Abstract
The transcriptional activation potential of proteins can be assayed in chimeras containing a heterologous DNA-binding domain that mediates their recruitment to reporter genes. This approach has been widely used in yeast and in transient mammalian cell assays. Here, we applied it to assay the transactivation potential of proteins in transgenic Drosophila embryos. We found that a chimera between the DNA-binding bacterial LexA protein and the transactivation domain from yeast GAL4 behaved as a potent synthetic activator in all embryonic tissues. In contrast, a LexA chimera containing Drosophila Fos (Dfos) required an unexpected degree of context to function as a transcriptional activator. We provide evidence to suggest that this context is provided by Djun and Mad (a Drosophila Smad), and that these partner factors need to be activated by signaling from Jun N-terminal kinase and decapentaplegic, respectively. Because Dfos behaves as an autonomous transcriptional activator in more artificial assays systems, our data suggest that context-dependence of transcription factors may be more prevalent than previously thought.
Transcriptional activators are modular, bearing separable DNA-binding and transactivation domains (1). The former allow them to bind to enhancers of target genes, the latter to recruit the basal transcription machinery to the promoter of these genes (2). The function of transactivation domains can be assayed if these are fused to a heterologous DNA binding domain, for example that of the bacterial regulator LexA. These chimeras reveal that typical yeast activators, such as GAL4 or GCN4, contain potent transactivation domains (1, 3). Although a single transactivation domain can suffice to activate transcription, synergy often is observed between multiple domains derived from the same, or from different, activators (e.g., refs. 4–6). This is thought to reflect multiple contacts with distinct components of the basal transcription machinery (ref. 7, and references therein).
In higher eukaryotes, transcriptional activation of target genes often is based on synergy between different enhancer-binding proteins (e.g., refs. 8–11). Typically, these enhancer-binding proteins can function autonomously to activate transcription of reporter genes from multimerized binding sites. However, there are exceptions to these “genuine” transcriptional activators, for example the high mobility group (HMG) proteins of the T cell factor (TCF) family. These factors can only activate linked genes in the presence of, and in cooperation with, adjacent DNA-binding proteins (12, 13). Therefore, TCFs act in a context-dependent way, and are thought to have architectural roles in the assembly of multiprotein enhancer complexes (14). Interestingly, TCFs serve as target transcription factors for Wnt-1/Wingless signaling (15, 16). Some of their DNA-binding partners are also signal response factors, others are tissue-specific proteins (17, 18). Context-dependence would seem to be a desirable property of signal response factors: obligatory partnerships with specific sets of other positionally activated proteins would enable a signal response factor to confer cell-type-specific responses to a canonical signaling cascade that acts on a multitude of tissues during development.
Endoderm induction in Drosophila is a developmental event during which Wingless, decapentaplegic (Dpp), and epidermal growth factor receptor (Egfr) signaling pathways synergize to stimulate the transcription of homeotic target genes (19). Drosophila Fos (Dfos) is a critical component of this inductive process: its expression is elevated locally in the endoderm in response to Dpp and Egfr signaling, and its function is required for the induction of the ultimate endodermal target gene labial (20). To test the transactivation potential of Dfos and its possible signal-dependence, we constructed chimeras between the whole or parts of Dfos and the DNA-binding LexA protein and expressed these in transgenic Drosophila embryos with the GAL4 system (21). We found that a GAL4-LexA chimera functions as an autonomous and efficient synthetic activator in all cells of transgenic embryos to stimulate transcription from a reporter containing multimerized LexA binding sites. Surprisingly, however, a Dfos-LexA chimera did not. Rather, Dfos-LexA requires a context of cooperating DNA sequences as well as inputs from different extracellular signals to function as a transcriptional activator.
Experimental Procedures
Plasmids and P Element Transformation.
