Abstract
TNFα gene expression is silenced in the endotoxin tolerant phenotype that develops in blood leukocytes after the initial activation phase of severe systemic inflammation or sepsis. The silencing phase can be mimicked in vitro by LPS stimulation. We reported that the TNFα transcription is disrupted in endotoxin tolerant THP-1 human promonocyte due to changes in transcription factor binding and enrichment with histone H3 dimethylated on lysine 9 (H3K9). Here we show that the TNFα promoter is hypermethylated during endotoxin tolerance and that H3K9 methylation and DNA methylation interact to silence TNFα expression. Chromatin immunoprecipitation and RNA interference analysis demonstrated that, in tolerant cells, TNFα promoter is bound by the H3K9 histone methyltransferase G9a which dimethylates H3K9 and creates a platform for HP1 binding, leading to the recruitment of the DNA methyltransferase Dnmt3a/b and an increase in promoter CpG methylation. Knockdown of HP1 resulted in a decreased Dnmt3a/b binding, sustained G9a binding, and a modest increase in TNFα transcription, but had no effect on H3K9 dimethylation. In contrast, G9a knockdown-disrupted promoter silencing and restored TNFα transcription in tolerant cells. This correlated with a near loss of H3K9 dimethylation, a significant decrease in HP1 and Dnmt3a/b binding and promoter CpG methylation. Our results demonstrate a central role for G9a in this process and suggest that histone methylation and DNA methylation cooperatively interact via HP1 to silence TNFα expression during endotoxin tolerance and may have implication for proinflammatory gene silencing associated with severe systemic inflammation.
Epigenetic mechanisms generate heritable marks on DNA and N-terminal tails of histones that maintain stable patterns of gene expression and are crucial in regulating gene activity as they impact chromatin structure and dynamics. These chromatin-based modifications control the recruitment of specific transcription factors and/or chromatin effectors, thereby providing a mechanism by which histones and DNA modifications regulate gene transcription (reviewed in Refs. 1–3). Methylation of histone H3 on lysine 9 (H3K9) and DNA on 5-cytosine bases, within the context of CpG dinucleotides, are two epigenetic marks whose increased levels are associated with heterochromatin formation and transcriptional silencing of several gene promoters (4).
H3K9 can exist in mono-, di-, or trimethylated state. Mono- and dimethylation are catalyzed by the histone methyltransferase G9a, whereas trimethylation is catalyzed by the methyltransferase SUV39h and is predominant in pericentric (constitutive) heterochromatin domains (1, 5). While G9a can also trimethylate H3K9 in vitro (6), it is a major histone methyltransferase for mono- and dimethylation of H3K9 in euchromatic (regulated) domains (7). Methylated H3K9 serves as a docking site for chromatin modifiers such as the heterochromatin-binding protein 1 (HP1),2 which mediates heterochromatin formation and is implicated in gene silencing (4, 8, 9).
DNA methylation, on the other hand, occurs by adding a methyl group at 5-cytosine in CpGs and is often associated with transcriptional silencing, whereas absence of CpG methylation links to transcriptional activation (10, 11). This methylation is catalyzed by the DNA methyltransferases Dnmt1 and Dnmt3a/b. Dnmt1 is the primary methyltransferase for the maintenance of hemimethylated DNA and is found mainly in pericenteric heterochromatin (12), whereas Dnmt3a/b are considered to be the de novo DNA methyltransferases (13). The signals that determine whether a particular CpG becomes methylated are unknown, but interactions between methylated DNA and chromatin effectors, such as methyl-CpG-binding proteins and HP1 play an important role in chromatin condensation and gene repression (14–16).
Recent studies support a mechanistic connection between DNA and histone methylation and transcriptional silencing (1, 17, 18), wherein components of each of the two epigenetic pathways are coupled. For example, methyl-CpG-binding proteins may recruit and interact with histone deacetylases, methyltransferases and methylated DNA, thereby providing a link between DNA and histone methylation (17, 19, 20). This connection is further reinforced through interactions with other chromatin remodeling proteins such as HP1 (16, 21–23).
HP1 is a non-histone protein enriched in heterochromatin (24) through binding to methylated H3K9 in pericentric and euchromatic domains of chromatin and is an essential component of heterochromatic gene silencing (25). It acts as an adapter to transmit epigenetic information between histone and DNA (1). The three variants of mammalian HP1 (α, β, γ) localize to regions of constitutive heterochromatin and euchromatin (2). Targeting HP1 to euchromatic sites is sufficient to induce gene silencing and local condensation of chromatin in several experimental systems (2) while loss of HP1 results in derepression of silenced genes (24). Although the precise mechanisms by which HP1 contributes to gene silencing is not understood, it appears that HP1 links DNA and histone through its interaction with and recruitment of histone and DNA methyltransferases and other chromatin modifiers (2, 4, 15, 26, 27).
Regulation of TNFα expression in monocytes is complex and involves transcriptional and post-transcriptional mechanisms (28, 29). Methylation of H3K9 marks the TNFα promoter for transcription silencing during endotoxin tolerance (28). This epigenetic mark correlates with disruption of TNFα transcription due to diminished binding of active NF-κB RelA/p65 and increased binding of repressive RelB protein, as well as binding of HP1. Endotoxin tolerance is defined by reprogramming of gene expression, including silencing of acute proinflammatory mediators, such as TNFα and IL-1β, in response to the stimulation of the Toll-like receptor (TLR) 4 by bacterial endodoxin (LPS) (30). The silencing phase develops rapidly after an initial activation phase that generates a cytokine storm that initiates both systemic and local acute inflammation (31–33), and is observed in blood neutrophils (34) and monocytes (35) during severe systemic inflammation in animals and human (36, 37). This tolerant phenotype can be generated in cultured cell lines and primary monocytes by LPS stimulation (38, 39).
Here, we show that the TNFα promoter is hypermethylated in human endotoxin tolerant THP-1 monocytes. Chromatin immunoprecipitation and RNA interference analysis revealed that the H3K9 methyltransferase G9a interacts with the de novo DNA methyltransferase Dnmt3a/b to silence the TNFα transcription during endotoxin tolerance. This interaction is mediated by HP1. Dnmt3a/b and HP1 recruitment is dependent on G9a binding to the promoter, as loss of G9a results in a significant decrease in both DNA and H3K9 methylation at the promoter region and restores TNFα transcription. These findings suggest that H3K9 dimethylation by G9a is causative in the process of TNFα gene silencing and does so via HP1 and Dnmt3a/b. Our studies provide a functional connection between DNA methylation and histone methylation, in which HP1 plays an important role, and further support a model of interdependence between these two epigenetic pathways in proinflammatory gene silencing during endotoxin tolerance.
EXPERIMENTAL PROCEDURES
Cell Culture and LPS Tolerance—The human promonocytic cells, THP-1, obtained from the American Type Culture Collection, were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, and 10% fetal bovine serum (HyClone, Logan, UT) in a humidified incubator with 5% CO2 at 37 °C. For induction of tolerance (38), cells were incubated for 16 h with 1 μg/ml of Gram-negative bacterial LPS (Escherichia coli 0111:B4, Sigma). LPS-tolerant and LPS-responsive (normal) cells were washed with minimal medium, cultured at 1 × 106 cells/ml and stimulated with 1 μg/ml LPS for the indicated times.
Methylation-specific Genomic Sequencing—To assess DNA methylation at the TNFα promoter, ∼150 ng of genomic DNA isolated from normal and LPS tolerant cells were subjected to sodium bisulfite treatment using the methylSEQR Bisulfite Conversion Kit (Applied Biosystems) according to the manufacturer's protocol, to convert cytosine in the genomic DNA to uracil. Converted DNA was then purified using an affinity column.
Sequence-specific PCR of the bisulfite-treated DNA was performed using primers specific to the human TNFα promoter designed using the Methyl Primer Express v1.0 (Applied Biosystems). The primers were specifically designed to screen a 300-bp promoter fragment containing 8 CpG doublets surrounding the NF-κB binding site at –98 bp (see Fig. 1A), and their sequence coordinates are: forward 5′-3′ (–349 to –336) and reverse 5′-3′ (–50 to –71). PCR was carried out in a volume of 50 μl containing 1× PCR buffer (10 mm Tris-HCl, pH 8.3, 50 mm KCl), 2.5 mm MgCl2, 20–40 ng of bisulfite-converted DNA, 25 pmol of each primer, 200 μm of each dNTP, and 1.25 units of TaqDNA polymerase (PerkinElmer Life Sciences, Emeryville, CA). PCR conditions were: 35 cycles at 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 2 min.
