Summary
Fission yeast expresses two kinesin 8s, klp5+ and klp6+, which are important for diverse cellular functions: mitosis, meiosis, and the maintenance of normal cell morphology. During vegetative growth these motors display complex localization patterns, moving from the cytoplasm during interphase to the kinetochores in early mitosis, the interpolar spindle in anaphase B, and then back into the cytoplasm. We have expressed GFP-tagged alleles of domains from these motors, seeking the signals required for their localizations. The tail of Klp5p localized to the interphase nucleus, more specifically to telomeres. Addition of the neck re-directed this fragment to microtubules in the cytoplasm. Klp6-tail and the neck-tail domains of both motors localized at microtubule ends. Klp6-neck-tail localized to the spindle in early mitosis but to the pole-proximal ends of the spindle in anaphase B. The Klp5-motor and motor-neck localized to microtubules, often causing them to bundle. Over-expression of Klp6-motor or motor-neck resulted in shorter microtubules. These localization patterns were no different when constructs were expressed in strains lacking either or both of the endogenous, full-length proteins. Our results indicate that the localization signals for these kinesins are not derived from simple amino acid sequences but from complex interactions among multiple domains of each motor.
Keywords: microtubule, mitosis, membrane traffic, polarity, depolymerization
Introduction
Kinesin motor proteins are important components of many microtubule-based cellular functions, including the proper segregation of chromosomes and the establishment and maintenance of cell polarity. These enzymes can carry cargo along the surface of microtubules (MTs), organize MTs into higher ordered arrays, or alter MT dynamics (Howard and Hyman, 2007). Kinesins have been categorized into subfamilies, based on the primary sequences of their motor domains, which interact with MTs in an ATP-sensitive manner; this classification generally correlates with cellular function, even though the remainder of the molecules can be divergent (Lawrence et al., 2004; Wickstead and Gull, 2006). The “tail” domain of each kinesin is thought to interact with its cargo, which may be other MTs, membrane bound organelles, RNA molecules, or kinetochores. The motor domain is usually connected to the tail domain by a neck or stalk, which often contains an α-helical segment that can foster dimerization.
The fission yeast genome encodes 9 kinesins that represent 7 families; there are two motors of the kinesin 8 family: klp5+ and klp6+ (Lawrence et al., 2004; West et al., 2001). Members of this family have also been described in budding yeast (Cottingham and Hoyt, 1997), Drosophila (Gandhi et al., 2004; Pereira et al., 1997) and humans (Lawrence et al., 2004; Zhu et al., 2005). In fission yeast the localizations of these gene products are complex. They bind to cytoplasmic MTs in interphase then move into the nucleus at mitosis, where they associate first with pre-anaphase MTs and/or kinetochores. Just after anaphase A they move to the interpolar spindle where they are restricted to the spindle midzone as the spindle elongates during anaphase B. After mitosis they return to the cytoplasm.
The phenotypes of kinesin 8 disruption mutants from several organisms suggest that these enzymes destabilize MTs in vivo and are thus required to maintain a balance in tubulin dynamics. Disruption of the KIP3 gene in S. cerevisiae leads to abnormally long cytoplasmic and spindle MTs; moreover, the spindle persists after anaphase B is complete (Cottingham and Hoyt, 1997; Miller et al., 1998; Varga et al., 2006). Disruption of either klp5+ or klp6+ in fission yeast also produces unusually long MTs in both interphase and mitosis; these MTs are resistant to the MT poison, thiabendazole (TBZ) (Garcia et al., 2002a; Garcia et al., 2002b; West et al., 2002; West et al., 2001). Disruptions of Aspergillus KipB (Rischitor et al., 2004) or Drosophila KLP67A (Gatt et al., 2005; Savoian et al., 2004) produce similar phenotypes: spindles that contain an unusually large number of long and persistent MTs. The ability of these enzymes both to move on MTs and to promote their depolymerization has recently been confirmed in vitro for budding yeast kinesin 8(Gupta et al., 2006; Varga et al., 2006).
The phenotypes of vegetative klp5Δ and klp6Δ mutants are very similar, which is not surprising, given the similarity in the amino acid sequences of their motor domains (Garcia et al., 2002a; Garcia et al., 2002b; West et al., 2002; West et al., 2001). The phenotypes are, however, complex, suggesting that these motors play multiple roles in cell physiology. Deletion strains fail to properly align chromosomes at the metaphase plate; they show defects in interphase cell morphology, and they are unable to produce viable spores. The phenotypes of the double motor deletion are similar, suggesting that the motors function as a single complex, an hypothesis that is supported in part by yeast two-hybrid and co-immunoprecipitation experiments: Klp5p appears to homodimerize, and Klp5p/Klp6p can form heterodimers (Garcia et al., 2002b; Li and Chang, 2003). Cooperative function of these motors is also suggested by two sets of genetic interactions. Deletions of either the ras-GTPase guanine nucleotide exchange factor gene, scd1+ (Li and Chang, 2003; Li et al., 2000), or components from the kinetochore-associated Dam1 complex (aka DASH) (Sanchez-Perez et al., 2005) are lethal in combination with either klp5Δ or klp6Δ.
On the other hand, the localization and apparent motility of either kinesin is not dependent on the expression of the other (West et al., 2002). Moreover, in contrast to the essentially indistinguishable phenotypes of each deletion in vegetative cells, the meiotic phenotypes of these motor deletions suggest distinct functions for klp5+ and klp6+. For example, the asci from klp5Δ and klp6Δ have differently aberrant morphologies, and heterozygous crosses between klp5Δ and klp6Δ have an intermediate spore viability, indicating a partial loss of function in two different meiotic functions (West et al., 2001). Finally, it is important to note that the tail domains of these kinesins, in contrast to the motor domains, are highly divergent, and they constitute nearly half of the mass of each motor (West et al., 2001). The possibility that different kinesin 8's play different roles in the same cell is important, because many organisms carry genes for several members of this motor family, and most current work on this enzyme has neglected the possibility that individual members of the family are distinct.
We have investigated the roles of the motor and tail domains in determining the localization and dynamics of both Klp5p and Klp6p. Domains of each motor were expressed in cells as EGFP-tagged molecules, and their localizations and phenotypes were followed through the cell cycle. We have also investigated the role of potential dimerization domains by expressing the motor or tail domains with and without their corresponding α-helical neck domain. This approach has revealed some unusual interactions between the domains of each kinesin and some unexpected cellular targets. It has also demonstrated a significant role for the neck domain in directing parts of both Klp5p and Klp6p to distinct cellular localizations.
Materials & Methods
Plasmid Construction
Plasmids were made using standard molecular biology techniques (Sambrook et al., 1989). The parent plasmids employed to express domains of Klp5p and Klp6p came from the pEGFP-REP series (Craven et al., 1998).
Insert DNA molecules corresponding to specific domains of Klp5p and Klp6p were amplified from wild type S. pombe genomic DNA using the polymerase chain reaction (PCR) with the primers indicated in Table 1. The sequences used for cloning are in italics; the 5’ primer corresponds to the sense strand of each kinesin gene. Klp5-full-length was made using Klp5-motor-5’ and Klp5-tail-3’ primers. Each fusion protein contained three Glycine residues as a spacer between the kinesin domain and the EGFP or Pk tags. The PCR reactions were carried out with “Platinum” or “turbo” Pfx thermostable DNA polymerases using the manufacturer's specifications (Invitrogen, Inc., San Diego, CA). The recombinant plasmids were transformed into the competent E. coli strains “SURE” (klp5) (Stratagene, Inc., LaJolla, CA) or DH5α (klp6). The accuracy of the resulting expression plasmids was verified by DNA sequencing.
Table 1.