For the LL and MadL reporters, the following oligonucleotides were cloned into HZ50PL as XbaI/KpnI inserts as described (22): LL (TCGAGCTGTATATACATACAGTGCTCGA)4, MadL (CTGTATATACATACAGTAGCGCCGGCGCTTCCAG)4 (LexA binding site underlined). Full-length LexA protein was used for the LexA chimeras (23). An efficient translational initiation context and a nuclear localization sequence (TPPKKKRKVED) were provided by inserting 5′-AATTCGATGGCTCATATGACCCCCCCCAAGAAGAAGCGCAAGGTGGAGGACGGA-3′ as a double-stranded oligonucleotide (13). A flexible linker encoding KLGGGAPAVGGGPK was inserted between LexA and its fusion partner (5′-AATTGGGCGGCGGCGCCCCCGCCGTGGGCGGCGGCCCCGC-3′). Constructs were generated by standard PCR techniques and checked by sequencing or by in vitro translation using the TNT Coupled Reticulocyte Lysate System (Promega). The GAL4 activation domain (GAD) spans residues 768–881, whereas the Dfos full-length, N-terminal and C-terminal inserts span residues 1–595, 1–249, and 325–595, respectively (Fig. 1B). These constructs were subsequently cloned into pUAST (21).
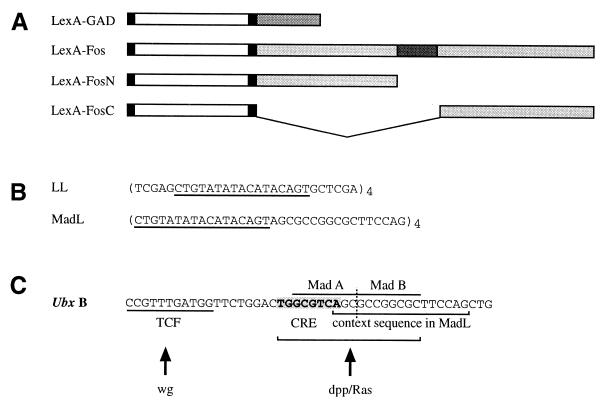
Figure 1.
Maps and sequences of constructs and of the signal-responsive module of the Ubx midgut enhancer. (A) Layout of the LexA chimeras. The LexA coding sequence (white) is flanked by translation initiation and nuclear translocation signals (left-hand black box) and flexible linker (right-hand black box). Dark gray top line, GAL4 activation domain (GAD); dark gray underneath, DNA-binding and dimerization domain of Dfos. (B) Sequences in the LL and MadL reporters; four tandem copies were inserted upstream of a canonical TATA box. The LexA binding site is underlined. The context sequence in MadL is derived from the Dpp/Ras responsive element in the Ubx midgut enhancer (bracketed in C). (C) The signal responsive module of the Ubx midgut enhancer contains a Wingless response sequence (binding site for TCF) and an adjacent bipartite Dpp/Ras response sequence (CRE, Dpp/Ras target; tandem Mad binding sites, Dpp target). Note that the Mad binding sites are also the target for a Wingless induced repressor (see text).
Transgenic flies were generated by standard techniques. For each construct, 4–8 lines were isolated and made homozygous for the transposon; each functional test in the embryo was carried out with at least two different lines.
Fly Strains and Phenotypic Analysis.
For transformation, cn, ry42 (lacZ constructs), and y w1118 flies (UAS constructs) were used as hosts. The following GAL4 driver and responder lines were used: arm.GAL4 (24) for ubiquitous expression; 24B.GAL4 (21) for mesodermal expression; 48Y.GAL4 (25) for endodermal expression; UAS.Djun and UAS.Dfos (26); UAS.junAsp, UAS.junAla (27); UAS.Drac1V12 and UAS.Dcdc42V12 (28); UAS.Dras1V12 (29).
The expression levels of different LexA constructs were tested (and compared between individual transgenic lines) when possible by α-Dfos antibody staining (20) after GAL4-mediated expression in the embryo. Also, they were overexpressed in the eye or wing disk [with GMR.GAL4 (30) and ms1096.GAL4 (31), respectively], which tends to cause phenotypic effects (eye roughening, wing crumpling, wing vein defects), probably because of the LexA chimeras acting through fortuitous genomic LexA binding sites near developmental control genes. On the basis of the severity of these phenotypes, individual lines were selected for each construct for further analysis. Their functionality was confirmed by their quenching effects on reporter-mediated background expression (see Results) in the embryo. Reporter gene expression was monitored by staining embryos with a mouse α-lacZ antibody (Promega) as described (32).
Results
We constructed chimeras that contain the LexA protein fused either to the whole of Dfos (LexFos), or to its N-terminal (LexFosN) or its C-terminal part (LexFosC). Only the former, but neither of the latter, contains the Dfos leucine zipper, which is known to mediate dimerization with Dfos itself and also with Djun (33). As a control, we fused the same part of LexA to the transactivation domain of GAL4 (LexGAD). These chimeras were expressed in transformed embryos by the GAL4 system (21) (Fig. 1A).
