Abstract
Bacterial chondroitin polymerase K4CP is a multifunctional enzyme with two active sites. K4CP catalyzes alternative transfers of glucoronic acid (GlcA) and N-acetylgalactosamine (GalNAc) to elongate a chain consisting of the repeated disaccharide sequence GlcAβ1–3GalNAcβ1–4. Unlike the polymerization reactions of DNA and RNA and polypeptide synthesis, which depend upon templates, the monosaccharide polymerization by K4CP does not. To investigate the catalytic mechanism of this reaction, we have used isothermal titration calorimetry to determine the binding of the donor substrates UDP-GlcA and UDP-GalNAc to purified K4CP protein and its mutants. Only one donor molecule bound to one molecule of K4CP at a time. UDP-GlcA bound only to the C-terminal active site at a high affinity (Kd = 6.81 μm), thus initiating the polymerization reaction. UDP-GalNAc could bind to either the N-terminal or C-terminal active sites at a low affinity (Kd = 266–283 μm) but not to both sites at the same time. The binding affinity of UDP-GalNAc to a K4CP N-terminal fragment (residues 58–357) was profoundly decreased, yielding the average Kd value of 23.77 μm, closer to the previously reported Km value for the UDP-GalNAc transfer reaction that takes place at the N-terminal active site. Thus, the first step of the reaction appears to be the binding of UDP-GlcA to the C-terminal active site, whereas the second step involves the C-terminal region of the K4CP molecule regulating the binding of UDP-GalNAc to only the N-terminal active site. Alternation of these two specific bindings advances the polymerization reaction by K4CP.
Chondroitin sulfate is a glucosaminoglycan that is a polysaccharide chain composed of the sugar molecules glucuronic acid (GlcA)2 and N-acetylgalactosamine (GalNAc). A disaccharide unit (GlcAβ1–3GalNAcβ1–4) continually repeats to form a nonbranching polysaccharide chain that can consist of over a hundred individual monosaccharides, each of which can be sulfated in various positions and quantities (1). Chondroitin chains are usually found attached to proteins, forming proteoglycans, and are present in the extracellular matrix and cell surfaces of various human tissues including the cartilage, aorta, skeletal muscle, eye, lung, and brain tissues (2–4). Through interaction with various extracellular proteins, chondroitin plays a critical role in the regulation of a variety of cellular activities including growth, development, and response to injury in the nervous system (5, 6). For example, the loss of chondroitin sulfate from the cartilage is a major cause of osteoarthritis and intervertebral disc and cartilage degeneration (7, 8). Multifunctional polymerases responsible for the synthesis of chondroitin chains have been cloned and characterized for their transfer reactions (9–12). However, the molecular mechanism of the polymerization reaction catalyzed by these enzymes remains virtually unknown.
Bacterial cells also synthesize a chondroitin chain. Although mammalian chondroitin polymerases are Golgi membrane-bound enzymes, those of bacterial polymerases are soluble enzymes. For instance, the K4 strain of Escherichia coli contains an enzyme, encoded by the KfoC gene of the K4 gene cluster, that catalyzes the synthesis of the chondroitin chain, named K4 chondroitin polymerase or K4CP (9). The reduced amino acid sequence of K4CP revealed two conserved UDP-sugar binding motifs (DXD), one located in the N-terminal region and the other located in the C-terminal region of the molecule. Comparative sequence analysis shows that the system of two active sites is conserved in the other bacterial and mammalian chondroitin polymerases as well as the other polysaccharide polymerases such as the heparan polymerase Exostosins (10, 13–16). We have previously employed isothermal titration calorimetry (ITC) to investigate the specific binding of donor substrate to UDP-N-acetylhexosaminyltransferase Exostosin Like-2 that possesses only one active site (17). Using the water solubility of the bacterial K4CP enzyme to our advantage, we have now extended our study to include the ITC measurement of binding specificity of donor substrates to the two active sites of K4CP to explicate the catalytic mechanism of how this enzyme executes the polymerization process.
We bacterially expressed a truncated enzyme of K4CP (from residues 55 to 686) that retained all of the original polymerase activity, from which the DXD motifs were mutated to generate various mutant enzymes. In addition, a deletion mutant lacking the C-terminal half of the K4CP was constructed. Wild type K4CP and these mutants were expressed in bacterial cells, purified, and subjected to ITC analysis to determine the nature of binding for the donor substrates UDP-GlcA, UDP-GlcNAc, and UDP-GalNAc. UDP binding was also investigated. Here we now present key elements of the specific donor bindings that are consistent with the catalytic mechanism of the alternative transfer reactions by K4CP.
EXPERIMENTAL PROCEDURES
Expression and Purification of Proteins—E. coli BL21 (DE3) cells transformed with pGEX K4CP were grown and harvested as previously reported (17). The cells were then disrupted in a French Press at room temperature under pressures varying between 8,000 and 24,000 p.s.i. Following ultracentrifugation, the K4CP protein was then purified from the supernatants by absorption onto glutathione-Sepharose resin (GE Healthcare) and eluted via thrombin cleavage overnight at 4 °C. The eluted protein was then dialyzed overnight into 25 mm HEPES (pH 7.5), 100 mm NaCl, 20 mm MnCl2, and 1 mm CaCl2. Purification of protein was confirmed by taking 10 μl of eluted protein and combining it with 4 μl of NuPage 4× LDS sample buffer (Invitrogen), 2 μl of 2-mercaptoethanol (Sigma-Aldrich) and then running the sample on a NuPage 4–12% Bis-Tris gel (Invitrogen). The gel was then stained with Coomassie Brilliant Blue G-250 (Fluka) and then destained to make the protein bands more clearly visible.
