Abstract
Genes coding for proteins involved in lipid metabolism and, in women, menopausal status are independently associated with high-density lipoprotein cholesterol (HDL-c) and low-density lipoprotein cholesterol (LDL-c) levels. We examined whether the association between common functional genetic polymorphisms of apolipoprotein E (apoE Cys112Arg and Arg158Cys) gene and LDL-c levels, as well as the associations between the cholesteryl ester transfer protein (CETP TaqIB), hepatic lipase (LIPC C-514T), and lipoprotein lipase (LPL Ser447Stop) genes and HDL-c levels are significantly modified by menopausal status. Plasma lipid concentrations, genotype, and menopausal status were assessed across four examinations in a sample of Caucasian and African-American women (n=4652 to 4876) who were aged 45-64 years at baseline from the Atherosclerosis Risk in Communities Study. The association between LDL-c levels and the apoE gene, and HDL-c levels and the LIPC and LPL genes were not modified by menopausal status. The only statistically significant gene by menopause interaction was with the CETP gene on HDL-c concentrations (p=0.04). However, the significant CETP gene by menopause interaction was possibly due to chance because of multiple testing. Postmenopausal women who were carriers of the A allele of the CETP gene had approximately 0.7 mg/dL lower HDL-c levels than pre-/perimenopausal counterparts, whereas the opposite pattern of HDL-c (0.4 mg/dL higher HDL-c postmenopausally) was observed for the GG genotype. Overall, our data suggest that the decrease in endogenous estrogen as a result of menopause may independently affect lipoprotein concentration, but does not alter the effect on plasma lipids of some common genetic polymorphisms that regulate lipoprotein metabolism.
Keywords: LDL, HDL, apolipoprotein E, cholesteryl ester transfer protein, hepatic lipase, lipoprotein lipase, menopause
Introduction
Epidemiologic research has provided strong evidence that low plasma concentrations of high-density lipoprotein cholesterol (HDL-c) and high plasma concentrations of low-density lipoprotein cholesterol (LDL-c) are associated with increased risk of coronary heart disease (CHD).(1,2) Many factors have been found to affect HDL-c and LDL-c levels, including specific genetic variants(3,4) and postmenopausal status in women.(5-8)
Apolipoprotein E plays an important role in lipid metabolism by serving as a receptor-binding ligand on the surface of very-low-density lipoproteins (VLDL), HDL, and chylomicrons, which mediate the clearance of these lipoproteins from plasma.(9) The apoE polymorphism is located on chromosome 19, exon 3 and has three alleles, ε2, ε3, and ε4. The ε2 and ε4 variants result from a C to T mutation at position 158 and a T to C mutation at position 112, respectively. Compared with the ε3 allele, the ε2 allele is associated with lower levels and the ε4 allele is associated with increased levels of LDL-c.(10) Further, the association of apoE allele with LDL-c has been reported to be significantly greater among postmenopausal women than premenopausal women.(11)
CETP plays a key role in metabolism of HDL by transferring cholesteryl esters from HDL to triglyceride-rich lipoproteins,(12) and thus, CETP activity is inversely related to HDL-c levels.(13) The TaqIB polymorphism of CETP is a noncoding mutation in intron 1 of the TaqI restriction site located on chromosome 16q21. The B2 (A) allele of the TaqIB polymorphism of CETP is associated with higher HDL-c levels than is the B1 (G) allele.(13) Hepatic lipase activity is inversely correlated with HDL-c levels, and is involved in the metabolism of HDL-c through selective uptake of HDL-cholesterol esters.(14) The C-514T polymorphism is located on chromosome 15q21-q22 in the promoter region of the LIPC gene, and the variant allele is the result of a C to T mutation at position -514. Postmenopausal women who are carriers of the variant T allele in the C-514T polymorphism of LIPC have higher HDL-c levels than postmenopausal women who are C/C for this polymorphism.(15) LPL is responsible for hydrolysis of triglyceride molecules into free fatty acids, conversion of chylomicrons to chylomicron remnants that play a role in conversion of VLDL to LDL, and regulation of HDL.(16) The Ser447Stop polymorphism of LPL is located on chromosome 8p22. A C to G substitution in exon 9 results in a premature stop codon and the deletion of the last two amino acids in the protein. Carriers of the variant 447X (G) allele of the Ser447Stop polymorphism of LPL have higher HDL-c than the S/S (CC) genotype.(17,18)
Independent of age and weight gain, menopause is associated with changes in lipid levels, specifically increases in total cholesterol, LDL-c, and triglycerides (TG),(5-7) and decreases in HDL-c.(8,19) The changes in LDL-c during menopause are thought to occur from reduced metabolism of LDL particles as a result of postmenopausal decreases in estrogen levels.),(5-7) Increased hepatic lipase activity resulting from reductions in estrogen in menopause may play a role lowering of HDL-c.(20).