To test these chimeras, we generated a reporter gene (LL) that contains four tandem LexA binding sites linked to a canonical TATA box and the β-galactosidase (lacZ) coding sequence (Fig. 1B). We also designed an alternative reporter gene (MadL; Fig. 1B) that is similar to LL, but in which the LexA binding sites are interspersed with a context sequence derived from the midgut enhancer of Ultrabithorax (Ubx). This context sequence includes a Mad (Mothers against Dpp) binding site, which, in the Ubx enhancer, cooperates with an adjacent CRE-like sequence to mediate the response to Dpp and Egfr signaling (Fig. 1C) (19, 26). Our evidence suggested that Dfos may be able to act through this CRE-like sequence (unpublished observations). Hence, the MadL reporter was designed to test whether the Dfos chimeras required context sequences for function.
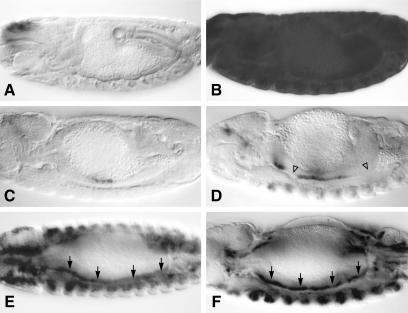
Individual transformants were established and tested for expression levels and functionality of their LexA chimeras (see Experimental Procedures). Embryos from LL and MadL reporter transformant lines were stained with an antibody against lacZ, revealing in both cases a distinct background pattern of lacZ staining that barely varied between lines. LL-mediated staining was very low (Fig. 2A). MadL-mediated background staining was more noticeable, especially in the endoderm primordia, and later in the middle midgut (Fig. 2C), presumably reflecting the activity of an endodermal factor binding to the context sequence in MadL.
Figure 2.
LexGAD is a potent autonomous activator in transgenic embryos. Side views of ≈14-h-old embryos, bearing lacZ reporter and expressing various LexA chimeras, stained with α-lacZ antibody. (A) LL reporter only. (B) LL with ubiquitous LexGAD; LexGAD activates LL throughout the embryo. (C) MadL reporter only. (D) MadL with mesodermal LexGAD; LexGAD cannot activate MadL in regions of the visceral mesoderm that do not experience Dpp stimulation (open triangles). (E) LL with mesodermal LexGAD; LexGAD activates LL throughout the visceral mesoderm (arrows). (F) MadL with mesodermal LexGAD and Dpp; in the presence of Dpp, LexGAD activates MadL throughout the mesoderm (arrows). Anterior to the left, dorsal up in all panels.
The Function of LexGAD as a Synthetic Activator.
We combined expression of individual chimeras and reporters in transgenic embryos. This revealed that LexGAD potently and ubiquitously activated transcription from the LL reporter (Fig. 2B). The lacZ staining levels were not completely uniform in some of the tissues, and were lower in some tissues than in others. For example, staining was very strong in the mesoderm, but weaker in the central nervous system. Nevertheless, we found LexGAD to be a potent synthetic activator that can function efficiently in all embryonic tissues.
Interestingly, when we tested LexGAD with the MadL reporter, the lacZ staining was more restricted. For example, when expressed in the mesoderm, LexGAD produced lacZ staining exclusively in the middle midgut and in a region around the gastric caeca (Fig. 2D), both areas which experience Dpp and Egfr signaling (19). This is in contrast to the test with LL in which mesodermal LexGAD produced essentially pan-mesodermal staining (Fig. 2E). This suggested that LexGAD activity can be restricted by a repressor that binds to the context sequence in MadL in the absence of Dpp/Egfr signaling. If so, this repression might be relieved by more widespread signaling.