Isothermal Titration Calorimetry—Isothermal titration calorimetry measurements were carried out in HEPES buffer using a VP-ITC MicroCalorimeter (Micro Cal, Inc.) at 20 °C. Substrate solution was injected into a reaction cell containing the protein. For substrate solutions containing UDP-GlcA, thirty injections of 3 μl at 180-s intervals were performed. For substrate solutions containing UDP, UDP-GlcNAc, or UDP-GalNAc, 30 injections of 3 μl at 300-s intervals were performed. Data acquisition and analysis were performed by the MicroCal Origin software package. Data analysis was performed by generating a binding isotherm and best fit using the following fitting parameters: n (number of sites), ΔH (cal/mol), ΔS (cal/mol/deg), and K (binding constant in m–1), and the standard Levenberg-Marquardt methods (18). Following data analysis, K (m–1) is then converted to Kd (μm).
Site-directed Mutagenesis—Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) following the protocols described in the accompanying instruction manual. The pairs of primers used to mutate the DCD-binding motif found in the N terminus region to ACA, DCE, and DCK were: TGGGTTCGGAGCCATAGCACAAGCCAGAATTGCAACATA and TATGTTGCAATTCTGGCTTGTGCTATGGCTCCGAACCCA; TGGGTTCGGAGCCATTTCACAATCCAGAATTGC and GCAATTCTGGATTGTGAAATGGCTCCGAACCCA; and TGGGTTCGGAGCCATTTTACAATCCAGAATTGC and GCAATTCTGGATTGTAAAATGGCTCCGAACCCA, respectively. The pairs of primers used to mutate the DSD-binding motif found in the C terminus region to ASA, DSE, DSK, DSA, ASD, and KSD were: TGGTTCAAGAAAGTCAGCAGAGGCTAACTGACCTATATA and TATATAGGTCAGTTAGCCTCTGCTGACTTTCTTGAACCA; TTCAAGAAAGTCTTCAGAGTCTAACTGACCTAT and ATAGGTCAGTTAGACTCTGAAGACTTTCTTGAA; TTCAAGAAAGTCTTTAGAGTCTAACTGACCTAT and ATAGGTCAGTTAGACTCTAAAGACTTTCTTGAA; TTCAAGAAAGTCAGCAGAGTCTAACTGACCTAT and ATAGGTCAGTTAGACTCTGCTGACTTTCTTGAA; TTCAAGAAAGTCATCAGAGGCTAACTGACCTAT and ATAGGTCAGTTAGCCTCTGATGACTTTCTTGAA; and TTCAAGAAAGTCATCAGATTTTAACTGACCTAT and ATAGGTCAGTTAAAATCTGATGACTTTCTTGAA, respectively. The primers used to generate the ACA and ASA mutants were also used to generate the double mutant, thereby mutating the DXD motifs found in both regions to AXA. The primers used to generate the ACA and DSA mutants were used to create the double mutant in which the N terminus-binding site was mutated to ACA and the C terminus-binding site was converted to DSA. The primers used to delete the C-terminal region of the enzyme were: CGCGGATCCAAAGCTGTTATTGATATTGAT and CGGAATTCTTAATGCGTAAACTCTTCATCAAA. The mutations were confirmed by sequencing with the Big Dye terminator cycle sequencing reaction kit (Applied Biosystems).
Enzyme Assays—Hydrolysis was determined as follows. 53 μg of protein were tested in an assay mixture containing 25 mm HEPES (pH 7.5), 100 mm NaCl, 20 mm MnCl2, 1 mm CaCl2, and 1 μl of either UDP-[3H]GlcNAc (36.0 Ci/mmol; PerkinElmer Life Sciences), UDP-[3H]GlcA (20.0 Ci/mmol; American Radiolabeled Chemicals, Inc.), or UDP-[3H]GalNAc (20.0 Ci/mmol; American Radiolabeled Chemicals, Inc.). Assay mixtures containing radioactive plus nonradioactive substrates at final concentrations of 1.6, 3.2, 6.4, 9.6, 16.0, 32.0, 64.0, 96.0, 128.0, and 200.0 μm were incubated for 1 h at 30 °C. The incubation mixtures were boiled for 2 min, and the precipitated protein was removed by centrifugation at 20,400 × g for 5 min. The resulting supernatants were subjected to a Hi-Load 16/60 Superdex 30pg column (GE Healthcare) equilibrated with 250 mm NH4HCO3 containing 7% 1-propanol using theÄKTA Purifier (GE Healthcare). Fractions were collected at a rate of 1 ml/min, and the amount of the hydrolyzed product UDP was quantified with a scintillation counter. Data analysis was performed using the Origin version 7.0 software programmed to fit data to an s/v-s plot to calculate the Km and Vmax values using Michealis-Menten kinetics.