Many studies have shown associations of specific genetic polymorphisms with HDL-c and LDL-c levels. However, the impact of these genes on lipids in women during and after the menopausal transition has not been thoroughly studied. As a result of decreasing estrogen levels, progression through the menopause transition may act synergistically with gene variation to alter lipoprotein levels. This longitudinal study aims to determine if there is an interaction between menopausal status and specific gene polymorphisms to influence lipoprotein levels beyond the independent contributions of menopause or genotype.
Methods
Study Population
The ARIC study is a prospective cohort of African-American and Caucasian adults aged 45-64 years gathered from four communities in the United States: Forsyth County, North Carolina; Jackson, Mississippi; Minneapolis suburbs, Minnesota; and Washington County, Maryland.(21) A total of 15,792 participants (8710 women) were enrolled by probability sampling from 1987 to 1989, and completed a home interview and clinic visit. Sampling differed by recruitment area, and only African-Americans were recruited from Jackson, Mississippi. A total of four visits were conducted, each spaced three years apart.
Participants were excluded if they were men, Asian or American Indian, had a history of cancer or developed cancer during study follow-up, had prevalent CHD or stroke at baseline, did not consent to DNA analysis, had never menstruated, were postmenopausal due to radiation, or who had inconsistent menopausal status over the four visits. Missing genotype values were also excluded for analysis of each gene, and those who had apolipoprotein E genotype ε24 were excluded from the apoE analysis due to limited frequency and inconsistent allelic effects. Women who had TG levels greater than 400 mg/dL were set to missing for LDL-c concentration for a specific visit, and those who were in the upper and lower 1% of the LDL-c or HDL-c distributions were also set to missing for that visit. Those who had 2 or more visits with missing LDL-c or HDL-c were excluded from our study sample. After these restrictions, the sample sizes ranged from 4652 to 4876 for our analyses.
Menopausal Status Definition
Based on the mean age of the women participating in this study, menopausal status was dichotomized into two groups: pre-/perimenopausal and postmenopausal. The groups were determined by questionnaire and self-reported menstrual status during the clinic interviews. A woman was categorized as pre-/perimenopausal if she (a) had menstruated in the last two years or (b) had a hysterectomy without total oophorectomy, but at the time of the interview was younger than age 55. A woman was defined as postmenopausal if she (a) had at least 24 consecutive months of amenorrhea, (b) had a bilateral oophorectomy, or (c) had a hysterectomy and was 55 years or older at the interview.
HDL and LDL Cholesterol Determination
Blood collection and processing techniques for the ARIC study have been previously described.(22) Enzymatic methods were used to measure total cholesterol(23) and TG levels.(24) Reagents were supplied by Boehringer-Mannheim Biochemical (Indianapolis, IN), and adapted for analysis using the Cobas-Bioanalyzer from Roche (Montclair, NJ). HDL cholesterol levels were determined enzymatically after dextran sulfate-Mg2+ precipitation of other lipoproteins.(25) LDL cholesterol levels were estimated with the Friedewald formula.(26)
Accuracy of lipoprotein measurements was assessed using standards from the Centers for Disease Control and Prevention (CDC). Repeatability of cholesterol measurements was tested by duplicate blood samples in 5% of study participants shipped to the laboratory one week after the original sample. The within-person and method coefficients of variation for these samples were 6.8% and 5.3%, respectively, for LDL-c and 5.2% and 4.4% for HDL-c.(27) Intraindividual reliability was assessed as being greater than 0.90 (correlation between 1-week apart measurements of cholesterol) for both LDL-c and HDL-c.(27)
Genotyping
Genotyping of the apoE (rs429358 and rs7412), CETP (rs708272), LIPC (rs1800588), and LPL (rs328) genes was conducted using the TaqMan assay from Applied Biosystems. Probe sequences were also obtained from Applied Biosystems and primer sequences were obtained from Genosys. The ABI 770 and the Sequence Detection System software from Applied Biosystems were used to perform allele detection and genotype calling. Details on primers and probes are available from the ARIC website, http://www.cscc.unc.edu/ARIC/.