Primers used to construct expression plasmids
| 5tail 5′A: gtcgtcgacgggtggtggttctgatcaggacgctgtaac |
| 5tail-3′A: caggatccctattaggtggctttctcttcttcg (N) |
| 5tail-3′B: caggatccaccaccggtggctttctcttcttcg (C) |
| 5neck-tail-5′: gtcgtcgacgggtggtttaaggaacatgatcagtgtagatcg |
| 5motor-5′: gagtcgtcgacgatgtcaagacagtcgtccattac |
| 5motor-3′: caggatccaccacttaacacttcggttttgatattc |
| 5motor-neck-3′: caggatccaccaccatttgattgcgatg |
| 6tail-5′A: gtcgtcgacgggtggtatttcttctcgagaaattaaaatg |
| 6tail-3′A: caggatccctataaagcattaggagtattctcagtccc (N) |
| 6tail-3′B: caggatccaccaccagcattaggagtattctcagtccc (C) |
| 6neck-tail-3′: gtcgtcgacgggtggtgtaagtgaatatgtacgcaccatttac |
| 6motor-5′: gagtcgtcgacgatgtcaagacagtcgtccattac |
| 6motor-3′: caggatccaccacttaacacttcggttttgatattc |
| 6motor-neck-3′: caggatccaccaccatttgattgcgatg |
| 6full5′: gggaatgaaagaagggtcttcaatttcc |
| 6full3′: gggagtccaccagcattaggagtattctcagtcccgc |
Strains and Cell Culture
All strains used in this study are listed in Table 2. The klp5Δ, klp6Δ, and klp5Δ;; klp6Δ double mutants are collectively referred to as klpΔ when the point being made is equally true for all of these strains. Cell culture and genetic manipulations were carried out using standard techniques (Moreno et al., 1991). Cells were transformed using a polyethylene glycol procedure (Elble, 1992) and plated on medium containing 5μg/ml thiamine to suppress expression of the trans-gene (Forsburg, 1993). Strains that contained the EGFP and Pk1 plasmids (described below) were maintained on defined medium (EMM, Moreno, et al., 1991) containing 5μg/ml thiamine. Expression from the pREP constructs was induced at 32°C by removal of thiamine from the culture medium. Expression was not detectable 8hrs after induction, so induction was continued for 16−20hrs, using cultures that were never allowed to grow beyond early log phase (∼105cells/ml). Cells that lacked birefringence and/or appeared dark under phase contrast were presumed dead, and not scored. Temperature sensitive strains were maintained at the permissive temperature of 25°C; the restrictive temperature was 36°C. Strains #99 and #100 were used as wild type, #381 as klp5Δ, #392 as klp6Δ, and #394 as klp5Δ; klp6Δ, unless otherwise noted.
Table 2.
S. pombe strains used in this study.
| Strain | genotype | source |
|---|---|---|
| 1 | 968, h90 | P. Nurse (#2) |
| 99 | ade6-M210, his3-D1, leu1−32, ura4−18, h− | H. Browning |
| 100 | ade6-M216, his3-D1, leu1−32, ura4−18, h+ | H. Browning |
| 376 | cdc25−22, ade6-M216, his3-D1, leu1−32, ura4−18, h− | this study |
| 381 | klp5Δ::ura4+, ade6-M216, his3-D1, leu1−32, ura4−18, h+ | R. West |
| 392 | klp6Δ::ura4+, ade6-M216, his3-D1, leu1−32, ura4−18, h− | R. West |
| 394 | klp5Δ::ura4+, klp6Δ::his3+, ade6-M210, his3-D1, leu1−32, ura4−18, h− | R. West |
| 467 | taz1:GFP:ura4+, leu1−32, ura4−18, h− | J.P. Cooper |
| 485 | klp5:GFP:ura4+, ade6-M210, his3-D1, leu1−32, ura4−18, h+ | R. West |
| 486 | klp6:GFP:ura4+, ade6-M210, his3-D1, leu1−32, ura4−18, h− | R. West |
| 610 | mal3Δ::ura4+, ade6-M216, his3-D1, leu1−32, ura4−18, h+ | U. Fleig |
| 620 | mis12:HA:LEU2, his3-D1, leu1−32, ura4−18, h+ | M. Yanagida |
| 622 | mis12:GFP:LEU2, ade6-M216, his3-D1, leu1−32, ura4−18, h− | M. Yanagida |
| 624 | klp5:GFP:ura4+, ade6-M210, his3-D1, leu1−32, ura4−18, h− | this study |
| 625 | klp6:GFP:ura4+, ade6-M216, his3-D1, leu1−32, ura4−18, h+ | this study |
| 626 | klp5:GFP:ura4+, ade6-M210, his3-D1, leu1−32, ura4−18, h− | this study |
| 627 | klp6:GFP:ura4+, ade6-M216, his3-D1, leu1−32, ura4−18, h+ | this study |
Immunocytochemistry
Cells used for immunofluorescence were grown to early log phase (1−3 × 105cells/ml) at 32°C in selective medium then collected by centrifugation for aldehyde fixations or by filtration for methanol fixation. Aldehyde fixation was done with a final concentration of 4% formaldehyde for 15 min, followed by repeated washes as described by (Hagan and Hyams, 1988). For methanol fixation, cells were collected by gentle suction onto a filter (Durapore 0.45μm HV, Millipore, Bedford, Mass.), which was then immersed with adhering cells in 100% methanol at −20°C for 15 min or −70°C for 1 hr. The cells were then collected by centrifugation and washed three-times with PEM (Hagan and Hyams, 1988), before being processed as described for aldehyde fixed cells. Primary and secondary antibody incubations were each done overnight in PEMBAL (Hagan and Hyams, 1988). EGFP signals were detected by their fluorescence, while immunoepitopes were detected with secondary antibodies conjugated to Alexa-Red (Molecular Probes/Invitrogen, Inc., Calsbad, CA). Sad1 polyclonal antibodies were the generous gift of I. Hagan (Hagan and Yanagida, 1995) (University of Manchester, Manchester, United Kingdom). Monoclonal antibodies against the Pk1 epitope were purchased from Serotec (Oxford, U.K.). Tubulin was stained with either the 4A1 or TAT1 monoclonal antibodies as previously described (West et al., 1998). DNA in fixed cells was stained with 1μg/ml 4’,6-diamino-2-phenylindole (DAPI) (Sigma, St. Louis, MO).
Microscopy
Live cells were stained for DNA with Hoechst 33342 as previously described (West, et al., 2002). Cells were mounted on glass slides, and images were collected with a Cooke SensiCam slow-scan CCD camera on a Zeiss Axioplan II fluorescence microscope with a 100X (n.a., 1.4) PlanApo objective lens, as previously described (West et al., 2001). Deconvolution and 2-D rendering were done with the SlideBook software package (v. 4.2.1) (Intelligent Imaging Innovations, Inc., Denver, CO). Figures were prepared with CorelDraw (Ottawa, Canada).