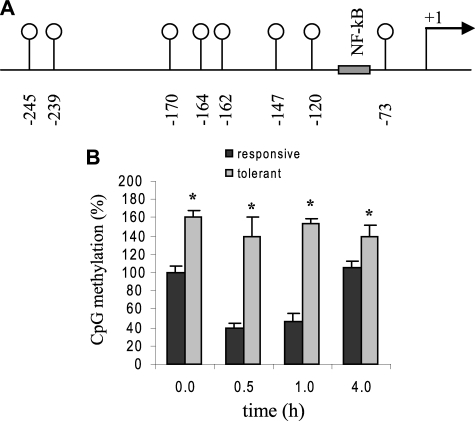
FIGURE 1.
TNFα promoter is hypermethylated in endotoxin-tolerant THP-1 cells. A, a schematic diagram representing the proximal promoter region of the human TNFα gene analyzed by bisulfite sequencing. The eight CpG dinucleotides located in this region are represented by vertical lines with open circles, and their locations relative to the transcription start site are indicated. B, bisulfite sequencing results. Endotoxin tolerance was induced by pretreatment with 1 μg/ml LPS for 16 h. Endotoxin responsive and tolerant cells were then stimulated with 1 μg/ml LPS for the indicated times. Genomic DNA was extracted and subjected to sodium bisulfite conversion, followed by PCR amplification and sequencing. The total numbers of methylated CpGs in responsive untreated cells (0 h) was set at 100%, and all other values were calculated relative to this value. The data represent the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05) between responsive versus tolerant cells.
DNA sequencing was performed using the POP7 sequencing polymer and ABI 3730XL Capillary DNA Analyzer as described previously (40). In this CpG methyl-specific sequencing, cytosine that is protected by methylation is read as “C” in the DNA sequence while unmethylated cytosine is read as “T”, the structural analog of uracil. All sequencing data were analyzed using the DNA analysis program Seqencher v8 (Gene Codes, Corp., Ann Arbor, MI) and compared with reference sequences representing methylated and unmethylated DNA.
ChIP Assays—Chromatin immunoprecipitation (ChIP) assay was performed to assess H3K9 dimethylation and chromatin protein interactions around the NF-κB binding site of the TNFα proximal promoter, using ChIP kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. Briefly, cells were harvested and proteins were cross-linked with DNA by the fixation in 1% formaldehyde for 10 min at room temperature. After washing with cold phosphate-buffered saline, cells were lysed in 1% SDS for 30 min on ice. The lysates were sonicated to shear DNA using Branson 250 sonicator (two 15-s pulses at 40% power in an ice bath, with 1 min between each pulse). These shearing conditions generate DNA fragments ranging in size from 500–1000 bp. Chromatin solution was pre-cleared with protein G-coated magnetic beads for 2 h at 4 °C. 10 μl of the chromatin solution was reserved as “input” sample. The remaining chromatin was immunoprecipitated overnight at 4 °C with 3 μg of antibody specific to HP1, Dnmt3a, Dnmt3b, RelB, IgG (Santa Cruz Biotechnology, Santa Cruz, CA), SUV39h1, G9a, or di-methylated histone H3 lysine 9 (H3K9me2) (Upstate Biotechnology, Lake Placid, NY). The chromatin/antibody complexes captured on the beads were washed several times and then eluted in 50 μl elution buffer. The immunoprecipitated and “input” sample cross-links were reversed by incubation for 2.5 h at 65 °C. After treatment with proteinase K at for 1 h at 37 °C, the reaction was stopped and the resulting DNA was stored at –20 °C until analyzed by standard and real-time PCR as described below. For ChIP reimmunoprecipitation (ChIP-ReIP) experiment, chromatin was first immunoprecipitated with the primary antibody (αHP1) and washed. Complexes were then eluted from the primary immunoprecipitates by incubation with 10 mm dithiothreitol at 37 °C for 25 min, diluted in IP buffer, and reimmunoprecipitated with the second antibodies (αSUV39h1, αDnmt3a/b). The remaining ChIP procedures were followed to reverse the cross-links and recover the DNA, as described above.
Semiquantitative PCR—PCR was performed in a 50-μl volume containing 5 μl of the ChIP DNA, 1 μm of each primer, 2 mm MgCl2, 0.2 μm dNTPs, and 0.04 units/μl AmpliTag Gold DNA polymerase (Applied Biosystems). The PCR conditions were as follows: 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C, 58 °C, and 72 °C for 30 s each, and a final cycle at 72 °C for 5 min. Equal amounts of PCR products were run on 1.2% ethidium bromide-stained agarose gel and images were captured using Quantity One Imager (Bio-Rad). The primers used in PCR were designed to amplify a sequence in the human TNFα proximal promoter region containing the κB3 site at –98 bp relative to the transcription start site (41) and were as follows: TNFα forward (5′-TACCGCTTCCTCCAGATGAG-3′) and TNFα reverse (5′-TGCTGGCTGGGTGTGCCAA-3′).
Quantitative PCR for Immunoprecipitated DNA—Real-time PCR was performed to precisely quantify the TNFα promoter sequence in the ChIP DNA. The same primers described above, labeled with an internal fluorogenic probe (5′-FAM-CTTGGTGGAGAAACC-TAMRA-3′), were used (Applied Biosystems, Foster City, CA). The PCR reaction (25 μl) contained 5 μl of ChIP DNA, 12.5 μl of 2× TaqMan Universal Master Mix containing DNA polymerase and dNTPs, 300 nm of each primer, and 100 nm internal probe. Reactions were run in duplicates at 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min (combined annealing and extension), using ABI Prism 7000 Sequence Detection System (Applied Biosystems). A IgG-immunoprecipitated DNA sample was also amplified as a negative control (data not shown). Sample data were normalized to the input DNA and are presented as fold change relative to DNA from unstimulated cells (0 h).
RNA Interference—Exponentially growing cells were harvested and washed with sterile PBS. Control (nonspecific), or HP1 or G9a-specific small interfering RNAs (siRNA) (Santa Cruz Biotechnology) with a 3′-dTdT overhang were transfected by electroporation with 5 μl of (0.5 μm) siRNA in 100 μl of Nucleofector (AMAXA, Gaithersburg, MD), according to the manufacturer's protocol. Immediately after transfection, cells were left unstimulated or stimulated with 1 μg/ml LPS, to induce tolerance. After 36 h, cells were harvested, washed with minimal medium, and left unstimulated or stimulated for 3 h with 1 μg/ml LPS. G9a was silenced by a pool of three target-specific 20–25 nucleotides siRNAs. HP1 was silenced by three pools of siRNAs.
mRNA Analysis—Expression of TNFα was evaluated by quantitative real-time PCR. Total RNA was isolated using RNA STAT-60 extraction kit, according to the manufacturer's protocol (Tel-Test, Friendswood, TX)). Two micrograms RNA were reverse-transcribed to cDNA in a 25-μl volume containing 0.2 μm dNTPs, 2.5 μm oligo d(T), 5 mm MgCl2, and 0.25 units/μl of murine leukemia reverse transcriptase (Applied Biosystems). The RT reaction was incubated for 1 h at 42 °C and 5 min at 99 °C. The PCR was performed using 5 μl of cDNA and TNFα and GAPDH predesigned TaqMan primer/probe sets (Applied Biosystems). The PCR conditions were as described above. Sample data were normalized to GAPDH mRNA values and are presented as percentage change relative to mRNA from endotoxin-responsive cells (set as 100%).