To test this, we coexpressed LexGAD with Dpp, or with an activated form of Dras1 (Dras*), in the mesoderm. Indeed, coexpression with Dpp enabled LexGAD to be active throughout the mesoderm when tested on MadL: we observed strong and uniform lacZ staining from the anterior to the posterior end of the visceral midgut mesoderm (Fig. 2F). On the other hand, whereas Dras* showed a noticeable stimulatory effect on MadL-mediated staining in the somatic mesoderm, and also a mild one in the visceral mesoderm, it did not lead to uniformly high expression in this tissue (not shown). These activating effects were specific for the MadL reporter, and not visible with LL. We conclude that Dpp signaling relieves the repressive effect of the MadL context sequence on LexGAD. Recall that this context sequence contains a Mad binding site (ref. 19; see Fig. 1C). Because Dpp signaling triggers the nuclear import of Mad (34), it is possible that Mad, on nuclear entry, binds to MadL and displaces a repressor from it, thus allowing full function of LexGAD. In support of this, we previously found that the Mad binding site in the Ubx enhancer is a target for a repressor whose activity is relieved by Dpp signaling (35).
The Context-Dependence of LexFos Chimeras.
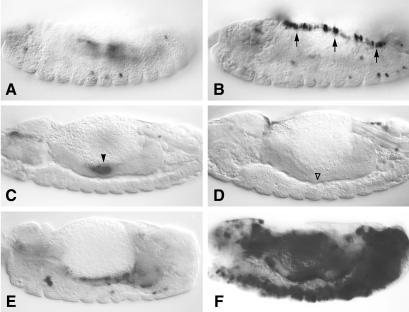
In contrast to LexGAD, none of the Dfos-LexA chimeras showed any transcriptional activity when tested with LL (not shown). This was somewhat surprising, given that a similar LexA-Fos chimera can activate transcription in yeast from multimerized LexA binding sites (36) and that Dfos can activate transcription in vitro (33). However, when we tested LexFos with MadL, this chimera produced conspicuous lacZ staining in the dorsal region of embryos, along the leading edge (Fig. 3B, compare with A). Significantly, these cells are involved in the process of dorsal closure of the embryo (37), and endogenous Dfos is not only expressed at high levels in these cells, but also required for this process (38, 39). This is a strong indication that LexFos mimics the function of endogenous Dfos in these cells.
Figure 3.
LexFos is a context-dependent transcriptional activator. Side views of ≈14-h-old embryos bearing MadL, stained with α-lacZ antibody (A and B, surface views; C–F, focused on midgut). (A) MadL reporter only. (B) MadL with ubiquitous LexFos; note lacZ staining in the dorsal leading edge cells (arrows) because of LexFos. (C) MadL only; endodermal background staining is indicated by arrowhead. (D) MadL with ubiquitous LexFos; the background staining because of MadL is suppressed by LexFos (▵). (E) MadL with ubiquitous Jun*. (F) MadL with ubiquitous LexFos and Jun*; Jun* potently synergizes with LexFos in various embryonic tissues.
Neither LexFosN nor LexFosC produced any staining in the leading edge cells (not shown). However, all three chimeras, after ubiquitous expression, eliminated the endodermal background staining that we observe with MadL alone (Fig. 3D, ▵; compare with Fig. 3C). This quenching activity may reflect a displacement by the chimeras of the endogenous endodermal factor binding to MadL (see above). The quenching provides further evidence that all three chimeras are expressed and functional at a comparable level. However, the observation that only LexFos, but neither of the others, shows any transactivation potential suggests that this potential may reside not in Dfos itself, but in a dimerization partner of Dfos. The only presently known dimerization partner of Dfos is Djun (33). Notably, Djun is also required for dorsal closure of the leading edge cells (40–43), and its activity for this process depends on the Jun N-terminal kinase (JNK) signaling pathway, which is active in these cells (37). It thus seemed conceivable that LexFos recruits activated Djun to mediate transcriptional activation of MadL.
To test this, we coexpressed LexFos with an activated form of c-Jun [JunAsp (27); we shall refer to this as Jun*], with a control version of this (JunAla; ref. 27), and also with Djun or with Dfos throughout the embryo. When testing these pairs with MadL, we found that Jun*, but none of the other Jun proteins nor Dfos, were able to synergize with LexFos to produce strong and widespread lacZ staining in various embryonic tissues (predominantly in the mesoderm, but also in the dorsal epidermis; Fig. 3F, compare with E). This effect of Jun* very largely depended on LexFos (Fig. 3F) as very little extra staining was observed when Jun* was tested alone with MadL (Fig. 3E), implying that recruitment of Jun* by LexFos is necessary for the widespread activity of LexFos under these conditions. Interestingly, it also depended on the reporter because there was no synergy between Jun* and LexFos when tested with LL (see below). Because only the activated version of Jun was able to produce widespread lacZ staining, this suggested that JNK signaling may be required for the transactivation potential of the LexFos/Jun dimer. If so, LexFos might be expected to exhibit a more widespread activity if expressed under conditions of ubiquitous JNK signaling. Note that the only embryonic territory known to experience JNK signaling are the leading edge cells (44), whereas Djun is expressed fairly ubiquitously throughout the embryo (43).