RESULTS
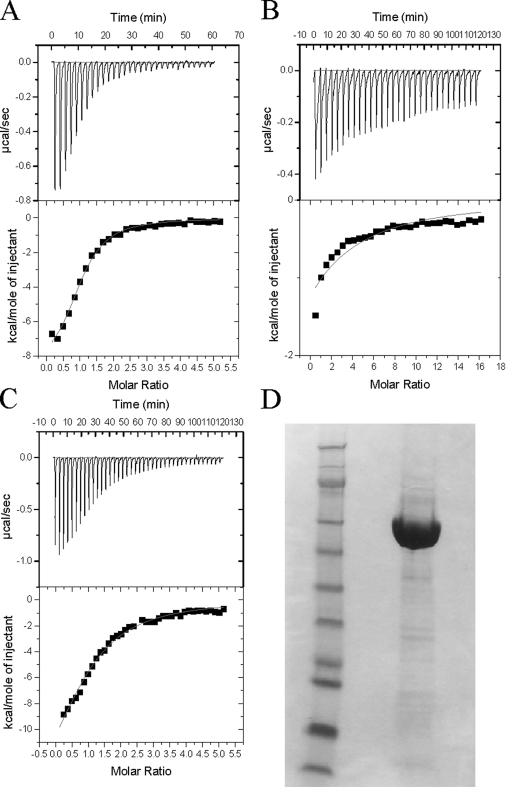
Binding of UDP-GlcA—ITC was performed to examine the binding of UDP-GlcA to the truncated K4CP (hereafter referred to as K4CP). The calorimetric profile showed that UDP-GlcA bound to K4CP at a ratio of one donor molecule to one enzyme molecule (Fig. 1). The binding of UDP-GlcA was tested at various temperatures ranging from 5 to 40 °C to ascertain the nature of donor binding (Table 1), from which the true values for the changes in entropy (ΔS) and enthalpy (ΔH) were directly determined by plotting Log K versus 1000/K°: ΔS = –25.18 ± 1.43 cal/mol/deg and ΔH =–14,000.45 ± 419.29 cal/mol (19). Using these values, it was found that the TΔS was lower in value than the ΔH value, suggesting that this binding is an enthalpy-driven reaction. In addition, the Gibbs free energy (ΔG) values remained constant over the various temperatures, implying that the nature of this binding reaction is spontaneous and that the maximum potential of K4CP to perform the binding remains the same over a range of temperatures. As expected, the Kd values varied from 3.26 to 55.09 μm depending upon the temperatures used for the assays, whereas the number of the UDP-GlcA molecules that bound to the K4CP molecule was the same at all temperatures (Table 1).
FIGURE 1.
A, calorimetric profile of UDP-GlcA binding to K4CP. The reaction cell contained a solution of K4CP (1. 4 ml and 124.8 μm) in 25 mm HEPES buffer (pH 7.5) containing 20 mm MnCl2, 0.1 m NaCl, and 1 mm CaCl2. The syringe contained 1 mm UDP-GlcA in the same buffer. Top panel, raw calorimetric data obtained from the injection of 3 μl aliquots of UDP-GlcA at 3-min intervals. The lower panel shows the integrated binding isotherm with the experimental points (▪) and best fit. B, calorimetric profile of UDP-GalNAc binding to K4CP. C, calorimetric profile of UDP-GlcA binding to the K4CP mutant construct 239ACA241. D, gel showing the purity of K4CP used in ITC experiments and enzyme assays. The first lane contains the SeeBlue Plus 2 prestained molecular marker (Invitrogen), and the second lane contains purified K4CP.
TABLE 1.
Thermodynamic parameters, ΔG, ΔH, ΔS, number of binding sites (n), and the binding constant, Kd for UDP-GlcA binding with wild type K4CP at five different temperatures
The substrate and K4CP concentrations were 0.8 mm and 42 μm, respectively.
| Temperature | ΔG | ΔH | ΔS | Kd | n |
|---|---|---|---|---|---|
| cal/mol | cal/mol | cal/mol/deg | μm | ||
| 5 °C | -6983.4 ± 1064 | -10,350 ± 1064 | -12.11 | 3.26 ± 1.78 | 1.05 ± 0.08 |
| 10 °C | -6815.3 ± 2557 | -12,280 ± 2557 | -19.31 | 5.55 ± 2.13 | 0.85 ± 0.14 |
| 20 °C | -6652.3 ± 1973 | -11,100 ± 1973 | -15.18 | 11.04 ± 3.33 | 0.82 ± 0.11 |
| 30 °C | -6331.4 ± 703.8 | -14,370 ± 703.8 | -26.53 | 27.32 ± 10.94 | 0.93 ± 0.25 |
| 40 °C | -6107.4 ± 4077 | -18,690 ± 4077 | -40.20 | 55.09 ± 8.24 | 0.97 ± 0.19 |
Binding of UDP-GalNAc and UDP—ITC was performed at 20 °C for determining the binding of UDP-GalNAc and UDP. Both UDP-GalNAc and UDP bound to K4CP at a one to one molecular ratio (Table 2). UDP-GalNAc was characterized by its extremely high Kd value, 358.17 μm at 20 °C. This value was more than 40-fold higher when compared with the Kd value of 8.5 μm at 20 °C yielded by UDP-GlcA binding and the averaged Kd value (6. 81 ± 1.72 μm) for UDP-GlcA, a number arrived at by calculating from five different binding assays. The Km values of UDP-GlcA and UDP-GalNAc transfer reactions were previously reported (9). Intriguingly, the Kd value of 358.17 μm was over 10-fold higher than the reported Km value (31.6 μm) of the UDP-GalNAc transfer reaction catalyzed by K4CP, whereas the corresponding Kd and Km values for UDP-GlcA were similar. The inactive donor substrate UDP bound to K4CP at a Kd value of 89.53 μm, which was higher than the Kd value of UDP-GlcA binding but was lower than that of UDP-GalNAc binding. Only one UDP molecule was found to bind to the K4CP molecule, which was unexpected given the fact that there are two binding sites for donor substrates. These experimental observations are at first glance puzzling but may provide the insightful clues necessary to understand the catalytic mechanism of the polymerization reaction; both UDP and UDP-GalNAc bind to K4CP at a one to one molecular ratio, and the Kd value of UDP-GalNAc was higher than the reported Km value.