Western Blot Analysis—Nuclear proteins were extracted by incubating cells on ice for 15 min in a buffer containing 10 mm HEPES (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 0.2 mm EDTA, 20 mm NaF, 1 mm Na4P2O7, 1 mm Na3VO4, 0.5 mm dithiothreitol, 0.1% Titon X-100, and 1× protease inhibitor mixture. Nuclei were pelleted by centrifugation at 5,000 rpm for 10 min at 4 °C and then resuspended in lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) and incubated on ice for 30 min. Extracts were cleared by centrifugation and protein concentration was determined. Whole cell extracts were prepared using the same nuclei lysis buffer. Equal amounts (50 μg) of protein were separated on SDS-PAGE and transferred to polyvinylidene difluoride membranes (Pierce). Membranes were blocked and probed overnight at 4 °C with appropriate dilutions of primary Abs against HP1, Dnmt3a/b, SUV39h1 (Santa Cruz Biotechnology), and G9a (Upstate). This was followed by incubation with appropriate horseradish peroxidase-conjugated secondary Abs (Santa Cruz Biotechnology). Proteins were visualized using high sensitivity ECL system (Pierce). Blots were stripped and reprobed with control Ab.
Statistical Analysis—Data were analyzed by Microsoft Excel 2003 and are presented as the mean (±S.E.) of three independent experiments. The Student's t test was used to determine significant differences between groups. p values <0.05 were considered statistically significant.
RESULTS
TNFα Promoter Is Hypermethylated in Endotoxin-tolerant THP-1 Cells—Our previous studies showed that the TNFα promoter nucleosome is dimethylated on H3K9 and bound by HP1 during transcriptional silencing in endotoxin-tolerant cells (28), a phenotype that naturally occurs in blood leukocytes during the course of severe systemic inflammation in animals and sepsis in human where the expression of many proinflammatory cytokines is silenced after the initial induction phase that follows LPS exposure (30).
We sought to determine the DNA methylation state of TNFα proximal promoter. This fragment contains 8 CpG dinucleotides around the NF-κB binding site and the TATA box (see Fig. 1). Bisulfite sequencing of the genomic DNA extracted from endotoxin-responsive (normal) and endotoxin-tolerant cells showed that most of these CpG doublets were methylated in greater numbers in tolerant cells versus responsive cells (Fig. 1). The total number of methylated CpGs in responsive untreated cells was assigned as 100% methylation and all other samples were calculated relative to this value. We observed a marked decrease in CpG methylation at 0.5 and 1 h after adding LPS to responsive cells. These two time points coincide with active TNFα transcription in endotoxin-responsive cells (28). By 4 h of stimulation, the methylation level retuned to near 0 h level. However, the level of CpG methylation was significantly higher in tolerant-untreated cells compared with responsive cells, and remained significantly high at all the time points tested. We also observed that CpGs at –239 and –245 were always methylated every time the experiment was repeated. These results demonstrate that the TNFα promoter is hypermethylated during endotoxin tolerance, which may contribute to TNFα gene silencing.
Histone and DNA Methyltransferases Bind to TNFα Promoter during Transcription Silencing—Our data indicated that, in tolerant cells, the TNFα promoter was hypermethylated on CpGs (Fig. 1) and dimethylated on H3K9 and bound by HP1 (28). These two epigenetic modifications contribute to gene silencing (1, 4). We hypothesized that histone methylation and DNA methylation may cooperatively interact to mediate TNFα silencing during endotoxin tolerance. In addition, HP1 may act as an adapter in mediating histone and DNA methylation through its binding to and recruitment of histone and DNA methyltransferases. If HP1 is indeed involved in the silencing of TNFα promoter, then its depletion will affects histone and/or DNA methylation. We used RNA interference and ChIP assays to test this hypothesis. Cells were transfected with a pool of siRNAs specifically designed to silence the three HP1 isoforms and then stimulated with LPS to induce tolerance. After 36 h, responsive and tolerant cells were harvested and stimulated for the indicated times. In the control experiment, cells were transfected with siRNAs not known to bind to HP1 sequences specifically. We first determined HP1 binding after HP1 knockdown. Fig. 3, A and B shows that HP1 binding to the TNFα promoter was markedly reduced after HP1-specific, but not control, siRNA transfection into responsive and tolerant cells. Also, HP1 nuclear protein level was markedly reduced after the knockdown (Fig. 3C). We next determined whether loss of HP1 would result in changes in TNFα transcription. As shown in Fig. 2, we observed ∼50% recovery of TNFα mRNA in endotoxin-tolerant cells after LPS stimulation compared with responsive cells. These data demonstrate that HP1 contributes to TNFα gene silencing.
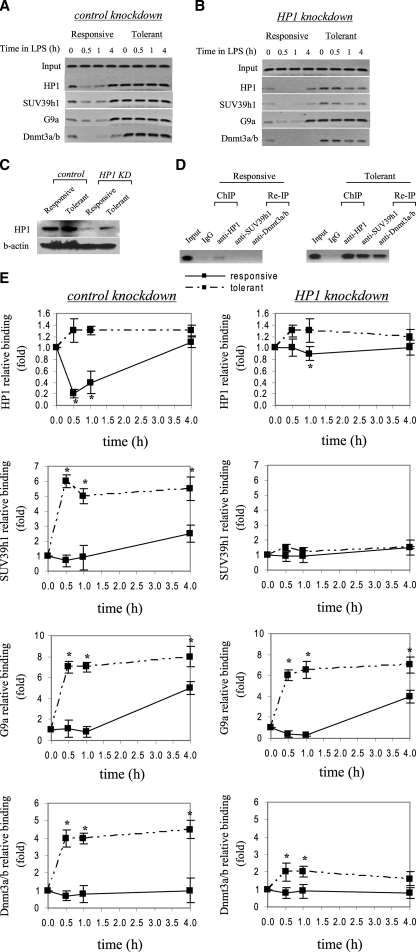
FIGURE 3.
HP1 gene silencing markedly reduces SUV39h1, Dnmt3a/b, but not G9a binding to the TNFα promoter. THP-1 cells were transfected with control or HP1-specific siRNA and then left unstimulated or stimulated with 1 μg/ml LPS for 36 h. Cells were then washed and left unstimulated or stimulated with 1 μg/ml LPS for the indicated times. ChIP assay was performed on cross-linked chromatin, immunoprecipitated with antibodies specific to HP1, SUV39h1, G9a, and Dnmt3a/b. The relative enrichment of TNFα promoter sequences in the immunoprecipitated DNA was analyzed by semiquantitative PCR (A and B) and real-time PCR (E) using primers that amplify the proximal promoter region. In A and B, representative results are shown. In E, data were normalized to input DNA and are presented as fold change relative to unstimulated cells (0 h) (set as 1-fold). Data are the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05). C, HP1 protein expression in the nucleus. THP-1 cells were transfected with control or HP1-specific siRNA and left unstimulated or stimulated with 1 μg/ml LPS for 36 h. Responsive (unstimulated) and tolerant (stimulated) cells were then washed and stimulated for 1 h with 1 μg/ml LPS, and nuclear extract was prepared as described under “Experimental Procedures.” Blotted proteins were probed with HP1 Ab. Blots were stripped and reprobed with control Ab. D, ChIP reimmunoprecipitation showing the recruitment of SUV39h1 and Dnmt3a/b to the TNFα promoter by HP1. Chromatin was first immunoprecipitated with HP1 Ab (ChIP). Immunoprecipitates were then eluted and reimmunoprecipitated (Re-IP) with SUV39h1 or Dnmt3a/b Ab.
FIGURE 2.
HP1 gene silencing modestly restores TNFα mRNA level in tolerant cells. THP-1 cells were transfected with nonspecific or HP1-specific siRNA and then left unstimulated or stimulated with 1 μg/ml LPS for 36 h. Responsive (r) and tolerant (t) were washed and then left unstimulated or stimulated with 1 μg/ml LPS for 3 h. Total RNA was extracted and analyzed for TNFα mRNA expression by real-time PCR. Sample data were normalized to GAPDH data and are presented as percent change relative to mRNA level in LPS-responsive (r+) cells (set as 100%). Data are the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05), t+ versus t-.
We set out to determine the mechanism by which HP1 contribute to TNFα silencing. HP1 interacts with and recruits the histone methyltransferases SUV39h and G9a (4, 22, 42), both of which can direct H3K9 methylation (8, 43, 44). In addition, our data (not shown) indicated that both SUV39h1 and G9a were bound to the TNFα promoter in endotoxin-tolerant cells. HP1 knockdown resulted in a marked decrease in SUV39h1, but not G9a, binding (Fig. 3, A and B). In addition, ChIP analysis showed that the level of dimethylated H3K9 remained high in tolerant cells (data not shown). These results suggested that the promoter binding by SUV39h1 is a downstream event of HP1 binding and that SUV39h1 was not responsible for H3K9 dimethylation.