Results
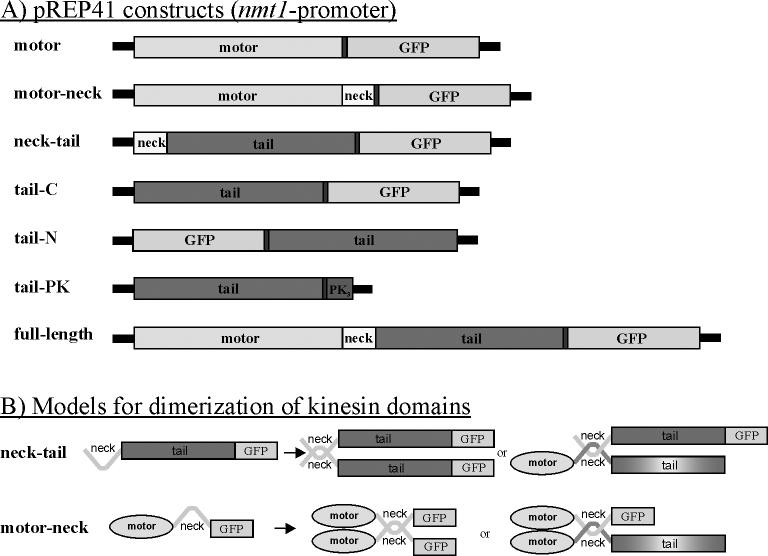
To identify the parts of Klp5 and Klp6 that are responsible for their complex patterns of localization we made plasmid constructs expressing the motor and/or tail domains of Klp5p and Klp6p as fragments tagged with the Enhanced Green Fluorescent Protein (EGFP) under a repressible promoter of moderate strength (See Materials & Methods and Figure 1). Data shown are from the pREP41-N, pEGFP-REP41-C, pEGFP-REP42-C, and pEGFP-REP41-Pk plasmids under induced conditions. Expression was never detected in repressed cultures. Expression was also undetectable from the ∼5X weaker pREP81 plasmids (Forsburg, 1993), even under induced conditions. Expression from the wild type nmt1+ promotor, which is much stronger, was generally undetectable when repressed but was lethal when induced. The motor and tail domains were also expressed with the addition of the short α-helical “neck” to test the role of potential dimerization in protein localization (Figure 1). All of these constructs were expressed in wild type, klp5Δ, klp6Δ, and klp5Δ;klp6Δ genetic backgrounds to see whether the localization of each domain was dependent on interactions with the endogenous Klp5 and Klp6 proteins (Figure 1B).
Figure 1. Klp5 and Klp6 plasmid constructs and dimerization models.
A) Diagrams of the major domains of Klp5p and Klp6p, cloned as Pk1- or GFP-tagged constructs and expressed under the regulation of the NMT41 S. pombe plasmid expression system (Forsburg, 1993). Each construct, name given on the left, was made with both klp5+ and klp6+, except the tail-Pk construct, which was only made with Klp5. The full-length construct was previously described and integrated (West, et al., 2001). B) Models of the potential dimerizations of various kinesin domain proteins (all shown with GFP) either with themselves or with the endogenously expressed, full length Klp5p or Klp6p (no GFP). In all cases, the Klp fragment could be from either Klp5p or Klp6p, and the endogenous protein could be either Klp5p or Klp6p.
Klp5-Motor and Klp5-Motor-neck
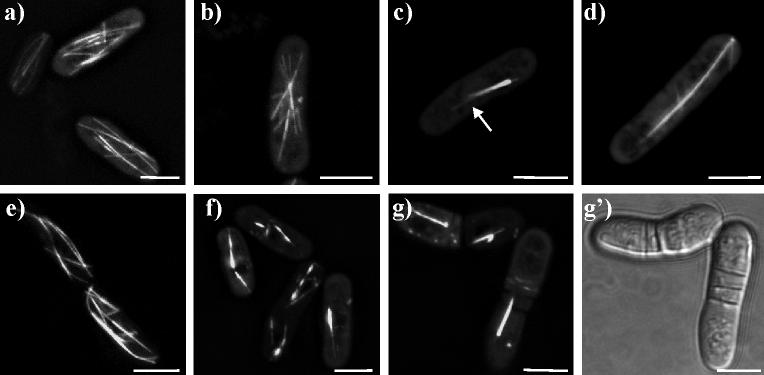
The motor domain of Klp5p is defined here as the amino-terminal 395 amino acids, ending just before the start of the predicted α-helical neck (West et al., 2001). Expression of this domain in otherwise wild type fission yeast cells gave a staining pattern that resembled the normal pattern of MT localization (Figure 2). In many of the cells, the pattern looked indistinguishable from that described for MTs in wild type cells expressing no plasmid. However, some cells gave an indication of juxta-nuclear bundling, with MT arrays resembling asters (Figure 2b). In more extreme cases the MTs appeared as a single, tight bundle, with very few single MTs emerging from the bundle (Figure 2c).
Figure 2. Klp5-motor-EGFP and Klp5-motor-neck-EGFP localize to microtubules.
a-d) live cells expressing Klp5-motor-EGFP. a) cells with nearly normal MT arrays; b) cell with MTs clustered in an aster-like array; c) interphase cell with a single large bundle of MTs; d) cell with late mitotic spindle. e-g) live cells expressing Klp5-motor-neck-EGFP. e) cells with nearly normal MT arrays; f) cells with several MT bundles; g) mitotic cells with late spindle defect; g') bright field image showing multiple septa defect. White bar = 5μm in all panels.
One feature of our experimental method is variability in the level of protein expression, which is most commonly a result of transfection by different numbers of plasmids (Moreno et al., 1991; Hayles & Nurse, 1992). Gene repression by the regulatable promoter was sufficiently strong that in 5 mM thiamine, no fluorescence was seen and no mutant phenotypes were observed. Removal of thiamine for 16−20hrs induced expression to detectable but variable levels. GFP fluorescence was not quantified, but qualitatively any phenotype seen was always stronger when fluorescence from the GFP-Klp chimera was greater. Cells with no visible fluorescence were not scored; rarely, a cell was brightly fluorescent, and these too were ignored. All cells used for the study of protein localization showed an intermediate level of fluorescence and appeared approximately equal in brightness. For example, modest expression of Klp5-motor gave a fluorescence image that resembled normal MT distributions (Fig. 2), but higher levels of expression induced bundles of fluorescent fibers and induced a slight bowing of the cells (not shown). The Klp5-motor domain bound to both mitotic spindles and astral MTs over a range of expression levels. However, unlike endogenous Klp5p, the Klp5-motor did not become restricted to the spindle midzone during anaphase B (Figure 2d).
The Klp5-motor-neck fragment included the motor plus the carboxy-proximal 40 amino acids, which are predicted to be in a coiled-coil confirmation (West et al., 2001). This fragment also bound MTs efficiently (Figure 2e), but in most cells fluorescent bundles, presumably MTs, were much more obvious (Figure 2f,g). Frequently, there were one, two or three short bundles lying close to the nucleus (Figure 2f). Like the Klp5-motor domain, the Klp5-motor-neck domain led to modest morphology defects. The Klp5-motor-neck localized to the mitotic spindle, and like the motor domain alone, did not become restricted to the spindle mid-zone. Additionally, the spindle appeared to persist through mitotic exit and septation, leading to cells with a spindle and multiple septa (Figure 2g,g').
To test for interactions between the Klp5-motor and the endogenous Klp5p and Klp6p, the Klp5-motor and Klp5-motor-neck constructs were also expressed in klp5Δ, klp6Δ, and klp5Δ; klp6Δ double mutants. The absence of either or both of these endogenous kinesins did not have a detectable effect on the Klp5-motor/Klp5-motor-neck localization pattern or phenotype (data not shown), suggesting that the behavior of these kinesin fragments is not significantly affected by the presence of the endogenous Klps.
Together, these data show that the motor and motor-neck domains of Klp5p bind MTs efficiently, as expected. Surprisingly, however, the expression of these domains generated significant levels of MT bundling. Addition of the neck with the motor also created a multi-septation defect.