TABLE 2.
Thermodynamic parameters, ΔG, ΔH, ΔS, number of binding sites (n), and the binding constant, Kd for donor binding with wild type K4CP in solution at 20 °C
UDP-GlcA and UDP-GalNAc concentrations were 1 mm; UDP and UDP-GalNAc concentrations were 4 mm. K4CP concentration was 124.8 μm. All of the experiments were conducted in triplicate to confirm the results obtained. Fresh enzyme was prepared for each experiment.
| Substrate | ΔG | ΔH | ΔS | Kd | n |
|---|---|---|---|---|---|
| cal/mol | cal/mol | cal/mol/deg | μm | ||
| UDP-GlcA | -7209.1 ± 1124 | -11,270 ± 1124 | -13.86 | 8.05 ± 1.52 | 0.95 ± 0.07 |
| UDP-GalNAc | -4779.2 ± 483.9 | -11,630 ± 483.9 | -22.61 | 358.17 ± 26.64 | 1.25 ± 0.32 |
| UDP | -5431.7 ± 1953 | -8643 ± 1953 | -10.96 | 89.53 ± 22.77 | 0.91 ± 0.09 |
Delineating the Specific Donor-binding Site—Site-directed mutagenesis was employed to mutate the aspartic acid residues of the K4CP DXD motifs: either the N-terminal 239DCD241 or the C-terminal 519DSD521 to 239ACA241 or 519ASA521. In addition, both 239DCD241 and 519DSD521 were simultaneously mutated to 239ACA241 and 519ASA521, hereafter just referred to as DXDdm. These mutants were subjected to ITC analysis (Table 3). UDP-GlcA bound to the 239DCD241 mutant but not to the 519DSD521 mutant, and moreover, the Kd value of this binding was close to that of the UDP-GlcA binding to K4CP. These results unequivocally identified the C-terminal DSD as the sole binding site of UDP-GlcA. UDP-GalNAc was found to bind to the both 239ACA241 and 519ASA521 mutants at similar affinities; their Kd values were within the range of the Kd value (358.18 μm) of UDP-GalNAc binding to K4CP. Thus, UDP-GalNAc is capable of binding to either the N- or C-terminal DXD motif. This tantalizing observation lead us to measure UDP-GalNAc transfer activity by the DXD mutants: K4CP and the 519DSD521 mutant catalyzed the reaction at 5.895 and 5.469 pmol/min/μg protein, respectively, but the 239ACA241 exhibited no transfer activity (Table 4). Because UDP-GalNAc was found to bind to K4CP at a one to one molecular ratio, the N-terminal 239DCD241 should be the binding site of UDP-GalNAc during the transfer reaction. However, the present binding assays using ITC could not determine the binding site of UDP-GalNAc. In a fashion similar to UDP-GalNAc, UDP bound to both the 239ACA241 and 519ASA521 mutants. Because the Kd value for the former mutant was closer to that for K4CP (89.53 μm) than that for the latter mutant, this one molecule of UDP appeared to bind to the C-terminal 519DSD521 of K4CP. As expected, when both 75DCD77 and 519DSD521 were mutated, the DXDdm possessed no capability to bind any of the donor substrates.
TABLE 3.
Comparison of the binding constants Kd of UDP, UDP-GalNAc, UDP-GlcNAc, and UDP-GlcA for wild type K4CP to the mutant constructs 239ACA241, 519ASA521, and DXDdm
In 239ACA241 the DXD-binding motif of the N-terminal region was eliminated by mutating the existing DCD amino acid sequence to ACA. In 519ASA521 the DXD-binding motif of the C-terminal region was eliminated by converting the existing DSD amino acid sequence to ASA. In DXDdm, both DXD-binding motifs were eliminated by mutating the N-terminal binding site to ACA and the C-terminal binding site to ASA. All of the ITC experiments were performed at 20 °C and in triplicate to confirm results using a fresh batch of enzyme produced for each experiment. ND, no detectable binding.
| Enzyme | UDP-GlcA | UDP-GalNAc | UDP |
|---|---|---|---|
| Wild | |||
| Kd | 8.05 ± 1.52 | 358.17 ± 26.64 | 89.53 ± 22.77 |
| n | 0.95 ± 0.07 | 1.25 ± 0.32 | 0.91 ± 0.09 |
| 239ACA241 | |||
| Kd | 14.12 ± 1.50 | 266.74 ± 57.58 | 54.23 ± 11.18 |
| n | 0.99 ± 0.08 | 0.94 ± 0.11 | 0.72 ± 0.34 |
| 519ASA521 | |||
| Kd | ND | 283.69 ± 30.76 | 16.70 ± 2.04 |
| n | ND | 1.15 ± 0.09 | 0.99 ± 0.08 |
| DXDdm | |||
| Kd | ND | ND | ND |
| n | ND | ND | ND |
TABLE 4.