Because HP1 interacts with Dnmt3a/b (21), we next investigated Dnmt3a/b binding after HP1 knockdown. Fig. 3, A and B shows that there is a marked decrease in Dnmt3a/b binding after HP1 depletion, suggesting HP1 participates in recruiting Dnmt3a/b methyltransferase activity to the TNFα promoter during transcriptional silencing in tolerant cells.
Depletion of HP1 resulted in the loss of promoter binding by SUV39h1 and Dnmt3a/b. To confirm whether HP1 is responsible for the recruitment of SUV39h1 and Dnmt3a/b, we performed ChIP reimmunoprecipitation (ChIP-ReIP) assay. Cross-linked chromatin was first immunoprecipitated with anti-HP1. The immunoprecipitated complexes were then washed and reimmunoprecipitated with anti-SUV39h1 or anti-Dnmt3a. As shown in Fig. 3D, promoter DNA fragments were enriched in both primary and secondary immunoprecipitates. This result demonstrates that HP1 plays a role in the simultaneous recruitment of SUV39ha and Dnmt3a/b to the TNFα promoter during transcription silencing.
Loss of G9a Binding and H3K9 Dimethylation from TNFα Promoter Reverses Transcription Silencing and Restores TNFα mRNA Levels in Tolerant Cells—The data presented in Fig. 3 suggested that SUV39h1 was not involved in H3K9 dimethylation and that G9a binding is an upstream event of HP1 and Dnmt3a/b recruitment. To test this, we next knocked down G9a and investigated downstream events by assessing H3K9 dimethylation and HP1 binding. As shown in Fig. 4, A and B, G9a binding markedly decreased after the knockdown. We also confirmed that G9a protein expression was markedly decreased after the knockdown (Fig. 4C). Loss of G9a resulted in a substantial decrease in H3K9 methylation and a decrease in HP1 binding. These results suggest that G9a directs H3K9 dimethylation at the TNFα promoter and that loss of this histone mark results in near loss of HP1 binding in tolerant cells. We next blocked H3K9 dimethylation to test whether HP1 binding is dependent on H3K9 dimethylation. We incubated tolerant cells with 6 μm BIX-01294, which has been shown to specifically inhibit G9a methyltransferase activity and H3K9 dimethylation (45). After 6 h, LPS was added, and the incubation continued for 1 h. At this time point, high levels of H3K9 dimethylation and HP1 binding are detected. The results (Fig. 4D) show that BIX-01294 decreased H3K9 dimethylation significantly, while cells incubated with LPS or DMSO alone retained high levels of H3K9 methylation. It should be noted that the decrease in H3K9 dimethylation was not due to toxic effects by DMSO or BIX-01294, as cells were 90–95% viable by the end of incubation, as determined by trypan blue exclusion. Also, a dose response experiment showed that BIX-01294 at 6 μm concentration was most effective in inhibiting H3K9 dimethylation (data not shown). The decrease in H3K9 methylation was accompanied by a marked decrease in HP1 binding, suggesting that H3K9 dimethylation is a primary target for HP1 binding.
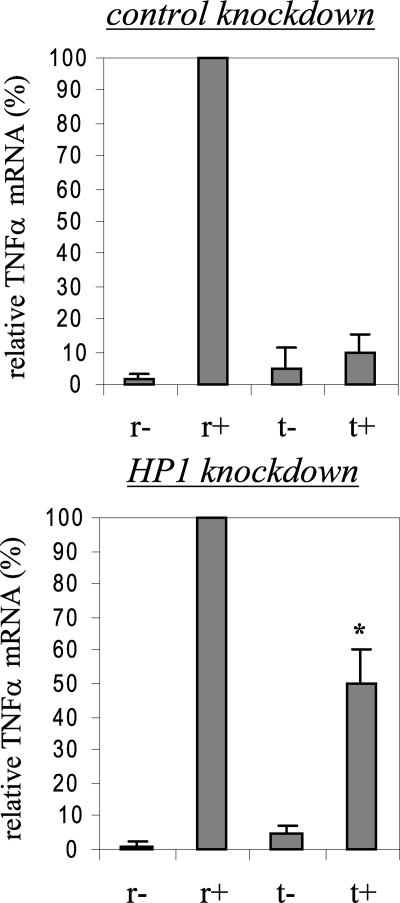
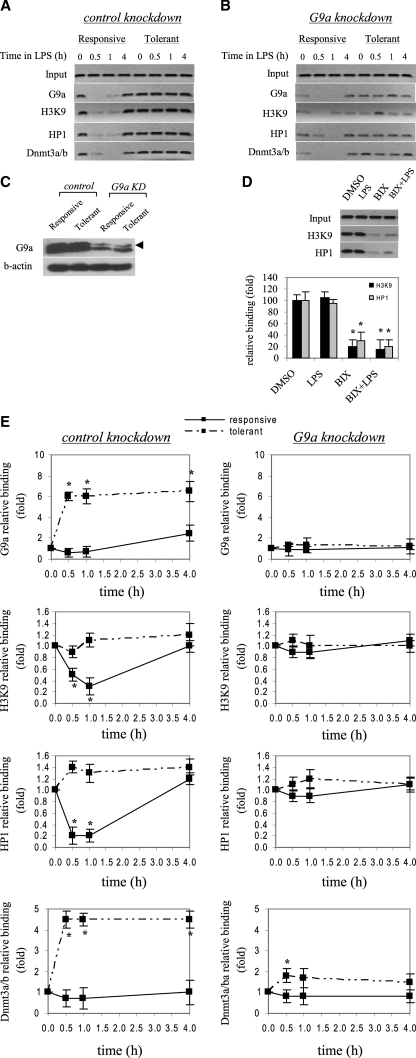
FIGURE 4.
G9a gene silencing significantly decreases H3K9 dimethylation and promoter binding by HP1 and Dnmt3a/b. Cells were transfected with control or G9a-specific siRNA and then left unstimulated or stimulated with 1 μg/ml LPS for 36 h. Cells were then washed and left unstimulated or stimulated with 1 μg/ml LPS for the indicated times. ChIP assay was performed on cross-linked chromatin, immunoprecipitated with antibodies specific to HP1, G9a, dimethyl H3K9, and Dnmt3a/b. The relative enrichment of TNFα promoter sequences in the immunoprecipitated DNA was analyzed by semiquantitative PCR (A and B) and real-time PCR (E), using primers that amplify the proximal promoter region. In A and B, representative results are shown. In E, data were normalized to input DNA and are presented as fold change relative to unstimulated cells (0 h) (set as 1-fold). Data are the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05). C, G9a protein expression in the nucleus. THP-1 cells were transfected with control or G9a-specific siRNA and left unstimulated or stimulated with 1 μg/ml LPS for 36 h (to induce tolerance). Responsive (unstimulated) and tolerant (stimulated) cells were then washed and stimulated for 1 h with 1 μg/ml LPS, and nuclear extract was prepared as described under “Experimental Procedures.” Blotted proteins were probed with G9a Ab. Blots were stripped and reprobed with control Ab. D, inhibiting H3K9 dimethylation prevents HP1 binding. LPS-tolerant cells were incubated for 6 h with 6μm of the G9a specific inhibitor BIX-012194. LPS was added for the last 1 h. Cells were harvested, and ChIP was performed with dimethyl H3K9 or HP1 Ab. Enrichment of TNFα promoter sequences in the immunoprecipitated DNA was assessed by semiquantitative PCR (upper panel) and real-time PCR (lower panel).