To mimic constitutive JNK signaling, we made use of an activated form of Drac (Drac*) that is thought to activate JNK (44). When LexFos was coexpressed with Drac* throughout the embryo and tested with MadL, we found very strong and widespread lacZ staining (not shown). However, the embryos were highly abnormal and it was difficult to assess in which cells reporter activity was observed. As it seemed that most activity was in the mesoderm, we coexpressed LexFos and Drac* in the mesoderm and tested them with MadL. This produced conspicuous staining throughout the embryonic mesoderm, including the visceral mesoderm (Fig. 4B, compare with A), but also in cells dotted around the embryo (possibly blood cells, mesodermal derivatives) and, curiously, within the yolk. A similar effect was observed after coexpression with an activated version of Drosophila cdc42 (Dcdc42*), also thought to activate JNK (44) (not shown). Clearly, LexFos was strongly responsive to Drac* (and Dcdc42*) in mesodermal cells. Neither LexFosN nor LexFosC were able to synergize with Drac*, once again indicating that the target of Drac* may not be LexFos itself, but an endogenous dimerization partner of LexFos. This partner could be Djun because Djun is known to be phosphorylated by JNK (45). The synergy between Drac* and LexFos is consistent with the synergy between Jun* and LexFos, and with the observation that LexFos is only active in cells which experience JNK signaling (i.e., the leading edge cells).
Figure 4.
LexFos synergizes with JNK and Dpp signaling. Side views of ≈14-h-old embryos bearing MadL, stained with α-lacZ antibody. (A) MadL with mesodermal Drac*. (B) MadL with mesodermal LexFos and Drac*; Drac* potently synergizes with LexFos in various embryonic tissues. (C) MadL with ubiquitous Dpp. (D) MadL with ubiquitous LexFos and Dpp; Dpp synergizes with LexFos in the anterior endoderm (arrows; lacZ staining in the salivary glands, arrowheads in C and D depends on Dpp, but not on LexFos).
Recall that the activating effect of Jun* or Drac* on LexFos was not observed in all embryonic cells. For example, the synergy between Jun* or Drac* and LexFos was predominantly seen in the mesoderm, whereas there was little if any synergy in the ventral ectoderm of the embryo. Furthermore, the synergy between Jun* or Drac* and LexFos was not observed if tested with the LL reporter (not shown). This suggested that the activity of LexFos depended on a context sequence, and possibly on a second context signal. Dpp seemed an obvious candidate because MadL contains part of a Dpp response element (the Mad binding site; Fig. 1). Also, the embryonic territories in which we observed the synergy between Jun* or Drac* and LexFos appear to correlate with territories that experience Dpp stimulation (e.g., the mesoderm and the dorsal epidermis). We thus coexpressed Dpp with LexFos and tested its activity in embryos with MadL. This revealed robust additional lacZ staining in the anterior endoderm, and some additional staining scattered throughout the embryo (Fig. 4D). Evidently, Dpp signaling can also synergize with LexFos. However, this synergy is less pronounced and less widespread than that between LexFos and Jun* or JNK signaling, being largely restricted to the endoderm. This suggests that the anterior endoderm is an embryonic territory that normally experiences JNK but no Dpp signaling. Conversely, the mesoderm, the dorsal epidermis, and the other tissues in which we observe synergy between LexFos and ectopic JNK signaling presumably experience Dpp but no JNK stimulation in normal development.
Discussion
LexA chimeras have been a potent tool for probing the transcriptional activation potential of proteins (2). These chimeras can reveal autonomous transcriptional activation domains that interact with, and probably recruit, the general transcription machinery. For example, yeast GAL4 contains an autonomous and potent transactivation domain that functions fairly universally (1, 46–48). We show here that a LexA chimera with this domain constitutes a synthetic activator (LexGAD), which, like GAL4 itself, functions autonomously and efficiently in all embryonic tissues of Drosophila. This opens up the possibility of using LexA chimeras for assaying the transcriptional activation potential of proteins during embryonic development of Drosophila. The same approach could also be applied for demonstrating and mapping transcriptional repression domains (see ref. 49). Finally, the synthetic activator LexGAD could be used as an alternative to, or in conjunction with, GAL4 itself (see ref. 21) for targeted expression of genes in transformed Drosophila.