Hydrolysis and transfer of donor substrates by wild type K4CP, and the mutant constructs 239ACA241 and 519ASA521
The hydrolysis of Km (μm) of UDP-GalNAc and UDP-GlcA and the transfer activity (pmol/min/μg of protein) of UDP-GalNAc and UDP-GlcNAc by wild type, 239ACA241 and 519ASA521 are shown. ND, no detectable transfer. Assay mixtures containing radioactive plus nonradioactive substrates at final concentrations of 1.6, 3.2, 6.4, 9.6, 16.0, 32.0, 64.0, 96.0, 128.0, and 200.0 μm were incubated with 53 μ g of protein for 1 h at 30 °C.
| Substrate | Wild | 239ACA241 | 519ASA521 | |
|---|---|---|---|---|
| Hydrolysis | UDP-GlcA | ND | ND | ND |
| UDP-GalNAc | 65.09 | 135.31 | 24.27 | |
| Transfer activity | UDP-GalNAc | 5.895 | ND | 5.469 |
| UDP-GlcNAc | 0.394 | ND | 0.534 |
To further elucidate the binding of UDP-GalNAc, we attempted to construct deletion mutants by separating the K4CP molecule into N- and C-terminal fragments (residues 58–357 and residues 358–686, respectively). Although the C-terminal fragment did not express in our bacterial system, we were able to express and purify the N-terminal fragment for ITC analysis (Table 5). Consistent with our previous data, the N-terminal fragment abrogated the binding of UDP-GlcA. Conversely, both UDP-GalNAc and UDP retained their binding to the fragment. Most interestingly, the Kd value of UDP-GalNAc binding became similar to the reported Km value of UDP-GalNAc transfer activity by K4CP. To reutilize the correlation of this Kd value with the catalytic activity of K4CP, we measured hydrolysis of K4CP and the DXD mutants using UDP-GlcA and UDP-GalNAc as substrates (Table 4). No hydrolysis activity was observed with UDP-GlcA with any of these enzymes. On the other hand, UDP-GalNAc was hydrolyzed by all enzymes. The Km value of the hydrolysis by the 519ASA521 mutant was nearly identical to the Kd value of UDP-GalNAc binding to the N-terminal fragment, and moreover, the corresponding value for hydrolysis by K4CP was closer to this Kd value. These results are consistent with the conclusion that the N-terminal 239DCD241 is the binding site of UDP-GalNAc. Furthermore, the relatively high affinity of UDP-GalNAc binding provides critical information for us to hypothesize about the molecular mechanism by which K4CP regulates the binding of UDP-GalNAc to N-terminal 239DCD241.
TABLE 5.
Thermodynamic parameters, ΔG, ΔH, ΔS, number of binding sites (n), and the binding constant Kd for donor binding with the K4CP mutant construct KctDel
The experiments were performed in solution at 20 °C and in triplicate to confirm the results using a fresh batch of enzyme produced for each experiment. In this mutant the entire C-terminal region is deleted, isolating the N-terminal region for analysis. The UDP, UDP-GlcA, UDP-GlcNAc, and UDP-GalNAc concentrations were 2 mm. KctDel concentration was 31 μm. ND, no detectable binding.
| Substrate | ΔG | ΔH | ΔS | Kd | n |
|---|---|---|---|---|---|
| cal/mol | cal/mol | cal/mol/deg | μm | ||
| UDP-GlcA | ND | ND | ND | ND | ND |
| UDP-GalNAc | -6184.3 ± 5325 | -7463 ± 5325 | -4.22 | 34.72 ± 9.14 | 0.82 ± 0.20 |
| UDP | -6361.4 ± 1721 | -9249 ± 1721 | -9.53 | 26.00 ± 6.36 | 0.96 ± 0.16 |
Defining the Functions of the C-terminal DSD Motif—Recent x-ray crystal structures of the glycosyltransferases GlcAT1 and EXTL2 have demonstrably shown that the first aspartic acid residue in the DXD motif interacts with the sugar moiety of the donor substrate and that the second aspartic acid residue interacts with the UDP portion of the donor substrate (20, 21). Using this information as a guide, the first aspartic acid of the DXD motif was substituted with a negatively charged glutamic acid and a positively charged lysine to produce the following mutants: 239DCE241, 239DCK241, 519DSE521, and 519DSK521. These mutants were subjected to ITC analysis to decipher the role of aspartic acid in the regulation of donor substrate binding (Table 6). UDP-GlcA was able to bind to all four mutants at a one to one molecular ratio. Thus, the GlcA moiety solely determined the binding of UDP-GlcA. Moreover, the results indicated that the mutations of the N-terminal 239DCD241 had no effect on the binding of UDP-GlcA to the C-terminal 519DSD521. UDP-GalNAc bound to the 239DCE241, 239DCK241, and 519DSE521 mutants (Table 6), which is consistent with the facts that the second aspartic acid does not directly interact with the sugar moiety and that UDP-GalNAc can bind to either the N-terminal or C-terminal DXD motif. A surprising finding was that the 519DSK521 mutant abrogated the bindings of UDP-GalNAc as well as that of UDP. Because the second aspartic acid is the primary residue responsible for the ability of the enzyme to interact with the UDP moiety, it may be reasonable to conclude that UDP does not bind to the C-terminal 519DSK521 site. UDP-GalNAc bound to the C-terminal 519DSD521 site weakly, and its sugar moiety may not be able to overcome the change to the second aspartic acid residue, thereby resulting in its inability to bind to the 519DSK521 site. The most surprising finding, however, was that no binding was detected for UDP-GalNAc and UDP despite the fact that the N-terminal 239DCD241 site remained unaltered, because both UDP-GalNAc and UDP bind to the N-terminal 239DCD241 site of the 519ASA521 mutant (Table 3).