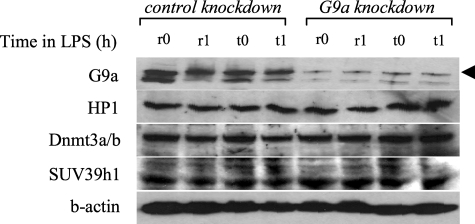
Because G9a loss reduced H3K9 methylation and HP1 binding, and since the data presented in Fig. 3 suggested that loss of HP1 binding resulted in a concomitant decrease in Dnmt3a/b binding, we hypothesized that absence of G9a may affect Dnmt3a/b binding and promoter CpG methylation. We observed a significant decrease in Dnmt3a/b binding after G9a knockdown (Fig. 4, A and B), suggesting that G9a is involved in targeting Dnmt3a/b to the TNFα promoter. G9a likely achieves this role through HP1, as HP1 binding was also reduced after G9a knockdown (compare Figs. 3 and 4). It must be noted that the decrease in promoter binding by HP1, SUV39h1, and Dnmt3a/b after G9a knockdown was not due to a decrease in their protein expression, because Western blot analysis of whole cell extracts showed that their total protein levels were nearly similar to those levels before G9a knockdown (Fig. 5), suggesting that level of protein expression is not responsible for the LPS-induced change in promoter binding by HP1, SUV39h1, or Dnmt3a/b.
FIGURE 5.
Western blot analysis of whole cell extract after G9a knockdown. Cells were transfected with control or G9a-specific siRNA and left unstimulated or stimulated with 1 μg/ml LPS for 36 h (to induce tolerance). Responsive (unstimulated) and tolerant (stimulated) cells were then washed and stimulated for 0.5 h and 1 h with 1 μg/ml LPS. Whole cell lysates were prepared and analyzed for the expression of G9a, HP1, Dnmt3a/b, and SUV39h1. Blots were first probed with G9a Ab, stripped, and reprobed with HP1, Dnmt3a/b, and SUV39h1 Abs.
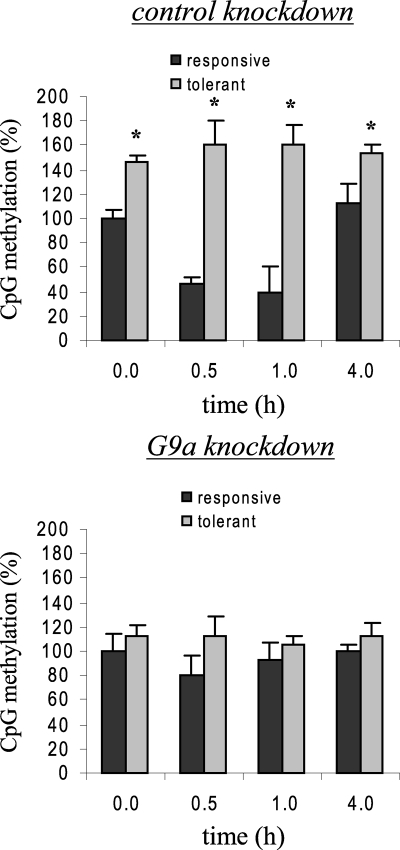
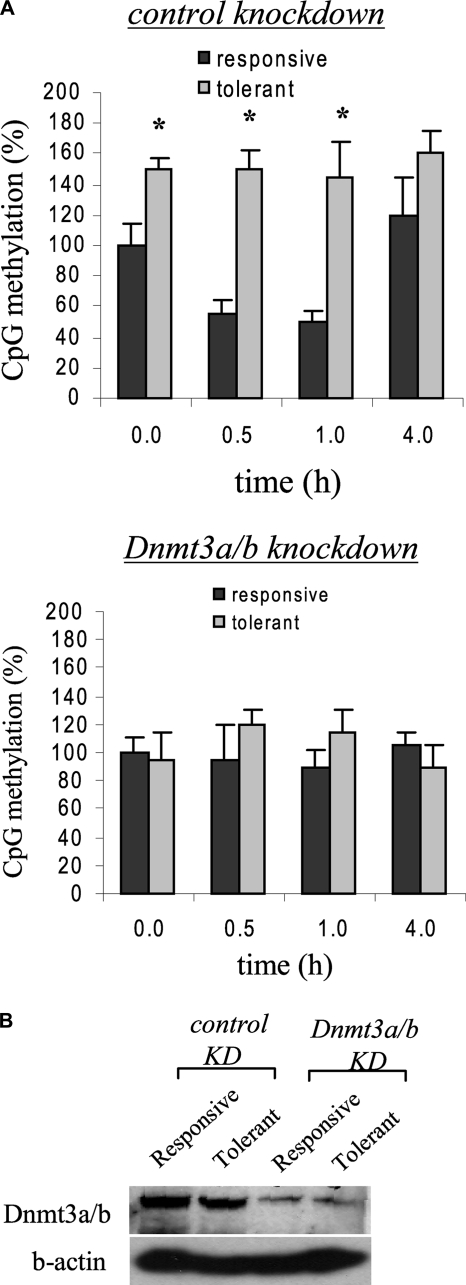
H3K9 methylation directs DNA methylation by recruiting CpG methyltransferase activities (14). We next examined CpG methylation status after G9a knockdown. Bisulfite sequencing revealed a comparable levels of CpG methylation in responsive and tolerant cells (Fig. 6), suggesting that G9a also direct CpG methylation at the TNFα promoter through Dnmt3a/b activity and that its loss results in a marked decrease in DNA methylation. To test whether Dnmt3a/b is responsible for CpG methylation, we performed bisulfite sequencing of promoter CpGs after Dnmt3a/b knockdown. Fig. 7A shows promoter CpG methylation was nearly diminished after Dnmt3a/b knockdown, demonstrating that Dnmt3a/b is directly involved in CpG methylation at the TNFα promoter. Decrease in Dnmt3a/b nuclear protein expression after knockdown was confirmed by Western blot (Fig. 7B).
FIGURE 6.
Loss of G9a binding prevents TNFα promoter CpG hypermethylation in tolerant cells. Cells were transfected with control or G9a-specific siRNA and then left unstimulated or stimulated with 1 μg/ml LPS for 36 h. Endotoxin-responsive and -tolerant cells were then stimulated with 1 μg/ml LPS for the indicated times. Genomic DNA was extracted and subjected to sodium bisulfite conversion, followed by PCR amplification and sequencing. The total numbers of methylated CpGs in responsive untreated cells (0 h) was set at 100%, and all other values were calculated relative to this value. The data represent the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05) between responsive versus tolerant cells. R, responsive; t, tolerant.
FIGURE 7.
Dnmt3a is responsible for TNFα promoter methylation. A, cells were transfected with control or Dnmt3a/b-specific siRNA and left unstimulated or stimulated with 1 μg/ml LPS for 36 h (to induce tolerance). Responsive (unstimulated) and tolerant (stimulated) cells were then washed and stimulated with 1 μg/ml LPS for the indicated times. Genomic DNA was extracted and analyzed by bisulfite sequencing. The total number of methylated CpGs in responsive untreated cells (0 h) was set at 100%, and all other values were calculated relative to this value. The data represent the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05) between responsive versus tolerant cells. B, Dnmt3a/b protein expression in the nucleus after the knockdown. Cells were transfected with Dnmt3a/b as described in A, and nuclear extract was prepared at 1 h after LPS stimulation and analyzed for Dnmt3a/b expression by Western blot.
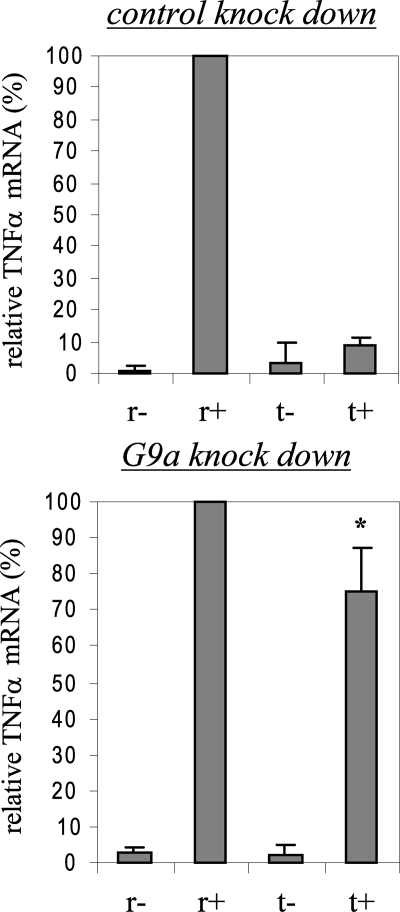
Finally, we measured TNFα RNA expression after G9a knockdown. Fig. 8 shows that the TNFα messages were restored in tolerant cells to about 80% of the original level seen in responsive cells. The level of TNFα transcripts was higher than that seen after HP1 knockdown (compare Figs. 2 and 8).