Statistical Analysis
All analyses were conducted using SAS version 8.2 (SAS Institute, Cary, NC). Participant baseline characteristics by menopausal status were compared using a chi-square for categorical measures and analysis of variance for continuous measures. Each gene was tested for allelic dominance or co-dominance by comparing statistical significance of least-squares means of lipid values between genotypes. Based on the results of the dominance or co-dominance evaluations, the genotypes were grouped for comparison as follows: ε22/ε23 vs. ε34/ε44 vs. ε33 for apoE (subjects with the ε24 genotype were excluded); AA vs. AG vs. GG for CETP; TT vs. CT vs. CC for LIPC; GG/CG vs. CC for LPL.
Associations of the genes across four exams with LDL-c and HDL-c, and the interaction between genotype and menopausal status were estimated using repeated measures regression with fixed effects (proc mixed procedure with compound symmetric covariance matrix) after adjustment for a combined race and center variable (African-Americans from Forsyth County, NC; Caucasians from Forsyth County, NC; African-Americans from Jackson, MS; Caucasians from Minneapolis suburbs, MN, and Caucasians from Washington County, MD) and the following time-varying covariates: visit (1 (referent), 2, 3, 4), age (years), body mass index (<25.0 (referent), 25.0 to 29.9, >29.9 kg/m2), cholesterol-lowering medications (current, never (referent)), smoking status (current, former, never (referent)), menopausal status (postmenopausal, pre/perimenopausal (referent)), and use of hormone replacement therapy (current estrogen only or estrogen plus progestin, former estrogen only or estrogen plus progestin, never (referent)).
We first assessed whether the effect of genotype on lipoprotein concentration was modified by race. We then conducted a sensitivity analysis on fasting (8 or more hours) participants and found no difference in mean LDL-c or HDL-c cholesterol level between fasting participants versus all participants (data not shown). Therefore, we included both fasting and non-fasting cholesterol measurements in our analyses. We also ran a sensitivity analysis excluding all individuals who did not have cholesterol measurements for all four visits. Compared to excluding those with any missing visits, inclusion of participants who had one missing visit did not vary the mean cholesterol values (data not shown). Thus, we included individuals who had either one or no missing cholesterol values in all analyses. We ran three models to test the main effects of genotype on cholesterol levels: an unadjusted model, a demographic model adjusted for age and a combined race/center variable, and a full model, which included the covariates from the demographic model plus body mass index, smoking, use of cholesterol-lowering medications, use of hormone replacement therapy, and menopausal status. Finally, we added a gene by menopausal status interaction to our full model to test for an additive effect on lipid levels as women progressed through menopause.
Results
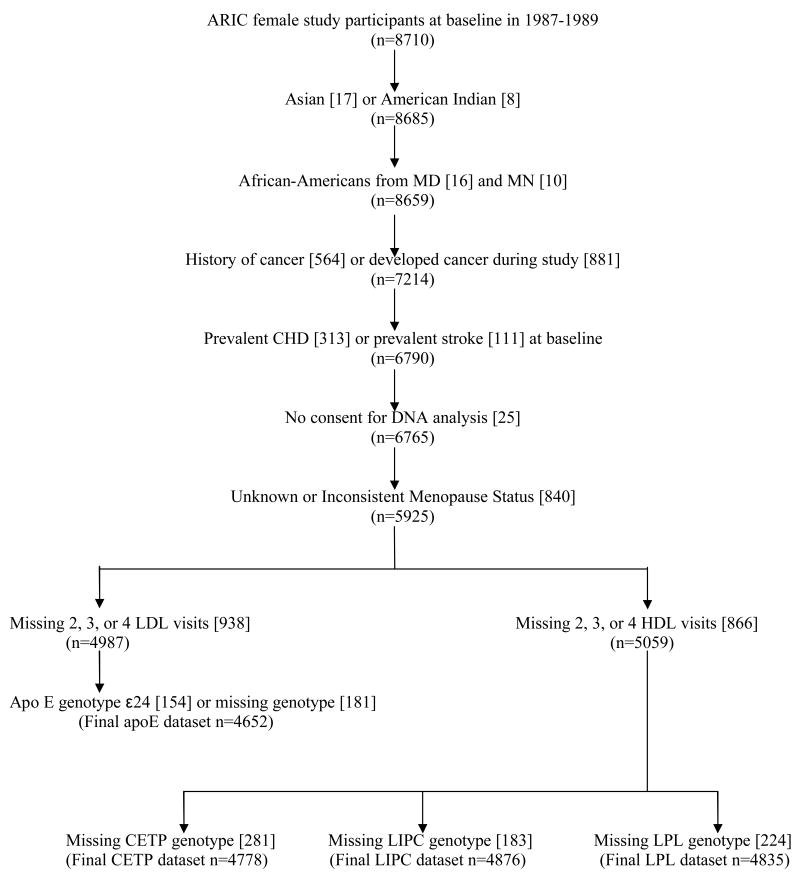
As shown in Figure 1, 4652, 4778, 4876, and 4835 ARIC female study participants at baseline in 1987-1989 met the study inclusion criteria for the apoE, CETP, LIPC, and LPL analyses, respectively. All women included in our analyses had repeated cholesterol measurements at either 3 or all 4 ARIC visits. Table 1 describes baseline characteristics of the study sample by menopausal status. Postmenopausal women were older, had a higher frequency of obesity and overweight, were more likely to be taking cholesterol-lowering medication and hormone replacement therapy, more commonly had diabetes, and were more frequently current smokers than pre-/perimenopausal women at baseline. Approximately three-fourths of the women in this dataset were Caucasian, but there was a higher percentage of African-Americans among the postmenopausal women than in the pre-/perimenopausal group.