We applied this approach of LexA chimeras to assess the transcriptional activation potential of Dfos in Drosophila embryos. To our surprise, we found that LexFos exhibited no activity on its own in stably transformed embryos. Rather, LexFos required a surprising degree of context as its function as a transcriptional activator depended on interaction with multiple signal-activated partner factors (see below). This contrasts with the autonomous function of Dfos in activating transcription in vitro (33). Furthermore, c-Fos behaves as an autonomous transcriptional activator in yeast (36) and in transfected mammalian cells (50–52). Bearing in mind that Dfos and c-Fos may not be true functional homologs and may thus not be entirely comparable regarding their transcriptional activation potential, we nevertheless think it possible that these differences in transactivation potential may reflect the different assay conditions used. In our assay system, LexFos is expressed at roughly the same levels as endogenous Dfos, and its target reporter is present as a single copy gene stably integrated into a chromosome and wrapped up in chromatin. Potentially, this assay is more stringent than a transient transfection assay, and is thus likely to reveal true functional requirements that are significant for normal development.
A precedent for a transcription factor showing variable functional autonomy depending on the assay conditions may be found in the case of TCFs. One member of this group, LEF-1, contains a context-dependent transcriptional activation domain that, together with a T cell-type specific partner transcription factor, recruits a context-dependent coactivator (called ALY) (ref. 53; see also Introduction). However, LEF-1 and other TCFs can also use an alternative coactivator to stimulate transcription, namely the Wnt-induced factor β-catenin/Armadillo (15, 16). Experiments with a synthetic target reporter containing multimerized TCF binding sites have revealed an autonomous transactivation potential of TCF/β-catenin (54–56). However, this potential is only apparent in transient transfection or injection assays, whereas on chromosomal integration in transgenic mice, the same reporter does not reliably respond to Wnt-induced TCF/β-catenin (H. Clevers, personal communication). Likewise, a similar reporter with multimerized TCF binding sites requires a context sequence to respond to TCF/Armadillo in transgenic Drosophila embryos (18). This strongly suggests that TCFs are genuinely context-dependent, regardless of which coactivator they recruit, and that the apparent discrepancies between autonomous vs. context-dependent behavior may reflect the different assay conditions.
Which factors provide the context for Dfos function? Several lines of evidence implicate JNK and Dpp signaling and their transcriptional target factors Djun and Mad as the essential context. First of all, the only embryonic cells in which LexFos functions reliably and robustly to stimulate transcription are the dorsal leading edge cells which experience both of these signals (see above; ref. 37). Second, neither of the LexFos derivatives (LexFosN, LexFosC) function in these cells, strongly implicating the basic leucine zipper domain of LexFos (the only domain absent from both derivatives) in its function. As this domain mediates dimerization with Djun, the only known dimerization partner of Dfos in Drosophila (33, 57), this indicates that the activity of LexFos depends on Djun. Recall that Djun is present and activated by JNK signaling in the leading edge cells (40–43). Third, JNK signaling as mimicked by overexpression of Drac* or Dcdc42*, potently synergizes with LexFos to mediate widespread transactivation in the embryo. A very similar widespread synergy was also seen between LexFos and Jun*, a mutant form of c-Jun that mimics signal-activation of this protein (27). The embryonic territories in which these synergies were observed appear to correspond to sites of Dpp stimulation. Consistent with this, we also observed a limited synergy between Dpp and LexFos in some embryonic cells. These synergies strongly implicate JNK and Dpp as necessary context signals for LexFos function. Last, LexFos activity strictly depended on the context sequence in the MadL target reporter; under no conditions did it transactivate the LL reporter (albeit LexGAD very efficiently did so; see above). The context sequence in MadL essentially consists of a binding site for the Dpp response factor Mad (Fig. 1), which is thus a likely partner for the putative LexFos/Djun* dimer.