FIGURE 8.
Loss of G9a binding restores TNFα mRNA expression in tolerant cells. THP-1 cells were transfected with nonspecific or G9a-specific siRNA and then left unstimulated or stimulated with 1 μg/ml LPS for 36 h. Responsive (r) and tolerant (t) were washed and then left unstimulated or stimulated with 1 μg/ml LPS for 3 h. Total RNA was extracted and analyzed for TNFα mRNA expression by real-time PCR. Sample data were normalized to GAPDH data and are presented as percent change relative to mRNA level in LPS-responsive (r+) cells (set as 100%). Data are the mean ± S.E. of at least three independent experiments. *, significant difference (p < 0.05), t+ versus t-.
Together, our results demonstrate that G9a functions upstream of HP1 binding, DNA methylation, and heterochromatin formation at the TNFα promoter and suggest that H3K9 dimethylation is a primary epigenetic signal recognized by HP1 and is sufficient for initiating transcription silencing in vivo.
DISCUSSION
Dysregulated transcription, a salient feature of endotoxin tolerance, results in reprogramming of gene expression with sustained silencing of LPS-induced proinflammatory genes, including TNFα and IL-1β (28, 30, 46). The tolerant phenotype is observed in blood neutrophils and monocytes during severe systemic inflammation in animal and sepsis in human (30). This phenotype can be generated in cultured cell lines and primary monocytes by LPS stimulation (30).
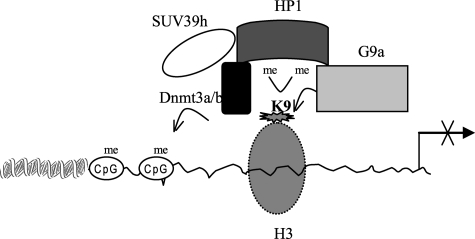
Both H3K9 and DNA CpG methylation regulate gene expression via controlling interactions with transcription activators/repressors or chromatin modifiers (1, 4, 47) and have been reported to contribute to silencing of many genes, including inflammatory mediators (48–54). In this study, we provide evidence that the TNFα promoter nucleosomes is enriched with H3K9 dimethyl marks and hypermethylated on DNA during endotoxin tolerance. The results show that the methyltransferase G9a plays a primary role in directing H3K9 dimethylation and promoter CpG methylation. We found that these two chromatin-based events are linked. HP1 acts as an adapter to link these two pathways by binding to H3K9 and by recruiting the DNA methyltransferase Dnmt3a/b. We found that loss of G9a binding resulted in loss of H3K9 and DNA methylation and restored TNFα transcription in tolerant cells. This is most consistent with G9a-dependent methylation of H3K9 acting as a nucleation point for HP1 recruitment. Subsequent spreading of H3K9 marks via HP1 binding (9) in conjunction with recruitment of additional methyltransferase activity (mediated in part by the interaction of HP1 with SUV39h1 (43)) recruits additional histone and DNA methyltransferase activities, thus reinforcing the silenced chromatin state. Our data support the model proposed in Fig. 7, where recruitment of G9a mediates dimethylation of H3K9, creating a binding site for HP1. HP1 would then interact with and recruits Dnmt3a/b methyltransferase to methylate nearby CpGs. Thus, HP1 mediates communication between H3K9 and DNA methylation to regulate TNFα gene silencing.
We observed a significant increase in G9a binding and H3K9 dimethylation associated with transcription silencing in tolerant cells. H3K9 methylation is involved in the transcriptional silencing of many euchromatic genes (49, 55, 56) Dimethyl H3K9 functions as a docking site for chromatin adapters, like HP1, that can translate H3K9 methylation signal into DNA methylation (1, 4). Genetic disruption of G9a reduced H3K9 dimethylation and DNA methylation in several euchromatic regions (7, 57). Our results showed that loss of G9a binding to the TNFα promoter resulted in a decrease in both H3K9 dimethylation and CpG methylation and reactivated TNFα expression. The loss of G9a correlated with loss of HP1, Dnmt3a/b, and SUV39h binding, demonstrating a central role for G9a in the expression silencing of TNFα in endotoxin-tolerant cells. SUV39h catalyzes H3K9 trimethylation at pericentric heterochromatic regions where its genetic disruption reduces H3K9 trimethylation (1, 6). Although both G9a and SUV39h can methylate H3K9, it is believed that one of the two proteins cannot rescue the function of the other in vivo. Our data suggested that SUV39h1 is downstream of G9a binding. SUV39h1 may thus play a secondary role in silencing TNFα transcription. It might trimethylate H3K9 after the transition of TNFα chromatin from euchromatic (active) to heterochromatic (silent) state, which has already been catalyzed by G9a and HP1 binding. This trimethylation may then create more binding sites for HP1, resulting in further condensation of the heterochromatin (see Fig. 9). Additionally, a recent study (5) has shown that G9a can methylate itself on a lysine residue and that this auto-methylation creates a binding site for HP1 proteins, suggesting that G9a can also participate in heterochromatin formation via protein-protein interactions.
FIGURE 9.
A model depicting a central role for G9a in proinflammatory gene silencing during endotoxin tolerance. G9a is first recruited to euchromatic regions, likely via a transcriptional repressor, where it dimethylates H3K9. The HP1 adaptor then binds to dimethylated H3K9 and recruits Dnmt3a/b to enhance cytosine methylation, resulting in transformation of euchromatin to silent heterochromatin and transcription silencing. In addition, HP1 may recruit SUV39h, which can trimethylate H3K9 on adjacent histone to reinforce the silent heterochromatin state.
It is unclear how G9a is targeted to the TNFα promoter in tolerant cells. We could not detect significant changes in G9a protein expression in responsive or tolerant cells. Also, there are no reports showing that G9a expression can be induced by a certain signal. However, G9a could be targeted to the TNFα promoter by a protein cofactor. We have previously shown that the NF-κB family protein RelB binds to the IL-1β and TNFα promoters and participates in the transcription silencing during endotoxin tolerance (28, 58). This is significant, because RelB is expressed only during endotoxin tolerance (58). It is possible that RelB participates in targeting G9a to the TNFα promoter in tolerant cells. In this regard, a previous study (56) has shown that the transcriptional repressor retinoblastoma (Rb) protein mediates cyclin E promoter silencing in fibroblasts by directing H3K9 methylation and heterochromatin formation through its interaction with SUV39h1 and HP1.
Our results showed that HP1 binding was dependent on G9a binding and H3K9 dimethylation and that HP1 might act to transmit a histone methylation signal to DNA methylation via the recruitment of Dnmt3a/b, supporting that HP1 is involved in the silencing activity of G9a and Dnmt3a/b. Blocking G9a methyltransferase activity and H3K9 dimethylation by the G9a specific inhibitor BIX-01294 resulted in a loss of HP1 binding. Several modes of actions are proposed for HP1 mediation of heterochromatin formation and gene silencing (3), and a model for spreading silent heterochromatin is proposed by Bannister et al. (9). In this model, HP1 proteins bind to methylated H3K9 and recruit histone methyltransferase activity, likely SUV39h that acts on adjacent histones to generate new binding sites, allowing HP1 to spread along the chromatin. Thus, HP1 propagates the epigenetic information. Our data showed that SUV39h1 binding was lost upon HP1 knockdown, further supporting the notion that SUV39h1 association with HP1 binding may, in some way, participate in the TNFα heterochromatin formation and silencing. Also, it has been demonstrated that HP1 exerts its silencing effects by associating with transcription co repressors and chromatin complexes, including Dnmt3a/b (3). Furthermore, HP1 binding may involve association with sequence specific transcription regulators. For example, many E2F- and Myc-regulated genes are repressed by HP1 via its interaction with an E2F-6 complex containing G9a (3). HP1 can also be targeted to cyclin E promoter by Rb protein (3). Interestingly, localization of HP1 proteins is disrupted in G9a-null mice (8). However, it still unclear as to what extent the localization of HP1 proteins to distinct regions of chromatin is determined solely by its binding to methylated H3K9 (9, 59), or requires auxillary factors (60), an RNA component (61) and HP1-specific modifications (62). A recent study shows that HP1 proteins can be extensively modified post-translationally, similar to histone, indicating that transcription silencing mediated by H3K9 via HP1 is a more complex process, suggesting the presence of an HP1-mediated “silencing subcode” within the “histone code” (62).