Figure 1. Study Sample (n) and Exclusions [n excluded at each step], ARIC.
Table 1. Baseline ARIC characteristics by menopausal status, 1987-1989*.
| Pre-/Perimenopausal
n=1665 |
Postmenopausal
n=3053 |
p-value | |
|---|---|---|---|
| Age, years | 49.1 ± 0.11 | 56.4 ± 0.08 | <0.0001 |
| Race | |||
| Caucasian | 1311 (78.7) | 2177 (71.3) | <0.0001 |
| African-American | 354 (21.3) | 876 (28.7) | |
| Smoking Status | |||
| Current | 297 (17.9) | 669 (21.9) | 0.003 |
| Former | 415 (24.9) | 693 (22.7) | |
| Never | 951 (57.2) | 1690 (55.4) | |
| BMI, kg/m2 | |||
| <25.0 | 734 (44.1) | 1136 (37.2) | <0.0001 |
| 25.0-29.9 | 492 (29.5) | 1009 (33.1) | |
| >29.9 | 439 (26.4) | 908 (29.7) | |
| Diabetes | 109 (6.6) | 319 (10.5) | <0.0001 |
| Lipid Lowering Medications | 8 (0.5) | 108 (3.6) | <0.0001 |
| HRT use | |||
| Current | 249 (15.5) | 654 (22.2) | <0.0001 |
| Former | 1216 (75.8) | 1780 (60.4) | |
| Never | 140 (8.7) | 514 (17.4) | |
| LDL-c, mg/dL | 124.54 ± 0.88 | 141.08 ± 0.65 | <0.0001 |
| HDL-c, mg/dL | 58.09 ± 0.38 | 58.03 ± 0.28 | 0.89 |
Values are N (%) for categorical variables and mean ± standard error for continuous variables.
We used the dataset for hepatic lipase (n=4876), and 158 women were missing on menopausal status at baseline in this dataset.
All four genes were in Hardy-Weinberg equilibrium within race groups (Table 2). For the CETP gene, Caucasians had much higher proportions of genotypes AA and AG, whereas over half of the African-Americans were genotype GG. The majority of Caucasians carried at least one C allele for LIPC, whereas the majority of African-Americans were carriers of the T allele. Less than 5% of Caucasians were TT for the LIPC gene, yet almost 28% of African-Americans were genotype TT.
Table 2. Genotype frequencies by race group, ARIC.
| Caucasian | African-American | |
|---|---|---|
| Apolipoprotein E | ||
| E22 | 28 (0.8) | 18 (1.4) |
| E23 | 443 (12.5) | 171 (13.7) |
| E33 | 2094 (58.9) | 575 (45.9) |
| E34 | 832 (23.4) | 358 (28.6) |
| E44 | 68 (1.9) | 65 (5.2) |
| E24* | 89 (2.5) | 65 (5.2) |
| n | 3554 | 1252 |
| Cholesteryl Ester Transfer | ||
| Protein | ||
| AA | 684 (19.4) | 73 (5.9) |
| AG | 1683 (47.6) | 484 (38.9) |
| GG | 1168 (33.0) | 686 (55.2) |
| n | 3535 | 1243 |
| Hepatic Lipase | ||
| TT | 172 (4.8) | 357 (27.8) |
| TC | 1196 (33.3) | 630 (49.1) |
| CC | 2225 (61.9) | 296 (23.1) |
| n | 3593 | 1283 |
| Lipoprotein Lipase | ||
| GG | 32 (0.9) | 9 (0.8) |
| CG | 715 (19.6) | 164 (13.7) |
| CC | 2894 (79.5) | 1021 (85.5) |
| n | 3641 | 1194 |
Values are N (%)
The ε24 genotype was included for determination of Hardy-Weinberg Equilibrium, but was excluded from analysis.