Klp6-Motor and Klp6-Motor-Neck
The Klp6p motor domain consists of the amino-terminal 403 amino acids of the protein; it ends at the beginning of the α-helical neck (West et al., 2001) (Figure 1). Expression of the Klp6p motor domain in wild type interphase cells gave diffuse cytoplasmic staining with a slight accumulation of fluorescence in the nucleus (Figure 3a,b). There was only a faint indication of MT staining (Figure 3a, arrows). Slight morphology defects were also observed, with cells appearing asymmetric or bowed. Despite the lack of strong MT binding, tubulin staining revealed that cells expressing Klp6-motor contained MTs that were significantly shorter than those in wild type cells (Figure 3 c,d). These short MTs were juxtanuclear, and were observed in approximately half the cells of a given culture. In mitotic cells the Klp6-motor localized to the spindle, but it neither shortened the spindle in an obvious way nor became restricted to the spindle midzone, as endogenous protein does (Figure 3b).
Figure 3. Klp6-motor-EGFP and Klp6-motor-neck-EGFP shorten microtubules.
a, b) live cells expressing Klp6-motor-EGFP. a) Interphase cells; arrow indicates cell where some fluorescence is concentrated on fibers that resemble MTs; b) mitotic cells. c, d) fixed cells stained for tubulin (red), and DNA (blue); c) cell with Klp6-motor-EGFP, repressed by the addition of thiamine; d) cell expressing Klp6-motor-EGFP. e,f) live cells expressing Klp6-motor-neck-EGFP; e) interphase cells; f) mitotic cells, inset; cell with a monopolar spindle. g-i) fixed cells with Klp6-motor-neck-EGFP (green), tubulin staining (red), and DNA staining (blue); g and h display results from same experiment as c and d. i) tubulin (Texas Red) channel; i') Klp6-motor-neck-EGFP (FITC) channel; i”) combined channels with DNA (DAPI) DNA. White bars = 5μm.
The Klp6p motor-neck includes the motor domain plus the 36 amino acid α-helical segment that is carboxy-proximal to the motor domain (Figure 1) (West et al., 2001). This fragment was somewhat more efficient at MT binding than the motor alone, producing a MT pattern that was clearly above general cytoplasmic staining (Figure 3e,f). However, there was still significant cytoplasmic and nuclear staining with this construct. Mitotic spindle staining was apparent both in early and late mitotic cells (Figure 3f,i), though once again, midzone staining was not apparent. Expression of the Klp6-motor-neck domain frequently produced spindle staining that was consistent with a monopolar spindle phenotype (Figure 3f, inset). Like the Klp6-motor domain, expression of the Klp6-motor-neck domain also led to significantly shortened juxtanuclear MTs, compared to wild type controls (Figure 3g,h). The Klp6-motor-neck fragment was somewhat more efficient at MT shortening than the Klp6-motor domain.
The localization and phenotypes described here for both Klp6-motor and Klp6-motor-neck were not significantly different following their expression in either wild type or in klp5Δ, klp6Δ, or klp5Δ;klp6Δ backgrounds. The klp5Δ and klp6Δ mutants have somewhat longer MTs when compared to wild type cells (West et al., 2001), but this phenotype was largely absent when Klp6-motor/motor-neck was expressed in them. These data indicate that the Klp6-motor and Klp6-motor-neck domains can destabilize MTs in the absence of either of the endogenous motor proteins, even under conditions that normally lead to longer MTs.
Collectively, these data show that the Klp6-motor/motor-neck domains are not efficient at MT binding but are nonetheless effective at MT depolymerization. The addition of the neck to the motor enhanced the MT depolymerization phenotype, which may be responsible for the appearance of monopolar spindles, but it did not have any other significant effect.
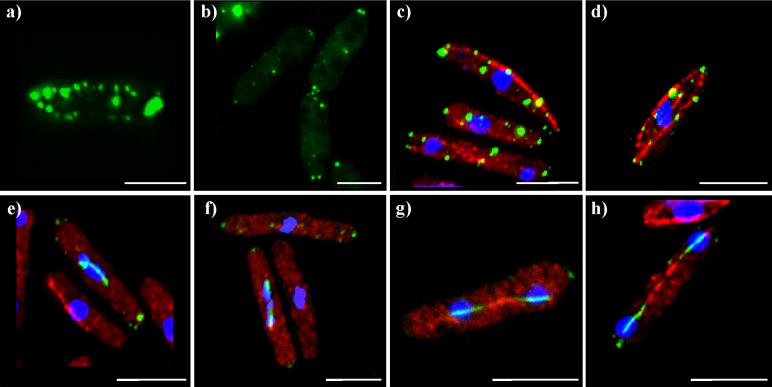
Over-expression Klp5-Full-length and Klp6-Full-length
Full-length Klp5p and Klp6p were expressed using the same expression system as was used for the individual domains. This approach helped to distinguish between phenotypes that were generated by domain-specific interactions and those created by the expression level of Klp5p or Klp6p in general.
Expression of the full-length Klp5p led to a high degree of MT bundling and to significant defects in cell morphology (Figure 4). Cells often contained a single MT bundle that originated near the nucleus (Figures 4a,b), frequently a short distance (0.2−0.5μm) from a single, brightly staining dot (Figure 4c). The fluorescent intensity along the length of each MT bundle decreased stepwise, consistent with the signal's arising from many (>12) MTs (data not shown). Individual MTs can also be seen extending from the tips of the main MT bundle (Figure 4c). Occasionally, cells contained large aggregates of fluorescence with individual fibers emerging from them (Figure 4d). Mitotic spindles also showed staining, though the spindles looked abnormal (Figure 4e).
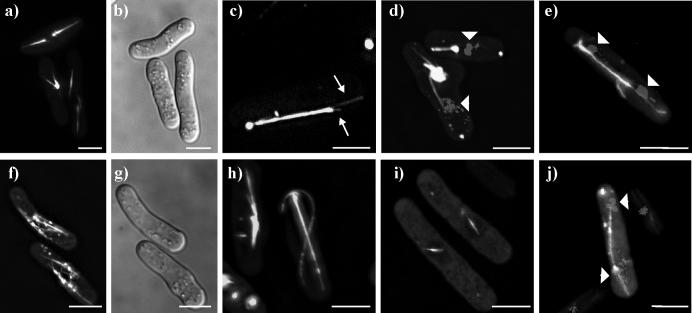
Figure 4. Localization of Klp5-full-length-EGFP and Klp6-full-length-EGFP.
a-e) Klp5-full-length-EGFP imaged in live cells. a) cells with one or two large bundles of MTs; b) bright field image of cells in first panel; c) cell with single large bundle of MTs, with individual MTs splaying out from the bundle (arrows); d) cells stained for DNA (Hoechst) show that bundles are not spindles; e) mitotic cell in late anaphase B. f-j) Klp6-full-length-EGFP imaged in live cells. f) two cells with shorter-than-normal peri-nuclear MTs and dots; b) bright field image of previous panel; h) cells with MT bundles and long, curled MTs; i) two cells with short spindles; j) cell stained for DNA (Hoechst) with a somewhat abnormal, late mitotic spindle. White bar = 5μm.
The Klp6-full-length protein localized efficiently to MTs, in contrast to either the Klp6-motor or the Klp6-motor-neck domains (Figure 4f-j). The MTs commonly looked shorter than normal; though positioned around the nucleus, they were somewhat disorganized (Figure 4f). Curiously, the Klp6-full-length protein was less efficient at MT depolymerization than either the Klp6motor or Klp6motor-neck domains. There were also several dots associated with the MTs, reminiscent of the dots observed for Klp6-tail/neck-tail (see also Figure 7 below). Significant defects in cell morphology were common in Klp6-full-length expressing cells (Figure 4g). Occasionally, the MTs looked over-grown, and they curled about the ends of the cells (Figure 4h), which is common in cells deleted for either Klp5 or Klp6. Mitotic spindles were labeled in both early and late mitosis, but late mitotic spindles looked abnormal (Figure 4i,j).