Dnmt3a/b are not known to bind to specific DNA sequences, suggesting that de novo DNA methylation mediated by DNA methyltransferases is controlled by recruiting factors. Our data showed that Dnmt3a/b binding during the silencing of TNFα transcription in tolerant cells was dependent on HP1 binding, as HP1 depletion resulted in loss of Dnmt3a/b binding. HP1-mediated recruitment of Dnmt3a/b has been reported (63). Our studies and those by others support the model of histone methylation-dependent DNA methylation (1, 9, 15, 50, 63). DNA methylation silences gene expression, at least in part, through the recruitment of methyl-CpG-binding proteins to methylated DNA (19, 64, 65). One such protein, MeCP2, silences gene expression partly by recruiting histone methyltransferase (19) and histone deacetylase (HDAC) activity (64, 65), thus reinforcing a repressive chromatin state. Dnmt3a can directly interact with HDAC1 (66). Our data3 supports that MeCP2 binds to the TNFα promoter during its expression silencing in tolerant cells.
Here, we showed an increase in TNFα promoter CpG methylation in tolerant cells is associated with Dnmt3a/b binding. This event was lost upon G9a knockdown, suggesting that DNA methylation by Dnmt3a/b is a secondary event to G9a binding and H3K9 methylation. This observation supports a recent study showing increased DNA methylation at Oct-3/4 promoter via Dnmt3a/b during silencing of embryonic cells (50). In addition, overexpression of Dnmt3b commonly occurs in human tumors, where it participates in hypermethylation of tumor suppressor genes (67, 68). Two recent studies show that developmental and TLR-mediated epigenetic changes are involved in the regulation of monocyte/macrophage TNFα gene expression (69, 70).
In summary, we show a functional connection between histone and DNA methylation in silencing TNFα expression during endotoxin tolerance. We demonstrate that these two pathways interact to remodel chromatin and disrupt TNFα transcription and that H3K9 dimethylation is a primary epigenetic signal recognized by HP1 and is sufficient for initiating gene repression pathway in vivo. Histone H3K9 dimethyltransferase G9a is first recruited to the promoter, possibly by a DNA binding transcriptional repressor(s) like RelB. HP1 recruitment follows. These chromatin modifications are linked to Dnmt3a/b recruitment, with de novo methylation of promoter CpG DNA. SUV39h1 recruitment may further enhance a switch from a euchromatin to heterochromatin state. Collectively, the epigenetic events result in a switch from euchromatin to heterochromatin, which with HP1 as a bridge results in sustained and hereditable gene silencing (see Fig. 9). Such coupling of transcription, histone, and DNA-based epigenetic processes cooperatively interact to silence proinflammatory gene expression during severe systemic inflammation.
Acknowledgments
We thank Sue Cousart (our laboratory) for administrative assistance and Abdoulaye Diallo (Center for Human genomics) for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI-09169 and R01AI-065791. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HP1, heterochromatin protein 1; ChIP, chromatin immunoprecipitation; TNFα, tumor necrosis factor α; siRNA, small interfering RNA; LPS, lipopolysaccharide.
M. El Gazzar, unpublished observations.
References
- 1.Brenner, C., and Fuks, F. (2007) Dev. Cell 12 843–844 [DOI] [PubMed] [Google Scholar]
- 2.Dormann, H. L., Tseng, B. S., Allis, C. D., Funabiki, H., and Fischle, W. (2006) Cell Cycle 5 2842–2851 [DOI] [PubMed] [Google Scholar]
- 3.Hiragami, K., and Festenstein, R. (2005) Cell Mol. Life Sci. 62 2711–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig, J. M. (2005) Bioessays 27 17–28 [DOI] [PubMed] [Google Scholar]
- 5.Chin, H. G., Esteve, P. O., Pradhan, M., Benner, J., Patnaik, D., Carey, M. F., and Pradhan, S. (2007) Nucleic Acids Res. 35 7313–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patnaik, D., Chin, H. G., Esteve, P. O., Benner, J., Jacobsen, S. E., and Pradhan, S. (2004) J. Biol. Chem. 279 53248–53258 [DOI] [PubMed] [Google Scholar]
- 7.Rice, J. C., Briggs, S. D., Ueberheide, B., Barber, C. M., Shabanowitz, J., Hunt, D. F., Shinkai, Y., and Allis, C. D. (2003) Mol. Cell 12 1591–1598 [DOI] [PubMed] [Google Scholar]
- 8.Tachibana, M., Ueda, J., Fukuda, M., Takeda, N., Ohta, T., Iwanari, H., Sakihama, T., Kodama, T., Hamakubo, T., and Shinkai, Y. (2005) Genes Dev. 19 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., and Kouzarides, T. (2001) Nature 410 120–124 [DOI] [PubMed] [Google Scholar]
- 10.Riggs, A. D., and Jones, P. A. (1983) Adv. Cancer Res. 40 1–30 [DOI] [PubMed] [Google Scholar]
- 11.Valinluck, V., and Sowers, L. C. (2007) Cancer Res. 67 5583–5586 [DOI] [PubMed] [Google Scholar]
- 12.Pradhan, S., Bacolla, A., Wells, R. D., and Roberts, R. J. (1999) J. Biol. Chem. 274 33002–33010 [DOI] [PubMed] [Google Scholar]
- 13.Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999) Cell 99 247–257 [DOI] [PubMed] [Google Scholar]
- 14.Klose, R. J., and Bird, A. P. (2006) Trends Biochem. Sci. 31 89–97 [DOI] [PubMed] [Google Scholar]
- 15.Li, H., Rauch, T., Chen, Z. X., Szabo, P. E., Riggs, A. D., and Pfeifer, G. P. (2006) J. Biol. Chem. 281 19489–19500 [DOI] [PubMed] [Google Scholar]
- 16.Smallwood, A., Esteve, P. O., Pradhan, S., and Carey, M. (2007) Genes Dev. 21 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobosy, J. R., and Selker, E. U. (2001) Cell Mol. Life Sci. 58 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, P. L., Veenstra, G. J., Wade, P. A., Vermaak, D., Kass, S. U., Landsberger, N., Strouboulis, J., and Wolffe, A. P. (1998) Nat. Genet. 19 187–191 [DOI] [PubMed] [Google Scholar]
- 19.Fuks, F., Hurd, P. J., Wolf, D., Nan, X., Bird, A. P., and Kouzarides, T. (2003) J. Biol. Chem. 278 4035–4040 [DOI] [PubMed] [Google Scholar]
- 20.Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001) Nature 410 116–120 [DOI] [PubMed] [Google Scholar]
- 21.Bachman, K. E., Rountree, M. R., and Baylin, S. B. (2001) J. Biol. Chem. 276 32282–32287 [DOI] [PubMed] [Google Scholar]
- 22.Fuks, F., Hurd, P. J., Deplus, R., and Kouzarides, T. (2003) Nucleic Acids Res. 31 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermann, A., Gowher, H., and Jeltsch, A. (2004) Cell Mol. Life Sci. 61 2571–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eissenberg, J. C., and Elgin, S. C. (2000) Curr. Opin. Genet. Dev. 10 204–210 [DOI] [PubMed] [Google Scholar]
- 25.Hediger, F., and Gasser, S. M. (2006) Curr. Opin. Genet. Dev. 16 143–150 [DOI] [PubMed] [Google Scholar]