Our results indicate that JNK-activated Djun and Dpp-activated Mad may be critical and widespread context partners of Dfos. Consistent with this, Dfos function is required for dorsal closure of the embryo (38, 39) and, by implication, functions normally in cells that experience JNK and Dpp signaling. In the embryonic midgut, Dfos functions in cells that experience Dpp and Egfr signaling (19, 20). Because the LexFos/JNK synergy in the mesoderm implies that JNK signaling is normally absent from this tissue, this suggests that the normal partner of Dfos in the midgut visceral mesoderm may be a factor, as yet unidentified, that is activated by Egfr signaling. Interestingly, synergy between the c-Jun/c-Fos dimer and TGF-β activated Smad has also been observed in mammalian cells (58–60). Furthermore, Jun proteins have recently been shown to bind directly to Smad3/4 (61). Thus, the partnership between signal-activated Jun/Fos dimers and Smads may be fairly widespread and fundamental.
Our work has revealed an unexpected degree of context-dependence of Dfos. As in the case of the well-documented context-dependence of TCF, the partners of Dfos appear to be signal-activated transcription factors, i.e., Djun and Mad. Smads themselves seem to function in a highly context-dependent way (62). And recently, a striking case of context-dependence has been discovered in the Drosophila wing disk in which the activity of the enhancer-binding protein Scalloped is determined by its obligatory partner factor Vestigial as well as by signal-activated factors (63, 64). Thus, although examples of context-dependent transcription factors are beginning to emerge, this mode of action still seems to be the exception rather than the rule. Interestingly, context-dependence appears prevalent among signal-responsive factors, perhaps reflecting a built-in versatility that this class of transcription factors need to have. Whatever the case, it is possible that context-dependence may have been overlooked in some cases, because of the assay systems used. If Dfos and TCFs were to be representative in this regard, this would suggest that there may be many more context-dependent transcription factors than previously thought.
Acknowledgments
We thank Lucas Waltzer and Rudi Grosschedl for helpful comments on the manuscript. D.S. was supported by a studentship from Trinity College, Cambridge, and currently holds a research fellowship from Peterhouse, Cambridge.
Abbreviations
- TCF
T cell factor
- JNK
Jun N-terminal kinase
- GAD
GAL4 activation domain
- Egfr
epidermal growth factor receptor
- Dfos
Drosophila Fos
- Dpp
decapentaplegic
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brent R, Ptashne M. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 3.Hope I A, Mahadevan S, Struhl K. Nature (London) 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 4.Lin Y S, Carey M, Ptashne M, Green M R. Nature (London) 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 5.Carey M, Lin Y S, Green M R, Ptashne M. Nature (London) 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M. Proc Natl Acad Sci USA. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 8.Arnosti D N, Barolo S, Levine M, Small S. Development (Cambridge, UK) 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 9.Szymanski P, Levine M. EMBO J. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 11.Carey M. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 12.Carlsson P, Waterman M L, Jones K A. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 13.Giese K, Grosschedl R. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 15.Bienz M. Curr Opin Cell Biol. 1998;10:366–372. doi: 10.1016/s0955-0674(98)80013-6. [DOI] [PubMed] [Google Scholar]
- 16.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 17.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 18.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 19.Szüts D, Eresh S, Bienz M. Genes Dev. 1998;12:2022–2035. doi: 10.1101/gad.12.13.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riese J, Tremml G, Bienz M. Development (Cambridge, UK) 1997;124:3353–3361. doi: 10.1242/dev.124.17.3353. [DOI] [PubMed] [Google Scholar]
- 21.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 22.Thüringer F, Bienz M. Proc Natl Acad Sci USA. 1993;90:3899–3903. doi: 10.1073/pnas.90.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golemis E A, Brent R. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanson B, White P, Vincent J P. Nature (London) 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 25.Martin-Bermudo M D, Dunin-Borkowski O M, Brown N H. EMBO J. 1997;16:4184–4193. doi: 10.1093/emboj/16.14.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eresh S, Riese J, Jackson D B, Bohmann D, Bienz M. EMBO J. 1997;16:2014–2022. doi: 10.1093/emboj/16.8.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treier M, Bohmann D, Mlodzik M. Cell. 1995;83:753–760. doi: 10.1016/0092-8674(95)90188-4. [DOI] [PubMed] [Google Scholar]