Together, these data showed that ectopically expressed Klp5-full-length had a localization and a phenotype that were similar to those found in cells expressing either Klp5-motor or Klp5-motor-neck. Klp6-full-length, on the other hand, produced results that were intermediate between Klp6-motor-neck and Klp6-tail-neck (see below).
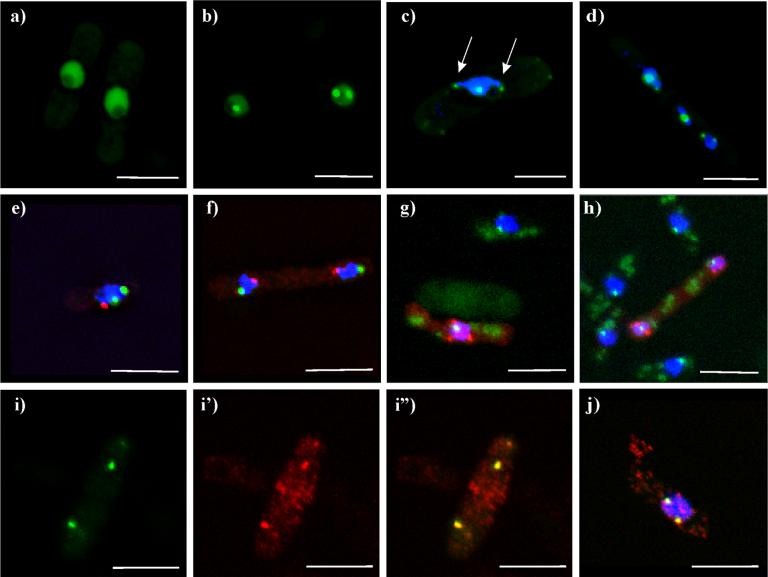
Klp5tail
The tail domain of Klp5p is the 447 amino acids on the carboxy-terminal side of the α-helical neck domain (Figure 1). Its sequence is unique, and its only recognizable motif is a nuclear localization signal (NLS) near its carboxy-terminal end (West et al., 2001).
The Klp5-tail protein localized predominately in the nucleus, with >5-fold enrichment over cytoplasmic staining (Figures 5a,b). Approximately 50% of these cells also showed one or more distinct dots within the nucleus, each with 3-to-5-fold further enrichment above the GFP signal in the nucleoplasm (Figures 5b,c). These dots were often associated with DNA-containing protrusions of the nucleus or with misaligned chromosomes during mitosis in klpΔ mutants (Figure 5d). Time-lapse microscopy indicated that these dots moved actively within the interphase nucleus, with a given dot often dividing into two smaller dots, then fusing back into one (data not shown). This pattern was seen throughout the cell cycle, including mitosis, and was not altered by deletion of either klp5+, klp6+, or both of these genes (data not shown).
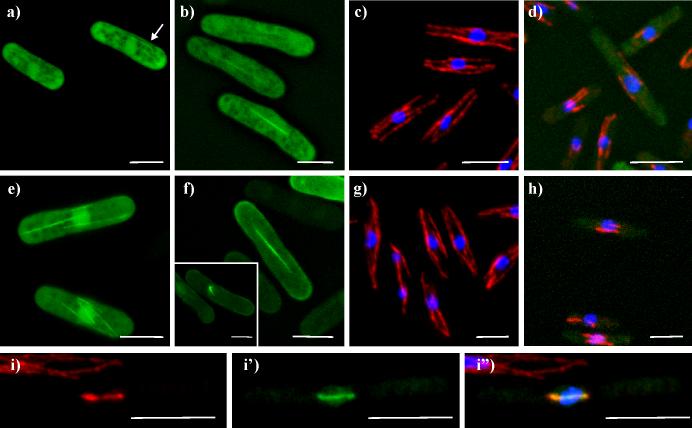
Figure 5. Klp5-tail-EGFP localization to telomeres.
a-d) live cells expressing Klp5-tail-EGFP. a) cells with nuclear staining, excluding the nucleolus; b) cell in late telophase with nuclear staining and dots in each nucleus; c-h,j) cells stained with Hoechst 33342 for DNA (blue); e-f) simultaneous staining with the spindle pole body marker, Sad1p (red). 5c shows nuclear dots associated with DNA protrusions (arrows); d) klpΔ cell with DNA staining, showing GFP dots associated with each chromosome. e) interphase cell; f) late mitotic cell. g, h) Klp5-tail-Pk1 (red) with the kinetochore marker Mis12-GFP (green), and DNA (blue). 5g and h) interphase cell and late mitotic cell, respectively, both with Klp5-tail-Pk1 stained red. i-j) Simultaneous staining of Klp5-tail-Pk1 (red) with the telomere marker Taz1-GFP (green). i) interphase cells, Taz1-GFP; i') Klp5-tail-Pk; i”) combined channels. j) mitotic cell, combined channels. White bar = 5μm.
While most of the cells had strong nuclear staining, about 10% of the cells showed cytoplasmic dots at the cell's ends or in linear tracks reminiscent of MTs. The cause for this variation in localization was not identified, but when it was seen, nuclear staining was usually not present. A few rare examples showed spindle mid body staining in addition to nuclear staining (data not shown).
The general nucleoplasmic staining and nuclear dot localization of the Klp5-tail are markedly different from the localizations of endogenous Klp5p and Klp6p previously described (West et al., 2002) or of any other motor protein. Given this contrast, it seemed possible that our observations might be due to effects from the carboxy-terminal EGFP tag, not from the Klp5-tail domain itself. We therefore made two additional constructs of the Klp5-tail domain: one with the Pk1 epitope at its carboxy-terminus, and one with EGFP at its amino-terminus (Figure 1A). The localizations of these new constructs were indistinguishable from those observed with the original Klp5-tail-EGFP construct (Figure 5, g-h), indicating that the tagging itself was not generating the localization to nuclear spots.
To identify the nuclear structure(s) to which the Klp5-tail was localizing, cells expressing this domain were co-stained with several markers for known nuclear structures. Previous work has localized several kinesin-like proteins to spindle pole bodies and kinetochores (Goldstein and Philp, 1999), so these organelles were of particular interest. Cells expressing Klp5-tail were fixed with methanol and stained with antibodies to the spindle pole body marker, Sad1p (Hagan and Yanagida, 1995) (Figure 5e,f). The Klp5-tail and Sad1p staining were clearly distinct in both interphase and mitotic cells (n= 80 cells), indicating that the Klp5-tail dots were not spindle pole bodies. Moreover, the single Sad1 dot in the presence of two Klp5-tail dots confirmed that these cells were not mitotic. The kinetochores were localized in cells expressing Klp5-tail-Pk1 using the kinetochore marker, Mis12-GFP (Goshima et al., 1999). These cells were formaldehyde fixed and stained for the Pk1 epitope, while GFP was visualized by its own fluorescence (Figure 5 g,h). The Klp5-tail dots were clearly distinct from the kinetochores in both interphase and mitosis (n= 18).
While kinesin motors have not previously been identified at telomeres, the arrangement of fission yeast telomeres was consistent with the localizations of the Klp5-tail chimeras. We explored this possibility using cells expressing the telomere marker, Taz1-GFP (Cooper et al., 1997). Surprisingly, the Klp5-tail-Pk1 and Taz1-GFP localizations overlapped and appeared coincident (Figure 5g-j). Further, the co-staining between the Klp5-tail and telomeres was independent of the numbers of Klp5-tail dots that were present in a given cell. A single Klp5-tail dot was the most common configuration, but as many as six dots could sometimes be resolved (average 2.0 ± 0.2 dots) (n=22). Wild type cells expressing just Taz1-GFP showed an average of 1.6 ± 0.9 dots (n=232 cells).