Before formal genotype-lipid and genotype-menopause interaction analyses, we tested an interaction between genotype and race. The interactions were not statistically significant for the apoE, CETP, or LPL genes (all interactions p>0.05). However, the effect of LIPC genotypes on HDL-c levels was modified by race (p=0.02). Although the genotype-race interaction was significant for LIPC, the mean HDL-c concentrations by menopausal status were similar to the means reported in Table 4 for all women except pre-/perimenopausal African Americans. Therefore, we report only pooled results. Additionally, we tested for a gene*visit interaction and found no significant interaction by visit for any of the genes (all interactions p>0.10). Since the effect of genotype on mean cholesterol level did not vary by visit, we report estimates pooled over all four ARIC visits.
Table 4. Adjusted Mean LDL-c and HDL-c Levels by Menopausal Status, ARIC.
| Pre-/Perimenopausal | Postmenopausal | Difference in Means | P-value for Interaction | |
|---|---|---|---|---|
| LDL-c, mg/dL | ||||
| Apolipoprotein E | ||||
| ε22/ε23 | 105.9 ± 1.75 | 116.0 ± 1.19 | 10.1 | 0.47 |
| ε34/ε44 | 130.6 ± 1.25 | 138.4 ± 0.86 | 7.8 | |
| ε33 | 124.1 ± 0.88 | 132.0 ± 0.60 | 7.9 | |
| HDL-c, mg/dL | ||||
| Cholesteryl Ester | ||||
| Transfer Protein | ||||
| AA | 61.2 ± 0.70 | 60.6 ± 0.52 | -0.6 | 0.04 |
| AG | 58.4 ± 0.42 | 57.6 ± 0.30 | -0.8 | |
| GG | 54.9 ± 0.45 | 55.3 ± 0.33 | 0.4 | |
| Hepatic Lipase | ||||
| TT | 59.5 ± 0.89 | 59.6 ± 0.65 | 0.1 | 0.80 |
| TC | 58.5 ± 0.46 | 58.2 ± 0.33 | -0.3 | |
| CC | 56.6 ± 0.39 | 56.2 ± 0.29 | -0.4 | |
| Lipoprotein | ||||
| Lipase | ||||
| GG/CG | 59.3 ± 0.63 | 59.6 ± 0.47 | 0.3 | 0.13 |
| CC | 57.2 ± 0.32 | 56.6 ± 0.23 | -0.6 | |
Values are mean (mg/dL) ± standard error across up to 4 ARIC visits.
Means are adjusted for visit, age, a combined center/race variable, body mass index, smoking, use of cholesterol lowering medications, and use of hormone replacement therapy.
The individual associations of genotypes with mean LDL-c and HDL-c, after accounting for time trend and the effect of aging, were consistent with findings from previous literature (Table 3). For example, compared to the ε33 genotype of apoE, the ε2 allele was associated with 16.5 mg/dL lower LDL-c and the ε4 allele was associated with 6.4 mg/dL higher LDL-c. Postmenopausal status was associated with an 8.2 mg/dL greater LDL-c compared to pre-/perimenopausal women (p<0.0001). There was no significant difference in HDL-c between the pre-/perimenopausal women and the postmenopausal women (p>0.10).
Table 3. Mean Difference in LDL-c and HDL-c by Genotype, ARIC.
| Model 1 | P-value | Model 2 | P-value | |
|---|---|---|---|---|
| LDL-c, mg/dL | ||||
| Apolipoprotein E | ||||
| ε22/ε23 | -15.7 | <0.0001 | -16.5 | <0.0001 |
| ε34/ε44 | 6.2 | <0.0001 | 6.4 | <0.0001 |
| ε33 | - | ref | - | ref |
| HDL-c, mg/dL | ||||
| Cholesteryl Ester | ||||
| Transfer Protein | ||||
| AA | 5.4 | <0.0001 | 5.5 | <0.0001 |
| AG | 2.2 | <0.0001 | 2.5 | <0.0001 |
| GG | - | ref | - | ref |
| Hepatic Lipase | ||||
| TT | 3.5 | <0.0001 | 3.3 | <0.0001 |
| TC | 2.2 | <0.0001 | 2.0 | <0.0001 |
| CC | - | ref | - | ref |
| Lipoprotein Lipase | ||||
| GG/CG | 2.8 | <0.0001 | 2.8 | <0.0001 |
| CC | - | ref | - | ref |
Values are mean differences between genotypes with 1 or 2 copies of the variant allele versus the wildtype over 4 ARIC visits.
Model 1 is adjusted for visit, age, and a combined center/race variable.