- 26.Fischle, W., Tseng, B. S., Dormann, H. L., Ueberheide, B. M., Garcia, B. A., Shabanowitz, J., Hunt, D. F., Funabiki, H., and Allis, C. D. (2005) Nature 438 1116–1122 [DOI] [PubMed] [Google Scholar]
- 27.Heo, K., Kim, B., Kim, K., Choi, J., Kim, H., Zhan, Y., Ranish, J. A., and An, W. (2007) J. Biol. Chem. 282 15476–15483 [DOI] [PubMed] [Google Scholar]
- 28.El, G. M., Yoza, B. K., Hu, J. Y., Cousart, S. L., and McCall, C. E. (2007) J. Biol. Chem. 282 26857–26864 [DOI] [PubMed] [Google Scholar]
- 29.Han, J., Brown, T., and Beutler, B. (1990) J. Exp. Med. 171 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCall, C. E., and Yoza, B. K. (2007) Am. J. Respir. Crit Care Med. 175 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Granowitz, E. V., Porat, R., Mier, J. W., Orencole, S. F., Kaplanski, G., Lynch, E. A., Ye, K., Vannier, E., Wolff, S. M., and Dinarello, C. A. (1993) J. Immunol. 151 1637–1645 [PubMed] [Google Scholar]
- 32.Vincent, J. L., and Abraham, E. (2006) Am. J. Respir. Crit Care Med. 173 256–263 [DOI] [PubMed] [Google Scholar]
- 33.Yoza, B. K., Hu, J. Y., Cousart, S. L., and McCall, C. E. (2000) Shock 13 236–243 [DOI] [PubMed] [Google Scholar]
- 34.McCall, C. E., Grosso-Wilmoth, L. M., LaRue, K., Guzman, R. N., and Cousart, S. L. (1993) J. Clin. Investig. 91 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz, C., Misset, B., Fitting, C., Bleriot, J. P., Carlet, J., and Cavaillon, J. M. (1991) Eur. J. Immunol. 21 2177–2184 [DOI] [PubMed] [Google Scholar]
- 36.Cavaillon, J. M., dib-Conquy, M., Fitting, C., Adrie, C., and Payen, D. (2003) Scand. J Infect. Dis. 35 535–544 [DOI] [PubMed] [Google Scholar]
- 37.Mathison, J. C., Virca, G. D., Wolfson, E., Tobias, P. S., Glaser, K., and Ulevitch, R. J. (1990) J. Clin. Investig. 85 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaRue, K. E., and McCall, C. E. (1994) J. Exp. Med. 180 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virca, G. D., Kim, S. Y., Glaser, K. B., and Ulevitch, R. J. (1989) J. Biol. Chem. 264 21951–21956 [PubMed] [Google Scholar]
- 40.Hawkins, G. A., Tantisira, K., Meyers, D. A., Ampleford, E. J., Moore, W. C., Klanderman, B., Liggett, S. B., Peters, S. P., Weiss, S. T., and Bleecker, E. R. (2006) Am. J. Respir. Crit. Care Med. 174 1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldfeld, A. E., Doyle, C., and Maniatis, T. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 9769–9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, M. D., Li, J., and Wong, J. (2005) Mol. Cell. Biol. 25 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aagaard, L., Laible, G., Selenko, P., Schmid, M., Dorn, R., Schotta, G., Kuhfittig, S., Wolf, A., Lebersorger, A., Singh, P. B., Reuter, G., and Jenuwein, T. (1999) EMBO J. 18 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters, A. H., Kubicek, S., Mechtler, K., O'Sullivan, R. J., Derijck, A. A., Perez-Burgos, L., Kohlmaier, A., Opravil, S., Tachibana, M., Shinkai, Y., Martens, J. H., and Jenuwein, T. (2003) Mol. Cell 12 1577–1589 [DOI] [PubMed] [Google Scholar]
- 45.Kubicek, S., O'Sullivan, R. J., August, E. M., Hickey, E. R., Zhang, Q., Teodoro, M. L., Rea, S., Mechtler, K., Kowalski, J. A., Homon, C. A., Kelly, T. A., and Jenuwein, T. (2007) Mol. Cell 25 473–481 [DOI] [PubMed] [Google Scholar]
- 46.Chan, C., Li, L., McCall, C. E., and Yoza, B. K. (2005) J. Immunol. 175 461–468 [DOI] [PubMed] [Google Scholar]
- 47.Li, B., Carey, M., and Workman, J. L. (2007) Cell 128 707–719 [DOI] [PubMed] [Google Scholar]
- 48.Boekhoudt, G. H., Guo, Z., Beresford, G. W., and Boss, J. M. (2003) J. Immunol. 170 4139–4147 [DOI] [PubMed] [Google Scholar]
- 49.Chan, G. C., Fish, J. E., Mawji, I. A., Leung, D. D., Rachlis, A. C., and Marsden, P. A. (2005) J. Immunol. 175 3846–3861 [DOI] [PubMed] [Google Scholar]
- 50.Feldman, N., Gerson, A., Fang, J., Li, E., Zhang, Y., Shinkai, Y., Cedar, H., and Bergman, Y. (2006) Nat. Cell Biol. 8 188–194 [DOI] [PubMed] [Google Scholar]
- 51.Fields, P. E., Kim, S. T., and Flavell, R. A. (2002) J. Immunol. 169 647–650 [DOI] [PubMed] [Google Scholar]
- 52.Goriely, S., Demonte, D., Nizet, S., De, W. D., Willems, F., Goldman, M., and Van, L. C. (2003) Blood 101 4894–4902 [DOI] [PubMed] [Google Scholar]
- 53.Rao, S., Gerondakis, S., Woltring, D., and Shannon, M. F. (2003) J. Immunol. 170 3724–3731 [DOI] [PubMed] [Google Scholar]
- 54.Weinmann, A. S., Plevy, S. E., and Smale, S. T. (1999) Immunity. 11 665–675 [DOI] [PubMed] [Google Scholar]
- 55.Hwang, K. K., Eissenberg, J. C., and Worman, H. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11423–11427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R. E., and Kouzarides, T. (2001) Nature 412 561–565 [DOI] [PubMed] [Google Scholar]
- 57.Fuks, F. (2005) Curr. Opin. Genet. Dev. 15 490–495 [DOI] [PubMed] [Google Scholar]
- 58.Yoza, B. K., Hu, J. Y., Cousart, S. L., Forrest, L. M., and McCall, C. E. (2006) J. Immunol. 177 4080–4085 [DOI] [PubMed] [Google Scholar]
- 59.Jacobs, S. A., and Khorasanizadeh, S. (2002) Science 295 2080–2083 [DOI] [PubMed] [Google Scholar]
- 60.Eskeland, R., Eberharter, A., and Imhof, A. (2007) Mol. Cell. Biol. 27 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maison, C., Bailly, D., Peters, A. H., Quivy, J. P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. (2002) Nat. Genet. 30 329–334 [DOI] [PubMed] [Google Scholar]
- 62.Lomberk, G., Bensi, D., Fernandez-Zapico, M. E., and Urrutia, R. (2006) Nat. Cell Biol. 8 407–415 [DOI] [PubMed] [Google Scholar]
- 63.Lehnertz, B., Ueda, Y., Derijck, A. A., Braunschweig, U., Perez-Burgos, L., Kubicek, S., Chen, T., Li, E., Jenuwein, T., and Peters, A. H. (2003) Curr. Biol. 13 1192–1200 [DOI] [PubMed] [Google Scholar]
- 64.Bird, A. P., and Wolffe, A. P. (1999) Cell 99 451–454 [DOI] [PubMed] [Google Scholar]
- 65.Wade, P. A. (2001) Oncogene 20 3166–3173 [DOI] [PubMed] [Google Scholar]
- 66.Fuks, F., Burgers, W. A., Godin, N., Kasai, M., and Kouzarides, T. (2001) EMBO J. 20 2536–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paz, M. F., Wei, S., Cigudosa, J. C., Rodriguez-Perales, S., Peinado, M. A., Huang, T. H., and Esteller, M. (2003) Hum. Mol Genet. 12 2209–2219 [DOI] [PubMed] [Google Scholar]
- 68.Robertson, K. D. (2001) Oncogene 20 3139–3155 [DOI] [PubMed] [Google Scholar]
- 69.Foster, S. L., Hargreaves, D. C., and Medzhitov, R. (2007) Nature 447 972–978 [DOI] [PubMed] [Google Scholar]
- 70.Sullivan, K. E., Reddy, A. B., Dietzmann, K., Suriano, A. R., Kocieda, V. P., Stewart, M., and Bhatia, M. (2007) Mol. Cell. Biol. 27 5147–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]