Klp5-neck-tail
The heterodimer model for Klp5/Klp6 predicts that addition of the neck domain to the Klp5-tail protein would facilitate dimerization of this Klp5p fragment with the endogenous Klp5, Klp6p, and/or itself (Garcia et al., 2002b; Li and Chang, 2003) (Figure 1B). We investigated whether adding the neck domain back to the tail would result in the Klp5-neck-tail protein having a wild type-like localization pattern that was, in turn, dependent on the expression of either Klp5p or Klp6p. A plasmid with the Klp5p-neck-tail domains fused to EGFP at the carboxy-terminus was constructed and transformed into wild type, klp5Δ, klp6Δ, and klp5Δ; klp6Δ strains.
The inclusion of the 40 amino acid α-helical neck had a significant effect on the localization of the Klp5-tail, re-directing it from the nucleus (Figure 5) to the cytoplasm (Figure 6). Most cells showed cytoplasmic dots that tended to lie towards the sides or ends of the cells. Nuclear staining was rarely observed, either in interphase or mitosis. Co-staining for tubulin showed that these dots were situated along MTs or at MT ends (Figure 6). This pattern persisted through the cell cycle, with neither the mitotic spindle nor kinetochores showing any staining (Figure 6). This pattern also did not change when the Klp5-neck-tail protein was expressed in klp5Δ, klp6Δ, or klp5Δ; klp6Δ backgrounds, indicating that the MT-end localization was not due to interactions with the endogenous full length kinesins. .
Figure 6. Klp5-neck-tail-EGFP on cytoplasmic dots and microtubules.
a) Live cell expressing Klp5-neck-tail-EGFP with cytoplasmic dots in linear arrays. b-d) methanol-fixed cells with Klp5-neck-tail-EGFP (green), tubulin staining (red), and DNA staining (blue); b) interphase cells; c) early mitotic cell; d) late mitotic cell. White bar = 5μm.
The localization of the Klp5-neck-tail to MT ends suggested that the neck domain might facilitate an interaction with a MT-TIP protein. A candidate for this interaction is the EB-1 homologue, mal3+ (Beinhauer et al., 1997). Mal3p is required for loading the fission yeast kinesin 7 protein, Tea2p, onto MT ends, and it stimulates Tea2p's ATPase activity (Browning and Hackney, 2005; Browning et al., 2003). We asked if the Klp5-neck-tail localization to MT-ends required Mal3p by expressing the Klp5-neck-tail plasmid in the mal3Δ strain. The localization of Klp5-neck-tail was not altered in the mal3Δ background, indicating that Klp5-neck-tail was not localizing to the MT ends through interactions with Mal3p (data not shown). We also noted that the klpΔ strains did not show genetic interactions with the mal3Δ allele, and the localization of the endogenous, full length Klp5p and Klp6p was not significantly altered in the mal3Δ background (data not shown).
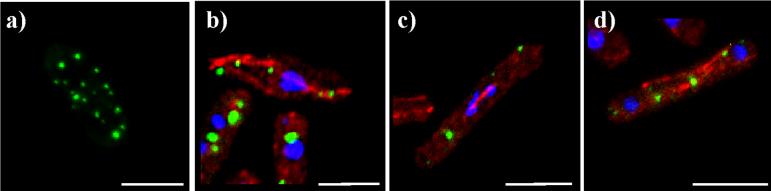
Klp6-tail
The tail domain of Klp6p is the 344 amino acids carboxy-terminal of the α-helix, and contrasts the Klp5-tail in its size, primary sequence, and predicted net charge (Klp5-tail, pI=5.5; Klp6-tail, pI=10) (West, et al., 2001) (Figure 1). To investigate the similarities and differences between these two kinesin tails, the Klp6-tail was expressed in yeast cells with EGFP fused to either its amino- or carboxy-terminus. These constructs were again expressed in four genetic backgrounds: wild type, klp5Δ, klp6Δ, and klp5Δ; klp6Δ.
The Klp6-tail localized primarily to dots in the cytoplasm, which were frequently at the tips of the cells, or in linear arrays reminiscent of MTs (Figure 7). A few of these dots moved over time but at rates no greater than the slowest kinesins (∼0.1 μm/min, data not shown). There was some general nuclear staining, but it was enriched only 1.5-fold over the cytoplasmic background. Nuclear dots like those seen with Klp5-tail were never observed. The Klp6-tail dots also did not show any association with the mitotic spindle or with other nuclear structures (Figure 7). The Klp6-tail localization pattern was unchanged by the use of amino-terminal and carboxy-terminal tags or by the genetic background in which it was expressed. Neither the Klp5-tail nor the Klp6-tail had an effect on the sensitivity of cells to the microtubule destabilizing drug, TBZ (data not shown).
Figure 7. Klp6-tail-EGFP and Klp6-neck-tail-EGFP form cytoplasmic dots on or near microtubules.
a) Live cell expressing Klp6-tail-EGFP with cytoplasmic dots in linear arrays; b) live cells expressing Klp6-neck-tail-EGFP with cytoplasmic dots at the cell ends. c,h) fixed cell with Klp6 constructs (green), tubulin staining (red), and DNA staining (blue); c) Klp6-tail-EGFP; d) Klp6-neck-tail-EGFP; e) Klp6-neck-tail-EGFP in early mitosis; f) Klp6-neck-tail-EGFP in mid mitosis; g) Klp6-neck-tail-EGFP in anaphase B; h) Klp6-neck-tail-EGFP in late mitosis. White bar = 5μm.
Klp6-neck-tail
As described for Klp5p, a heterodimer model predicts that the addition of the α-helical neck to the Klp6-tail should facilitate dimerization, particularly with the endogenous Klp5p (Garcia et al., 2002b; Li and Chang, 2003). A plasmid with the Klp6p-neck-tail domains was constructed and transformed into wild type, klp5Δ, klp6Δ, and klp5Δ; klp6Δ strains.
The Klp6p-neck-tail protein localized predominately as cytoplasmic dots, like those seen for the Klp6-tail construct (Figure 7). These dots were generally at the sides or tips of the cells, as described for the Klp6-tail. Two lines of evidence indicate that these dots lie along MTs. First, co-staining with tubulin antibodies showed that dots were either along, or at the ends of cytoplasmic MTs (Figure 7). The Klp6-neck-tail plasmid was also co-expressed with genomic copies of either Klp5GFPp or Klp6GFPp, both of which localize to cytoplasmic MTs (West, et al., 2001). Though all these molecules were tagged with GFP, the cells showed two distinct patterns; bright dots, and very faint MTs (data not shown). The bright dots are likely to be the Klp6-neck-tail, while the faint MT pattern is consistent with the whole motor GFP-tagged proteins. These data suggest that the Klp6-neck-tail localizes to the ends of MTs, and that this association does not displace the full length proteins from their normal MT pattern. As described for Klp5-neck-tail, the Klp6-neck-tail MT end staining was not dependent on the expression of Mal3p (data not shown).
The Klp6-neck-tail protein also localized to a unique pattern along the mitotic spindles, indicating that this protein is transported into the mitotic nucleus and can localize to spindle MTs (Figure 7e-h). This localization was consistent with the dimerization of the expressed protein with endogenous Klp5p during mitosis, but this spindle localization was also observed in strains that lacked Klp5p and Klp6p. There was no indication of kinetochore (or telomere) staining; pre-anaphase spindles showed uniform staining. During spindle elongation, the staining became restricted to the pole-proximal ends of the spindle (Figure 7f-h, 8b). The staining appeared along the entire spindle until its length averaged 3.7 ± 1.0 μm (n=23). As the spindle elongated further, each half spindle contained a pole-proximal region of staining that averaged 2.1 ± 0.5μm in length (n=117). The lengths of staining in each half spindle were equal within a factor of 1.2. These data suggest that the Klp6-neck-tail associates with the spindle early in mitosis, but this association takes on a rigor-like state, so the protein moves along with the interpolar MTs as the spindle elongates. This phenomenon was not changed in any of the klpΔ backgrounds. However, the expression of the Klp6-neck-tail plasmid in klp5Δ, klp6Δ double mutants did lead to a significant increase in cell length (16.1 ± 2.9μm, n=24 vs. 10.6 ± 2.4μm, n= 632).