Model 2 is model 1 plus the following time-dependent covariates: body mass index, smoking, use of cholesterol lowering medications, use of hormone replacement therapy, and menopausal status.
The models testing the interaction between genotype and menopause are found in Table 4. Menopausal status did not modify the association of apoE genotype on LDL-c levels (interaction p=0.47). HDL-c levels among genotypes for the LIPC and LPL genes were comparable between the pre-/perimenopausal group and the postmenopausal group (both interactions p>0.10). The only statistically significant gene by menopause interaction was with the CETP gene on HDL-c concentrations (p=0.04). Postmenopausal women who were carriers of the A allele of the CETP gene had approximately 0.7 mg/dL lower mean HDL-c levels than pre-/perimenopausal counterparts, whereas the opposite pattern of HDL-c (0.4 mg/dL higher HDL-c postmenopausally) was observed for the GG genotype.
Discussion
In this longitudinal analysis within the ARIC study, the associations of genotype alone with LDL-c and HDL-c were similar to associations reported previously.(10,13,15,17,18) The only statistically significant interaction was between the CETP gene and menopause on HDL-c levels, and it was of modest magnitude. The significant CETP gene by menopause interaction was possibly due to chance because of multiple testing.
As observed here, the ε2 allele is universally associated with lower LDL-c levels and the ε4 allele associated with higher LDL-c levels than the ε3 allele of apoE. The Framingham Offspring Study reported a significantly greater association of apoE with LDL-c among postmenopausal women compared to premenopausal women. The effect of the ε2 allele was to lower LDL-c by 8.2 mg/dL and 20.4 mg/dL, and the ε4 allele was associated with increases of 1.6 mg/dL and 7.1 mg/dL LDL-c among premenopausal and postmenopausal women, respectively.(7) The effect of apoE allele on LDL-c in our data was within the range reported in the Framingham Offspring Study, but we did not observe a statistically significant difference by menopausal status.
Women with one or more A alleles of the CETP gene had greater HDL-c levels than those who were GG. A meta-analysis of over 50 studies compared mean HDL-c levels in over 10,000 individuals homozygous for either the A (B2) or G (B1) allele of the TaqIB polymorphisms of CETP.(28) B2B2 individuals had significantly higher HDL-c (weighted mean difference of 4.64 mg/dL) than those with the B1B1 genotype. This meta-analysis included both male and female participants, but did not provide separate analysis among women based on menopausal status. Our CETP and HDL-c results showed a similar trend to the meta-analysis, but we also found modest evidence that menopausal status influences HDL-c concentrations differently by CETP genotype.
Women in our study with the T allele for the C-514T polymorphism of LIPC exhibited higher mean HDL-c levels than the CC group. Hepatic lipase activity has been shown to be inversely related to estradiol levels.(20) Additonally, HDL-c levels are positively associated with estradiol levels.(19) Mean HDL-c concentrations are significantly higher among carriers of the T allele and consistently highest among women with the TT genotype.(15) Our results for LIPC agree with previous literature with respect to mean HDL-c level differences by genotype. However, we did not find an interaction between LIPC genotype and menopausal status.
The Ser447Stop polymorphism of the LPL gene was associated with higher HDL-c levels among the GG/CG group compared to the CC group in our study. Our results are consistent with two meta-analyses that compared HDL-c among whites who carry the 447X (G) allele to those who are non-carriers of the 447X allele. Both concluded that individuals heterozygous and homozygous for the 447X allele had significantly higher HDL-c concentrations than those who were homozygous for the wildtype allele.(29,30) Out of almost 40 studies included in the meta-analyses, no study focused primarily on HDL-c levels among women; in fact, only half of the studies included women participants. Our study has shown the association of HDL-c with LPL is not modified by menopausal status.
Limitations of this study include some degree of misclassification of menopausal groups due to self-reported menopausal status, rather than a biological measure. However, self-report of menstrual patterns are likely to be more accurately reported than detailed information on cycle length and variability. Also, combining the pre- and perimenopausal women into the same comparison group may have lead to attenuated associations, although the age range of the younger ARIC women was consistent with the perimenopausal transition. Further investigation of the associations between menopausal transition and genes affecting lipoprotein concentrations may benefit from analyzing the perimenopausal women separately from premenopausal women in a younger cohort of women. Additionally, this study included only Caucasian and African-American women. Therefore, the results are not generalizable to women of other race groups.