TABLE 6.
Binding constants Kd (μm) of UDP, UDP-GalNAc, and UDP-GlcA
All of the ITC experiments were performed at 20 °C and in triplicate to confirm results using a fresh batch of enzyme produced for each experiment. ND, no detectable binding.
| Enzyme | UDP-GlcA | UDP-GalNAc | UDP |
|---|---|---|---|
| 239DCE241 | |||
| Kd | 5.68 ± 0.81 | 194.48 ± 54.74 | 106.49 ± 24.21 |
| n | 0.84 ± 0.03 | 0.81 ± 0.21 | 0.81 ± 0.19 |
| 239DCK241 | |||
| Kd | 3.57 ± 0.37 | 157.36 ± 39.91 | 45.64 ± 3.98 |
| n | 1.00 ± 0.02 | 0.96 ± 0.26 | 0.96 ± 0.05 |
| 519DSE521 | |||
| Kd | 4.42 ± 0.45 | 155.62 ± 26.67 | 75.13 ± 11.58 |
| n | 0.89 ± 0.01 | 0.81 ± 0.16 | 0.77 ± 0.24 |
| 519DSK521 | |||
| Kd | 12.09 ± 1.62 | ND | ND |
| n | 0.99 ± 0.03 | ND | ND |
| 519DSA521 | |||
| Kd | ND | 222.12 ± 70.26 | 9.56 ± 1.07 |
| n | ND | 0.75 ± 0.14 | 0.76 ± 0.02 |
| 519KSD521 | |||
| Kd | ND | ND | 35.77 ± 4.61 |
| n | ND | ND | 0.95 ± 0.22 |
| 519ASD521 | |||
| Kd | ND | ND | 23.61 ± 2.31 |
| n | ND | ND | 1.22 ± 0.09 |
| 239ACA241/519DSA521 | |||
| Kd | ND | ND | ND |
| n | ND | ND | ND |
The second alanine residue (Ala241) was responsible for binding the two donors to the N-terminal 239DCD241, because the 519DSA521 mutant retained the binding ability of UDP-GalNAc and UDP while losing its binding ability to UDP-GlcA (Table 6). These results suggested that, depending on the type of residue replacing the second aspartic acid, the C-terminal DSD motif differentially regulates the binding of UDP-GalNAc and UDP to the N-terminal DCD motif. Given this conclusion, the first aspartic acid residue of the C-terminal DSD motif was then mutated to lysine and alanine to produce the 519KSD521 and 519ASD521 mutants. This aspartic acid residue is the primary determinant for the binding of the sugar moiety of a donor substrate (20, 21). The subsequent loss of UDP-GlcA binding was consistent with the fact that the C-terminal DSD motif is the only site for which UDP-GlcA had an affinity (Table 6). It would be reasonable to observe no binding of UDP-GalNAc to these mutants, if indeed the C-terminal DSD was the only binding site in the K4CP molecule. However, the fact that UDP-GalNAc did not bind to the N-terminal DCD motif of the 519KSD521 and 519ASD521 mutants reiterated the possibility that the C-terminal DSD motif may regulate donor binding to the N-terminal DCD motif, thus becoming a primary regulator of enzyme function and activity of K4CP. It was reasonable that a continuous binding of UDP to these mutants is observed because UDP does not interact with the first aspartic acid residue, and the double mutant 239ACA241/519DSA521 did not bind any of the donor substrates.
DISCUSSION
The chondroitin polymerase K4CP is a multifunctional enzyme that transfers GlcA and GalNAc from the donor substrates UDP-GlcA and UDP-GalNAc to synthesize the (GlcAβ1–3GalNAcβ1–4)n chain. The K4CP molecule contains the two potential binding sites for donor substrates. We have used bacterially expressed and purified K4CP enzymes and have employed ITC to investigate the specific donor bindings. One insightful result is the fact that K4CP is capable of binding to only one donor molecule at a given time regardless of the type of donor substrates: UDP, UDP-GalNAc, or UDP-GlcNAc. Another significant result is the observation suggesting that the C-terminal 519DSD521 site provides K4CP with not only the sole binding site for UDP-GlcA but also the ability to regulate binding of UDP-GalNAc to the N-terminal donor-binding site 239DCD241. Thus, the chondroitin polymerase K4CP appears to utilize a fairly sophisticated mechanism to catalyze the polymerization reaction.
