Separation of Integral Membrane Proteins By Continuous Elution Electrophoresis With Simultaneous Detergent Exchange: Application to the Purification of the Fusion Protein of the Human Immunodeficiency Virus Type 1. Electrophoresis. 2002;23:1659–1666. doi: 10.1002/1522-2683(200206)23:11<1659::AID-ELPS1659>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 18.Raviv Y, Viard M, Bess J, Jr., Blumenthal R. Quantitative Measurement of Fusion of HIV-1 and SIV with Cultured Cells Using Photosensitized Labeling. Virology. 2002;293:243–251. doi: 10.1006/viro.2001.1237. [DOI] [PubMed] [Google Scholar]
- 19.Gallo SA, Puri A, Blumenthal R. HIV-1 gp41 Six-Helix Bundle Formation Occurs Rapidly after the Engagement of gp120 by CXCR4 in the HIV-1 Env-Mediated Fusion Process. Biochemistry. 2001;40:12231–12236. doi: 10.1021/bi0155596. [DOI] [PubMed] [Google Scholar]
- 20.Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 2000;74:3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veronese FD, Copeland TD, Oroszlan S, Gallo RC, Sarngadharan MG. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J. Virol. 1988;62:795–801. doi: 10.1128/jvi.62.3.795-801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami T, Ablan S, Freed EO, Tanaka Y. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J. Virol. 2004;78:1026–1031. doi: 10.1128/JVI.78.2.1026-1031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyma DJ, Jiang J, Shi J, Zhou J, Lineberger JE, Miller MD, Aiken C. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 2004;78:3429–3435. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic Structure of the Ectodomain from HIV-1 gp41. Nature. 1997;387:426–428. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 25.Chan DC, Fass D, Berger JM, Kim PS. Core Structure of gp41 from the HIV Envelope Glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 26.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry. 2007;46:1398–1401. doi: 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- 29.Dimitrov AS, Louis JM, Bewley CA, Clore GM, Blumenthal R. Conformational Changes in HIV-1 gp41 in the Course of HIV-1 Envelope Glycoprotein-Mediated Fusion and Inactivation. Biochemistry. 2005;44:12471–12479. doi: 10.1021/bi051092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion (review) Mol. Membr. Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 31.Nieva JL, Agirre A. Are fusion peptides a good model to study viral cell fusion. Biochim. Biophys. Acta. 2003;1614:104–115. doi: 10.1016/s0005-2736(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 32.Wexler-Cohen Y, Johnson BT, Puri A, Blumenthal R, Shai Y. Structurally altered peptides reveal an important role for N-terminal heptad repeat binding and stability in the inhibitory action of HIV-1 peptide DP178. J Biol. Chem. 2006;281:9005–9010. doi: 10.1074/jbc.M512475200. [DOI] [PubMed] [Google Scholar]
- 33.Delahunty MD, Rhee I, Freed EO, Bonifacino JS. Mutational analysis of the fusion peptide of the human immunodeficiency virus type 1: identification of critical glycine residues. Virology. 1996;218:94–102. doi: 10.1006/viro.1996.0169. [DOI] [PubMed] [Google Scholar]
- 34.Nieva JL, Suarez T. Hydrophobic-at-interface regions in viral fusion protein ectodomains. Biosci. Rep. 2000;20:519–533. doi: 10.1023/a:1010458904487. [DOI] [PubMed] [Google Scholar]
- 35.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 36.Blumenthal R, Dimitrov DS. Targeting the sticky fingers of HIV-1. Cell. 2007;129:243–245. doi: 10.1016/j.cell.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J. Biol. Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 38.Durrer P, Gaudin Y, Ruigrok RW, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 39.Layne SP, Merges MJ, Dembo M, Spouge JL, Nara PL. HIV requires multiple gp120 molecules for CD4-mediated infection. Nature. 1990;346:277–279. doi: 10.1038/346277a0. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. J. Virol. 2006;80:4388–4395. doi: 10.1128/JVI.80.9.4388-4395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 2002;76:2683–2691. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulligan MJ, Yamshchikov GV, Ritter GDJ, Gao F, Jin MJ, Nail CD, Spies CP, Hahn BH, Compans RW. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 1992;66:3971–3975. doi: 10.1128/jvi.66.6.3971-3975.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyss S, Dimitrov AS, Baribaud F, Edwards TG, Blumenthal R, Hoxie JA. Regulation of Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Fusion by a Membrane-Interactive Domain in the gp41 Cytoplasmic Tail. J. Virol. 2005;79:12231–12241. doi: 10.1128/JVI.79.19.12231-12241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubay JW, Roberts SJ, Hahn BH, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Yuan X, McLane MF, Lee TH, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen SS, Lee SF, Wang CT. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41. J. Virol. 2001;75:9925–9938. doi: 10.1128/JVI.75.20.9925-9938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujii G, Horvath S, Woodward S, Eiserling F, Eisenberg D. A molecular model for membrane fusion based on solution studies of an amphiphilic peptide from HIV gp41. Protein Sci. 1992;1:1454–1464. doi: 10.1002/pro.5560011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gawrisch K, Han KH, Yang JS, Bergelson LD, Ferretti JA. Interaction of peptide fragment 828–848 of the envelope glycoprotein of human immunodeficiency virus type I with lipid bilayers. Biochemistry. 1993;32:3112–3118. doi: 10.1021/bi00063a024. [DOI] [PubMed] [Google Scholar]
- 50.Srinivas SK, Srinivas RV, Anantharamaiah GM, Segrest JP, Compans RW. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J. Biol. Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]