In conclusion, we rejected our hypothesis that the association between LDL-c levels and the apoE gene, and HDL-c levels and the LIPC and LPL genes are modified by menopausal status. We did find that the association of the CETP gene with HDL-c levels was modestly modified by menopausal status, although this may have been due to chance because of multiple testing. Overall, our data suggest that the decrease in endogenous estrogen as a result of menopause may independently affect lipoprotein concentration, but does not alter the effect on plasma lipids of some common genetic polymorphisms that regulate lipoprotein metabolism.
Acknowledgments
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022. The authors thank the staff and participants of the ARIC study for their important contributions.
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Rosenson RS. Low HDL-C: a secondary target of dyslipidemia therapy. Am J Med. 2005 Oct;118(10):1067–1077. doi: 10.1016/j.amjmed.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Jones PH. Low-density lipoprotein cholesterol reduction and cardiovascular disease prevention: the search for superior treatment. Am J Med. 2004 Mar 22;116 6A:17S–25S. doi: 10.1016/j.amjmed.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Lusis AJ. Genetic factors affecting blood lipoproteins: the candidate gene approach. J Lipid Res. 1988 Apr;29(4):397–429. [PubMed] [Google Scholar]
- 4.Masson LF, McNeill G. The effect of genetic variation on the lipid response to dietary change: recent findings. Curr Opin Lipidol. 2005 Feb;16(1):61–67. doi: 10.1097/00041433-200502000-00011. [DOI] [PubMed] [Google Scholar]
- 5.Middelberg RP, Spector TD, Swaminathan R, Snieder H. Genetic and environmental influences on lipids, lipoproteins, and apolipoproteins: effects of menopause. Arterioscler Thromb Vasc Biol. 2002 Jul 1;22(7):1142–1147. doi: 10.1161/01.atv.0000022889.85440.79. [DOI] [PubMed] [Google Scholar]
- 6.Lewis-Barned NJ, Sutherland WH, Walker RJ, Walker HL, De Jong SA, Edwards EA, et al. Plasma cholesterol esterification and transfer, the menopause, and hormone replacement therapy in women. J Clin Endocrinol Metab. 1999 Oct;84(10):3534–3538. doi: 10.1210/jcem.84.10.6022. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer EJ, Lamon-Fava S, Cohn SD, Schaefer MM, Ordovas JM, Castelli WP, et al. Effects of age, gender, and menopausal status on plasma low density lipoprotein cholesterol and apolipoprotein B levels in the Framingham Offspring Study. J Lipid Res. 1994 May;35(5):779–792. [PubMed] [Google Scholar]
- 8.Guthrie JR, Taffe JR, Lehert P, Burger HG, Dennerstein L. Association between hormonal changes at menopause and the risk of a coronary event: a longitudinal study. Menopause. 2004 May-Jun;11(3):315–322. doi: 10.1097/01.gme.0000094208.15096.62. [DOI] [PubMed] [Google Scholar]
- 9.Davignon J, Cohn JS, Mabile L, Bernier L. Apolipoprotein E and atherosclerosis: insight from animal and human studies. Clinica Chimica Acta. 1999;286(12):115–143. doi: 10.1016/s0009-8981(99)00097-2. [DOI] [PubMed] [Google Scholar]
- 10.Foley SM. Update on risk factors for atherosclerosis: the role of inflammation and apolipoprotein E. Medsurg Nurs. 2005 Feb;14(1):43–50. [PubMed] [Google Scholar]
- 11.Schaefer EJ, Lamon-Fava S, Johnson S, Ordovas JM, Schaefer MM, Castelli WP, et al. Effects of gender and menopausal status on the association of apolipoprotein E phenotype with plasma lipoprotein levels. Results from the Framingham Offspring Study. Arterioscler Thromb. 1994 Jul;14(7):1105–1113. doi: 10.1161/01.atv.14.7.1105. [DOI] [PubMed] [Google Scholar]
- 12.Tall A. Plasma cholesteryl ester transfer protein. J Lipid Res. 1993 August 1;34(8):1255–1274. [PubMed] [Google Scholar]
- 13.Bernard S, Moulin P, Lagrost L, Picard S, Elchebly M, Ponsin G, et al. Association between plasma HDL-cholesterol concentration and Taq1B CETP gene polymorphism in non-insulin-dependent diabetes mellitus. J Lipid Res. 1998 Jan;39(1):59–65. [PubMed] [Google Scholar]