A critical question, the answer of which is required for understanding the catalytic mechanism of the polymerization, is whether K4CP has one active site at which both the UDP-GlcA and UDP-GalNAc transfer reactions take place or has two distinct active sites. K4CP catalyzes an inverting reaction, in which the α-configuration of the C1 bond of the donor sugar is inverted to the β-configuration in a product formed (9). X-ray crystal structures of various inverting glycosyltransferases have determined the conserved structural signatures and the unique conformation and orientation of substrates at the active site (21). The structures of mammalian glycosyltransferases are represented by the so-called GT-A fold that is a single globular protein consisting of two subdomains (21–24). The N-terminal subdomain has a Rossman fold and provides the binding site for the donor substrate, whereas the C-terminal subdomain consists of mixed β-sheets and constitutes the acceptor substrate binding site. At the active site that resides in the open cleft across the two subdomains, the leaving oxygen of the phosphate group and the C1 atom of the donor sugar and the acceptor OH group align to advance a bimolecular nucleophilic substitution type in line displacement reaction. Also the inverting glycosyltransferases are characterized by the presence of a catalytic base residue that deprotonates the acceptor OH group. The x-ray structure of K4CP has now been solved and has revealed two distinct domains. Both domains are characterized by having all of the structural features of the GT-A fold, and both can be perfectly superimposed with known structures of other glycosyltransferases. In addition, the revealed K4CP structure has the two GT-A folds that orient the two active site clefts at an 180° angle, suggesting that the two domains function independently, with nonsequential and random binding and release of donor substrates, as the catalytic mechanism of the polymerase reaction.
Our ITC study has concluded that UDP-GalNAc and UDP-GlcA specifically bind to the N- and C-terminal DXD motifs of the K4CP enzyme, respectively. The presence of two independent binding sites also been implicated in another glycosyltransferase, the Pasteurella multocida hyaluronan synthase (25). Consistent with this conclusion, the K4CP structure has led to the conclusion that UDP-GalNAc and UDP-GlcA are present at the N- and C-terminal GT-A domains, respectively.3 In our ITC study, however, only one molecule of UDP is found to bind to one molecule of a given K4CP enzyme. Although the binding of UDP to the 239ACA241 and 519ACA521 mutants indicates that UDP could bind to either the N- or the C-terminal DXD site, in fact UDP appears to bind to the C-terminal DSD of the wild type K4CP enzyme. During the GlcA transfer reaction, this binding of UDP can be transient because its Kd value is more than 10 times higher that the corresponding value of UDP-GlcA binding. Various experimental features associated with the use of protein crystals to solve structures, such as cocrystallization of K4CP with a high concentration of UDP, which may have led to UDP binding to both DXD sites. Intriguingly, structural analysis of multiple forms of the x-ray crystal structures found that either there is binding of two UDP molecules or no UDP binding at all. This finding may suggest that there is structural cross-talk between the N- and C-terminal DXD sites to regulate UDP binding.
Our binding assays demonstrated that UDP-GalNAc could bind to either the N-terminal or C-terminal DXD sites at similar affinities. Thus, during the polymerization reaction, UDP-GalNAc should preferentially bind to the N-terminal DCD site because the UDP-GalNAc transfer reaction is catalyzed by the N-terminal active site. We observed the selective binding of GalNAc to the N-terminal DCD only when the C-terminal DSD was mutated. Contrary to what analysis of the x-ray crystal structural data suggests, the C-terminal domain of K4CP appears to regulate donor binding and probably the transfer activity of the N-terminal domain. K4CP exhibited high affinity binding of UDP-GlcA to the C-terminal DSD motif only and did not hydrolyze UDP-GlcA, so that the enzyme can effectively convert the donor binding activity to its transfer activity. The inability of UDP-GlcA to bind to the N-terminal DCD motif and the constant occupation at the C-terminal DSD motif ensure that this enzyme will never transfer two glucuronic acids sequentially to an acceptor substrate and force UDP-GalNAc binding to the N-terminal DCD motif.
In conclusion, ITC analyses have now determined the specific binding of donor substrates to the bacterial chondroitin polymerase K4CP. In solution, only one donor substrate binds to K4CP at a given time. The C-terminal DSD motif binds to UDP-GlcA with a high affinity, apparently initiating the polymerase reaction and coordinating the N-terminal DCD motif to bind to UDP-GalNAc. The underlying molecular mechanism for control of the N-terminal domain by the C-terminal domain with subsequent transfer reactions is a subject ripe for further scrutiny and investigations.
This work was supported, in whole or in part, by the National Institutes of Health Grant Intramural Research Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GlcA, glucuronic acid; GalNAc, N-acetylgalactosamine; ITC, isothermal titration calorimetry.