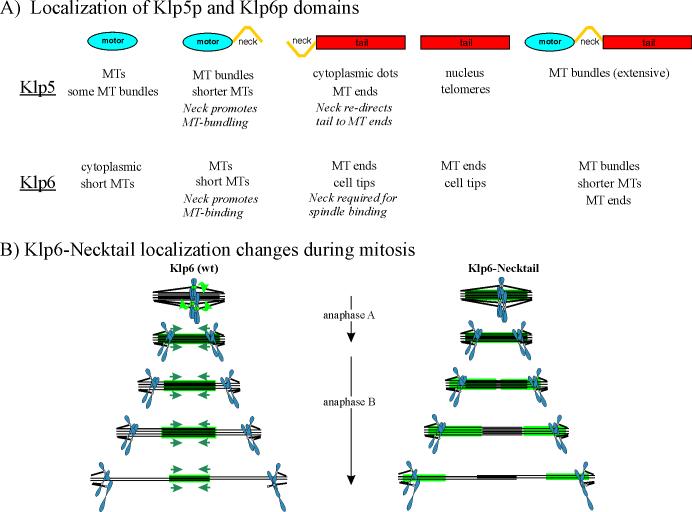
Figure 8. Diagrams summarizing the localization patterns of domains from Klp5p and Klp6p.
A) Kinesin domains expressed in fission yeast cells, with the primary localizations given below. B) Diagram of the changes in localization for endogenous Klp6p (left) and the Klp6-neck-tail domain (right) through mitosis. Green arrows indicate the relocation of Klp5p and Klp6p from kinetochores to the midregion of the interpolar spindle as the cell enters anaphase. Blue arrows indicate the motion of the Klp staining as anaphase B proceeds and the interpolar MTs slide in wild type cells. Klp-neck-tail is mislocalized during anaphase B, concentrating on the half spindles and not the zone of interdigitation. The length of the stained segment is constant as the spindle elongates.
Discussion
Expression of individual domains from both Klp5p and Klp6p has revealed a surprisingly complex set of cellular localizations. Moreover, the phenotypes of cells expressing these constructs are unique to each protein domain and to each kinesin (Figure 8A), even though they are insensitive to the amounts of endogenous Klp5/6 that are expressed. Current results have defied our efforts to propose a simple model, but the following statements summarize our observations:
Motor Domains
The motor domains of Klp5p and Klp6p are similar in primary sequence (66% identical/76% similar; West, et al., 2001), and the deletion of each gene produces nearly the same phenotype. However, current results demonstrate differences in the localizations and phenotypes of the motor and motor-neck fragments of these closely related kinesin 8s. This suggests that the limited primary structure differences do convey binding specificities that may help to define specific functions for each component of this dimeric motor enzyme. These different affinities may be the reason why the genomes of fission yeast and most other organisms so far studied encode at least two kinesin 8s. Budding yeast, which uses only one kinesin 8 that homodimerizes (Gupta et al., 2006; Varga et al., 2006), may restrict the function of this enzyme to simpler tasks.
Since deletion of either klp5+ or klp6+ leads to longer MTs and significant resistance to TBZ (West, et al., 2001; Garcia, et al., 2002), Klp5p, Klp6p, and other kinesin 8s, are thought to foster MT depolymerization in vivo. Similar results have now been reproduced in vitro (Gupta et al., 2006; Varga et al., 2006). Our data refine this picture by parsing the depolymerization activity into different parts of the heterodimeric kinesin 8 in fission yeast. Over-expression of either Klp5-motor or Klp5-motor-neck did not appear to shorten cytoplasmic MTs; it simply made them bundle. Klp5-motor-neck appeared to bind MTs strongly; the degree of MT bundling was greater with motor-neck than with either motor alone or with the full-length Klp5p. These data suggest that the neck promotes bundling, potentially through dimerization of the motor-neck domains (Figure 1B). This phenotype was independent of the expression of the endogenous Klp5p or Klp6p in the same cell. Presumably the monomeric Klp5-motor promotes MT bundling by a mechanism that is enhanced through dimerization with another Klp5-motor domain. The lack of apparent MT depolymerizing activity by the Klp5p constructs in the presence of Klp6p (wild type or klp5Δ backgrounds) also indicates that the Klp5-motor (and motor-neck) is not activated to depolymerize MTs by the presence of at least wild type levels of Klp6p. Perhaps the Klp5-motor (and motor-neck) domains do depolymerize MTs to some degree, but this effect is masked by the extensive bundling.
Klp6-motor-neck, on the other hand, bound MTs only weakly (as indicated by the distribution of fluorescence brightness), even though the expression of this allele led to significantly shorter MTs. We found no obvious structural motifs in the Klp6-motor domain that would indicate weaker MT binding. While addition of the neck did somewhat increase the efficiency of MT binding and depolymerization, possibly through fragment dimerization (Figure 1B), this does not provide a satisfactory explanation for the differences between Klp5 and Klp6. Moreover, the full-length Klp6p bound more efficiently to MTs, but it had only a modest effect on MT length. This suggests that the tail domain increases motor affinity for MTs but suppresses MT destabilization. A plausible hypothesis for all these data supposes that Klp5p is not very soluble unless it forms dimers with Klp6p, and full length Klp6p is similarly inclined. The motor and motor-neck fragments of Klp6p, on the other hand, are soluble and enzymatically active, so they can shorten even the over-length MTs found in klpΔ strains. These relative solubilities are consistent with the results from our efforts to express these motors in both bacteria and insect cells and to purify them in vitro (Fiedler et al., in preparation)
Klp5-motor/Klp5-motor-neck and Klp6-motor/Klp6motor-neck fail to become restricted to the spindle midzone in late mitosis. Endogenous, full length Klp5p/Klp6p remains in a region of constant size in the anaphase B midzone (∼3μm) while the spindle elongates from about 3μm to 12μm, suggesting that this motor displays a plus-end directed motility in vivo that works at the rate of spindle elongation (Figure 8B) (West, et al., 2001). The lack of spindle mid-zone localization for the motor and motor-neck fragments suggests that sequences within the tails are required for Klp5p/Klp6p to stay in the midzone. This behavior probably depends on controlled, plus-end directed motility in vivo. The fact that the abnormal anaphase localizations of full-length Klp5p/Klp6p constructs and motor or motor-neck fragments are not affected by the presence of the complementary motor suggests that the interactions between Klp5p and Klp6p that promote proper association with the anaphase interpolar spindle require a balance between the endogenous, full length polypeptides and the levels of expression that occur during their synthesis in a normal cell.
The septation defect reported here for the expression of Klp5-motor-neck is consistent with an interaction between the Klp5p and the regulatory machinery for septation reported by Li and Chang (2003). They found both genetic and physical interactions between Klp5p, Klp6p and the guanine nucleotide exchange factor Scd1p, a modifier of Ras1p activity. Furthermore, double mutants between sdc1 and either klp5 or klp6 showed defects in the localization of both actin and anillin to the equatorial band that anticipate the site of septation. Curiously, the phenotypes we have observed were specific for Klp5p, which is in contrast to the interactions described by Li and Chang. Gatt, et al. (2005) have reported that depletion of KLP67A is essential for proper spindle midbody formation and cytokinesis in Drosophila.