- 14.Perret B, Mabile L, Martinez L, Terce F, Barbaras R, Collet X. Hepatic lipase: structure/function relationship, synthesis, and regulation. J Lipid Res. 2002 Aug;43(8):1163–1169. [PubMed] [Google Scholar]
- 15.Yamakawa-Kobayashi K, Somekawa Y, Fujimura M, Tomura S, Arinami T, Hamaguchi H. Relation of the -514C/T polymorphism in the hepatic lipase gene to serum HDL and LDL cholesterol levels in postmenopausal women under hormone replacement therapy. Atherosclerosis. 2002 May;162(1):17–21. doi: 10.1016/s0021-9150(01)00675-x. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg IJ. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J Lipid Res. 1996 Apr;37(4):693–707. [PubMed] [Google Scholar]
- 17.Lopez-Miranda J, Cruz G, Gomez P, Marin C, Paz E, Perez-Martinez P, et al. The influence of lipoprotein lipase gene variation on postprandial lipoprotein metabolism. J Clin Endocrinol Metab. 2004 Sep;89(9):4721–4728. doi: 10.1210/jc.2003-031642. [DOI] [PubMed] [Google Scholar]
- 18.Brousseau ME, Goldkamp AL, Collins D, Demissie S, Connolly AC, Cupples LA, et al. Polymorphisms in the gene encoding lipoprotein lipase in men with low HDL-C and coronary heart disease: the Veterans Affairs HDL Intervention Trial. J Lipid Res. 2004 Oct;45(10):1885–1891. doi: 10.1194/jlr.M400152-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Sultan N, Nawaz M, Sultan A, Fayaz M, Baseer A. Effect of menopause on serum HDL-cholesterol level. Journal of Ayub Medical College, Abbottabad: JAMC. 2003 Jul-Sep;15(3):24–26. [PubMed] [Google Scholar]
- 20.Brinton EA. Oral Estrogen Replacement Therapy in Postmenopausal Women Selectively Raises Levels and Production Rates of Lipoprotein A-I and Lowers Hepatic Lipase Activity Without Lowering the Fractional Catabolic Rate. Arterioscler Thromb Vasc Biol. 1996 March 1;16(3):431–440. doi: 10.1161/01.atv.16.3.431. [DOI] [PubMed] [Google Scholar]
- 21.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989 Apr;129(4):687–702. [PubMed] [Google Scholar]
- 22.Papp AC, Hatzakis H, Bracey A, Wu KK. ARIC hemostasis study--I. Development of a blood collection and processing system suitable for multicenter hemostatic studies. Thrombosis & Haemostasis. 1989 Feb 28;61(1):15–19. [PubMed] [Google Scholar]
- 23.Siedel J, Hagele E, Ziegenhorn J, Wahlefeld A. Reagent for the enzymatic determination of serum total cholesterol with improved lipolytic efficiency. Clin Chem. 1983 June 1;29(6):1075–1080. [PubMed] [Google Scholar]
- 24.Nagele U, Hagele EO, Sauer G, Wiedemann E, Lehmann P, Wahlefeld AW, et al. Reagent for the enzymatic determination of serum total triglycerides with improved lipolytic efficiency. J Clin Chem Clin Biochem. 1984 Feb;22(2):165–174. doi: 10.1515/cclm.1984.22.2.165. [DOI] [PubMed] [Google Scholar]
- 25.Warnick GR, Benderson J, Albers JJ. Dextran sulfate-Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin Chem. 1982 Jun;28(6):1379–1388. [PubMed] [Google Scholar]
- 26.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Without Use of the Preparative Ultracentrifuge. Clin Chem. 1972 June 1;18(6):499–502. [PubMed] [Google Scholar]
- 27.Chambless LE, McMahon RP, Brown SA, Patsch W, Heiss G, Shen YL. Short-term intraindividual variability in lipoprotein measurements: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Epidemiol. 1992 Nov 1;136(9):1069–1081. doi: 10.1093/oxfordjournals.aje.a116572. [DOI] [PubMed] [Google Scholar]
- 28.Boekholdt SM, Thompson JF. Natural genetic variation as a tool in understanding the role of CETP in lipid levels and disease. J Lipid Res. 2003 July 1;44(6):1080–1093. doi: 10.1194/jlr.R200018-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Rip J, Nierman MC, Ross CJ, Jukema JW, Hayden MR, Kastelein JJP, et al. Lipoprotein Lipase S447X: A Naturally Occurring Gain-of-Function Mutation. Arterioscler Thromb Vasc Biol. 2006 June 1;26(6):1236–1245. doi: 10.1161/01.ATV.0000219283.10832.43. [DOI] [PubMed] [Google Scholar]
- 30.Wittrup HH, Tybjaerg-Hansen A, Nordestgaard BG. Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease. A meta-analysis. Circulation. 1999 Jun 8;99(22):2901–2907. doi: 10.1161/01.cir.99.22.2901. [DOI] [PubMed] [Google Scholar]