References
- 1.Wight, T. N., Heinegard, D. K., and Hascall, V. C. (1991) in Cell Biology of Extracellular Matrix (Hay, E. D., ed) pp. 45–78, Plenum Press, New York
- 2.Zako, M., Shinomura, T., Miyashi, O., Iwaki, M., and Kimata, K. (1997) J. Neurochem. 69 2155–2161 [DOI] [PubMed] [Google Scholar]
- 3.Oohira, A., Katoh-Semba, R., Watanabe, E., and Matsui, F. (1994) Neurosci. Res. 20 195–207 [DOI] [PubMed] [Google Scholar]
- 4.David, G., Lories, V., Heremans, A., Van der Schueren, B., Cassiman, J. J., and Van den Berghe, H. (1989) J. Cell Biol. 108 1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitch, M. T., Doller, C., Combs, C. K., Landreth, G. E., and Silver, J. (1999) J. Neurosci. 19 8182–8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caggiano, A. O., Zimber, M. P., Ganguly, A., Blight, A. R., and Gruskin, E. A. (2005) J. Neurotrauma 22 226–239 [DOI] [PubMed] [Google Scholar]
- 7.Sakai, K., Kimata, K., Sato, T., Gotoh, M., Narimatsu, H., Shinomiya, K., and Watanabe, H. (2007) J. Biol. Chem. 282 4152–4161 [DOI] [PubMed] [Google Scholar]
- 8.Roughley, P., Martens, D., Rantakokko, J., Alini, M., Mwale, F., and Antoniou, J. (2006) Eur. Cell. Mater. 11 1–7 [PubMed] [Google Scholar]
- 9.Ninomiya, T., Sugiura, N., Tawada, A., Sugimoto, K., Watanabe, H., and Kimata, K. (2002) J. Biol. Chem. 277 21567–21575 [DOI] [PubMed] [Google Scholar]
- 10.Jing, W., and DeAngelis, P. L. (2003) Glycobiology 13 661–671 [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa, H., Uyama, T., and Sugahara, K. (2001) J. Biol. Chem. 276 38721–38726 [DOI] [PubMed] [Google Scholar]
- 12.Sato, T., Gotoh, M., Kiyohara, K., Akashima, T., Iwasaki, H., Kameyama, A., Mochizuki, H., Yada, T., Inaba, N., Togayachi, A., Kudo, T., Asada, M., Watanabe, H., Imamura, T., Kimata, K., and Narimatsu, H. (2003) J. Biol. Chem. 278 3063–3071 [DOI] [PubMed] [Google Scholar]
- 13.Liu, J., and Mushegian, A. (2003) Protein Sci. 12 1418–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotting, C., Muller, S., Schottler, M., Schon, S., Prante, C., Brinkmann, T., Kuhn, J., and Kleesiek, K. (2004) J. Biol. Chem. 279 42566–42573 [DOI] [PubMed] [Google Scholar]
- 15.Breton, C., Bettler, E., Joziasse, D. H., Geremia, R. A., and Imberty, A. (1998) J. Biochem. (Tokyo) 123 1000–1009 [DOI] [PubMed] [Google Scholar]
- 16.Busch, C., Hofmann, F., Selzer, J., Munro, S., Jeckel, D., and Aktories, K. (1998) J. Biol. Chem. 273 19566–19572 [DOI] [PubMed] [Google Scholar]
- 17.Sobhany, M., Dong, J., and Negishi, M. (2005) J. Biol. Chem. 280 23441–23445 [DOI] [PubMed] [Google Scholar]
- 18.Press, W. H., Flannery, B. P., Teukolsky, S. A., and Vetterling, W. T. (1989) Numerical Recipes in FORTRAN: The Art of Scientific Computing, Cambridge University Press, Cambridge
- 19.Segel, I. H. (1976) Biochemical Calculations, 2nd Ed., pp. 145–278, John Wiley & Sons, Inc., New York
- 20.Pedersen, L. C., Dong, J., Taniguchi, F., Kitagawa, H., Krahn, J. M., Pedersen, L. G., Sugahara, K., and Negishi, M. (2003) J. Biol. Chem. 278 14420–14428 [DOI] [PubMed] [Google Scholar]
- 21.Negishi, M., Dong, J., Darden, T. A., Pedersen, L. G., and Pedersen, L. C. (2003), Biochm. Biophys. Res. Commu. 303 393–398 [DOI] [PubMed] [Google Scholar]
- 22.Bounrne, Y., and Henrissat, B. (2001) Curr. Opin. Struct. Biol. 11 593–600 [DOI] [PubMed] [Google Scholar]
- 23.Berton, C., Mucha, J., and Jeanneau, C. (2001) Biochemie 83 713–718 [DOI] [PubMed] [Google Scholar]
- 24.Tarbouriech, N., Charnock, S. J., and Davies, G. J. (2001) J. Mol. Biol. 314 655–661 [DOI] [PubMed] [Google Scholar]
- 25.Williams, K. J., Halkes, K. M., Kamerling, J. P., and DeAngelis, P. L. (2006) J. Biol. Chem. 281 5391–5397 [DOI] [PubMed] [Google Scholar]
- 26.Osawa, T., Sugiura, N., Shimada, H., Hirooka, R., Tsuji, A., Shirakawa, T., Fukuyama, K., Kimura, M., Kimata, K., and Kakuta, Y. (2009) Biochem. Biophys. Res. Commun., in press [DOI] [PubMed]