- 28.Luo L, Liao Y J, Jan L Y, Jan Y N. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 29.Lee T, Feig L, Montell D J. Development (Cambridge, UK) 1996;122:409–418. doi: 10.1242/dev.122.2.409. [DOI] [PubMed] [Google Scholar]
- 30.Freeman M. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 31.Capdevila J, Guerrero I. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szüts D, Freeman M, Bienz M. Development (Cambridge, UK) 1997;124:3209–3219. doi: 10.1242/dev.124.16.3209. [DOI] [PubMed] [Google Scholar]
- 33.Perkins K K, Admon A, Patel N, Tjian R. Genes Dev. 1990;4:822–834. doi: 10.1101/gad.4.5.822. [DOI] [PubMed] [Google Scholar]
- 34.Raftery L A, Sutherland D J. Dev Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Riese J, Eresh S, Bienz M. EMBO J. 1998;17:7021–7032. doi: 10.1093/emboj/17.23.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lech K, Anderson K, Brent R. Cell. 1988;52:179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- 37.Noselli S, Agnes F. Curr Opin Genet Dev. 1999;9:466–472. doi: 10.1016/S0959-437X(99)80071-9. [DOI] [PubMed] [Google Scholar]
- 38.Riesgo-Escovar J R, Hafen E. Science. 1997;278:669–672. doi: 10.1126/science.278.5338.669. [DOI] [PubMed] [Google Scholar]
- 39.Zeitlinger J, Kockel L, Peverali F A, Jackson D B, Mlodzik M, Bohmann D. EMBO J. 1997;16:7393–7401. doi: 10.1093/emboj/16.24.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou X S, Goldstein E S, Perrimon N. Genes Dev. 1997;11:1728–1737. doi: 10.1101/gad.11.13.1728. [DOI] [PubMed] [Google Scholar]
- 41.Riesgo-Escovar J R, Hafen E. Genes Dev. 1997;11:1717–1727. doi: 10.1101/gad.11.13.1717. [DOI] [PubMed] [Google Scholar]
- 42.Glise B, Noselli S. Genes Dev. 1997;11:1738–1747. doi: 10.1101/gad.11.13.1738. [DOI] [PubMed] [Google Scholar]
- 43.Kockel L, Zeitlinger J, Staszewski L M, Mlodzik M, Bohmann D. Genes Dev. 1997;11:1748–1758. doi: 10.1101/gad.11.13.1748. [DOI] [PubMed] [Google Scholar]
- 44.Noselli S. Trends Genet. 1998;14:33–38. doi: 10.1016/S0168-9525(97)01320-6. [DOI] [PubMed] [Google Scholar]
- 45.Sluss H K, Han Z, Barrett T, Davis R J, Ip Y T. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 46.Ma J, Przibilla E, Hu J, Bogorad L, Ptashne M. Nature (London) 1988;334:631–633. doi: 10.1038/334631a0. [DOI] [PubMed] [Google Scholar]
- 47.Fischer J A, Giniger E, Maniatis T, Ptashne M. Nature (London) 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 48.Kakidani H, Ptashne M. Cell. 1988;52:161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- 49.Wang H, Stillman D J. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucibello F C, Neuberg M, Jenuwein T, Muller R. New Biol. 1991;3:671–677. [PubMed] [Google Scholar]
- 51.Sutherland J A, Cook A, Bannister A J, Kouzarides T. Genes Dev. 1992;6:1810–1819. doi: 10.1101/gad.6.9.1810. [DOI] [PubMed] [Google Scholar]
- 52.Jooss K U, Funk M, Muller R. EMBO J. 1994;13:1467–1475. doi: 10.1002/j.1460-2075.1994.tb06401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruhn L, Munnerlyn A, Grosschedl R. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 54.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 55.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 56.Hsu S C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perkins K K, Dailey G M, Tjian R. EMBO J. 1988;7:4265–4273. doi: 10.1002/j.1460-2075.1988.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Derynck R. Trends Cell Biol. 1999;9:274–279. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]
- 59.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X F. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J M, Wang X F. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liberati N T, Datto M B, Frederick J P, Shen X, Wong C, Rougier-Chapman E M, Wang X F. Proc Natl Acad Sci USA. 1999;96:4844–4849. doi: 10.1073/pnas.96.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Feng X H, Derynck R. Nature (London) 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 63.Halder G, Polaczyk P, Kraus M E, Hudson A, Kim J, Laughon A, Carroll S. Genes Dev. 1998;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bray S. Curr Biol. 1999;9:R245–R247. doi: 10.1016/s0960-9822(99)80154-7. [DOI] [PubMed] [Google Scholar]