Expression of Klp5-motor, Klp5-motor-neck, and Klp5-full-length all produce similar phenotypes, with the exception that the motor alone produces a septation defect. These results suggest that most of the interactions of the whole protein are also achieved by the motor domain itself. The motor domain of Klp6p, on the other hand, has MT-depolymerizing activity that is affected, be it directly or indirectly, by elements in the tail domain. The absence of short MTs in cells over-expressing the full-length Klp6p, compared to just motor-neck, suggests that in the absence of Klp5p, the presence of the Klp6p tail inhibits the depolymerization capability of the motor domain.
Klp5 and Klp6 Tails
Cargoes for the kinesin 8 motors have not yet been identified, though their tail domains comprise nearly half of each protein. Each kinesin 8 tail is unique in its sequence, with the exception of a fission yeast-specific, 28 amino acid “tail box” and a putative NLS motif (West, et al., 2001). The “tail box” itself has no obvious homology with known protein motifs, and its functional significance remains unknown.
The localization of Klp5-tail to the telomeres has no precedent in the kinesin literature. While we have not detected telomere binding for the whole Klp5p in vegetative cells, an association of Klp5p with telomeres may be an important aspect of meiosis. Perhaps the expression of the Klp5-tail truncation mutant has revealed a normal meiotic function of this enzyme. Unfortunately, attempts to localize the endogenous, full length protein in meiotic cells have so far failed to produce interpretable images.
These results also show that the tail of Klp5p contains sufficient information for nuclear localization, consistent with the putative NLS motif. However, they also suggest that full length Klp5p negatively regulates this signal, perhaps through the motor domain; the whole protein was nuclear only in mitosis, while the tail was nuclear throughout the cell cycle.
Motor Complexes and Their Function
Evidence reviewed above suggests that Klp5p and Klp6p form a heterodimer in vivo. Nonetheless, we have previously shown that the dynamic localization pattern observed for either Klp5p or Klp6p is not dependent on the presence of its complementary motor (West, et al., 2002). Therefore, the interactions that determine motor localization and MT-based motility for both Klp5p and Klp6p can operate independently of heterodimer formation. On the other hand, deletion of either motor results in the loss of an apparent MT-depolymerizing activity, as indicated by the presence of similarly exaggerated MT arrays and TBZ resistance in all the klpΔ mutants (West, et al., 2001; Garcia, et al., 2002). Therefore, each motor contains sufficient information for localization and motility, but only together do these full length motors produce a depolymerizing activity. This raises the question, does each single motor lack depolymerase activity on its own, or is this activity differentially regulated in heterodimer versus single motor protein containing complexes? Our observations here favor a solubility/activity-dependent explanation for Klp6p, as this full-length construct does have some depolymerization activity in the absence of Klp5p. The more robust depolymerizing activity of Klp6 motor, and particularly its motor-neck, suggests that these fragments are more readily maintained in solution than the full length molecule. The increase in MT depolymerization with plasmid-expressed Klp6p, relative to that protein expressed from the endogenous gene in klp5Δ strains, may be simply an effect of over-expression. The same considerations might pertain to Klp5p, but the MT bundling of this motor, expressed on its own, may mask the effect. Klp5p appears to form homodimers in vivo, but this complex may have a different activity and regulation than the Klp5/6 heterodimer.
Kinesin 8 Neck Interactions
The results presented here have uncovered several interactions, unique for each kinesin 8, that were not apparent when the whole protein was observed (Figure 8A). We find an unexpected role for the short (predicted) α-helical coiled-coil domain of both Klp5p and Klp6p; several new potential interactions have been described. In the case of Klp5p, the neck domain re-directs the tail from the nucleus to the ends of MTs. This suggests that the Klp5-neck interacts with a MT-TIP protein, but this is neither the endogenous Klp5p, Klp6p, nor Mal3p. The Klp6-tail also interacts with MT ends, but this localization is independent of the neck. It remains to be determined if the Klp5-neck-tail and Klp6-tail (and Klp6-neck-tail) MT bindings share interaction partners.
The neck domain of Klp6p is required for the association of Klp6-tail with the mitotic spindle. Again, this interaction is independent of the presence endogenous, full length Klp5p or Klp6p. This suggests that the Klp6-neck interacts with a spindle component at the time when endogenous Klp6p is moving from kinetochores to the spindle. It is curious that the Klp6-neck-tail is not moved to the spindle midzone, even when the endogenous Klp6p and Klp5p are being expressing (experiments in the wild type background). The extent of Klp6-neck-tail spindle staining is similar to the length of MT interdigitation in early mitosis (Ding et al., 1993). However, the endogenous Klp6p continues to move towards the center of the spindle, maintaining its association with the interdigitating MT zone, while the Klp6-neck-tail remains stuck to the same MT region that it bound during its first association with the spindle (Figure 8b) (West, et al., 2002). The constant length of the Klp6-neck-tail band in each half spindle is also consistent with the reported lack of poleward flux of MTs in fission yeast (Mallavarapu et al., 1999).
Conclusion
The factors that regulate localization of kinesin 8s in fission yeast are complex. Our data highlight the importance of the α-helical neck domain, suggesting that both homo- and hetero-dimerizations of the two isoforms found in this one cell are possible. Other factors yet to be identified, such as interacting proteins or post-translational modifications are likely to play a role in the places these motors bind. While budding yeast has only a single gene for kinesin 8, many organisms have several. Students of this important motor enzyme in higher eukaryotes should be mindful of the complexities they may be over-looking by working with simply one isoform at a time. While our work has not spelled out the rules that govern localization of these motors, it has defined a problem that merits careful attention.
Acknowledgements
We thank Terra Malmstrom for help with plasmid construction and cell transformation, Thomas Fiedler and Katya Grishchuk for discussions and criticism of the manuscript, and Julie Cooler for the Taz1-GFP strain. This work was supported in part by NIH grant R01-033787 to JRM.
References
- Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–28. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H, Hackney DD. The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J Biol Chem. 2005;280:12299–304. doi: 10.1074/jbc.M413620200. [DOI] [PubMed] [Google Scholar]
- Browning H, Hackney DD, Nurse P. Targeted movement of cell end factors in fission yeast. Nat Cell Biol. 2003;5:812–8. doi: 10.1038/ncb1034. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–7. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J.Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic.Acids.Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell. 2004;15:121–31. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Spindle-kinetochore attachment requires the combined action of Kin I- like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. Embo J. 2002a;21:6015–24. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002b;12:610–21. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Gatt MK, Savoian MS, Riparbelli MG, Massarelli C, Callaini G, Glover DM. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J Cell Sci. 2005;118:2671–82. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Philp AV. The road less traveled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol. 1999;15:141–83. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–77. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr., Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–23. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–47. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–57. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–5. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chang EC. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics. 2003;165:477–88. doi: 10.1093/genetics/165.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Chen CR, Chang EC. Fission yeast Ras1 effector Scd1 interacts with the spindle and affects its proper formation. Genetics. 2000;156:995–1004. doi: 10.1093/genetics/156.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–6. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisen L, Wallack DL, Loayza D, Gammie AE, Rose MD. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;9:2051–68. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Pereira AJ, Dalby B, Stewart RJ, Doxsey SJ, Goldstein LS. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J Cell Biol. 1997;136:1081–90. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischitor PE, Konzack S, Fischer R. The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot Cell. 2004;3:632–45. doi: 10.1128/EC.3.3.632-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: a Laboratory Manual. Second Edition Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005;24:2931–43. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–77. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–62. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–40. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, Troxell CL, McIntosh JR. Two Related Kinesins, klp5+ and klp6+, Foster Microtubule Disassembly and Are Required for Meiosis in Fission Yeast. Mol Biol Cell. 2001;12:3919–32. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR. cut11+: A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:2839–55. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–43. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–99. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]