Abstract
We determined the effect of the N-terminal histone tails on nucleosome traversal by yeast and human RNA polymerase II (pol II). Removal of H2A/H2B tails, H3/H4 tails, or all tails increased complete traversal of the nucleosome by human pol II, although the increase varied considerably depending on the template and on which tails were removed. Human pol II achieved >80% traversal of one nucleosomal template lacking the H2A/H2B tails, but even in those reactions, the transcript elongation rate was lower than the rate on pure DNA templates. For yeast pol II, transcription proceeded much farther into the nucleosome in the absence of tails, but complete read-through was not substantially increased by tail removal. Transcription factor IIS provided roughly the same level of read-through stimulation for transcript elongation in the presence or absence of tails. FACT also stimulated elongation on nucleosomal templates, and this effect was similar regardless of the presence of tails. For both polymerases, removal of the H2A/H2B tails reduced pausing throughout the nucleosome, suggesting that histone tails affect a common step at most points during nucleosome traversal. We conclude that histone tails provide a significant part of the nucleosomal barrier to pol II transcript elongation.
It has long been appreciated that nucleosomes form a strong blockade to transcript elongation by pol II in vitro (1, 2). It has not been established what role, if any, the N-terminal tails of the histones play in this blockade. The core structure of the nucleosome does not depend on the tails (3–7). However, the tails are strongly positively charged, and they could associate nonspecifically with the DNA, thereby impeding polymerase access to the template (8–10). The tails could influence traversal in other ways, e.g. by affecting the ability of the H2A/H2B dimer to exchange, which is likely to be involved in at least some traversal events (2). The N-terminal tails could also affect more complex unfolding transitions in the nucleosome, which would facilitate traversal (11, 12). Higher order chromatin structure, which could affect the efficiency of transcription through chromatin, can be influenced by the N-terminal tails (13–16). Covalent modifications of the tails, especially of the H3 tail of promoter-proximal nucleosomes, are well known to be correlated with transcriptional activity in vivo (reviewed in Refs. 17–19). A recent survey of transcriptionally active human genes revealed that most of these genes contain a high level of RNA polymerase II (pol II)3 immediately downstream of the transcription start and a strongly positioned nucleosome with a promoter-proximal edge at about position +40 (20). This suggests that the ability of pol II to cross the first nucleosome it encounters after transcript initiation is a major and general control point for gene expression. In light of all of these findings, we decided to explore directly the effect of the histone tails on the ability of pol II to traverse single nucleosomes. As an initial test of the effect of the tails, we assembled nucleosomes that lack either subsets of the tails or all of the tails. We found that the nucleosomal barrier to transcript elongation by pol II is lowered but not eliminated by tail removal.
EXPERIMENTAL PROCEDURES
Protein Purification—Yeast pol II with a hexahistidine-tagged RPB3 subunit was purified as described (2). Full-length recombinant hexahistidine-tagged yeast transcription factor (TF) IIS was expressed and purified as described (21). For transcription studies with yeast pol II, native core histones were purified from chicken erythrocytes (22). Trypsinized chicken core histones were obtained using a very similar protocol. In brief, H1-stripped chromatin was treated with trypsin as described (23), and trypsinized H2A/H2B dimers and H3/H4 tetramers were purified on a hydroxylapatite column (22). Recombinant human FACT (a heterodimeric factor that facilitates chromatin transcription) was kindly provided by Dr. D. Reinberg.
Human pol II and TFIIH were purified from HeLa cell nuclear extracts, and recombinant human TATA box-binding protein, TFIIB, TFIIE, TFIIF, and TFIIS were prepared as described (24). Plasmids expressing native and N-terminally deleted (tailless) Xenopus histones were kindly provided by Dr. Karolin Luger. Tailless histones contained only the trypsin-resistant globular domains, consisting of amino acids 13–118 for H2A, amino acids 24–122 for H2B, amino acids 27–135 for H3, and amino acids 20–102 for H4 (25). Expression of recombinant histones and preparation of histone dimers, tetramers, and octamers were performed as described (26, 27).
DNA Templates, Nucleosome Assembly, and Transcription Complex Assembly—The 603 nucleosome positioning sequence was originally identified by Lowary and Widom (28); the exact sequence of the 603 segment used in these studies is given in supplemental Fig. S1. Nucleosome assembly and transcription by yeast pol II were conducted as described (24). In short, mononucleosomes were reconstituted on DNA fragments bearing the 603 or 603R elements by decreasing salt dialysis using different combinations of intact or trypsinized chicken core histones. (Note that 603 and 603R are identical sequences; they were constructed to ligate to the transcription complex in opposite orientations.) Yeast pol II elongation complexes were assembled on a 50-bp DNA fragment from 9-nucleotide RNA and separate template and non-template strands. These complexes were immobilized on nickel-nitrilotriacetic acid-agarose beads, washed, eluted from the beads, and ligated to nucleosomal templates or the equivalent non-nucleosomal DNA. pol II was advanced to position +45 using [α-32P]NTPs to label the RNA. Transcription was resumed from position +45 by addition of a large excess of unlabeled NTPs in the presence of KCl at the concentrations indicated in the figures and figure legends. FACT (∼0.15 μm) or yeast TFIIS (0.1 μm) was added to the +45 complexes as indicated in the figures; factor additions were made along with the chase NTPs. Transcript elongation reactions with yeast pol II were normally run for 10 min except as indicated for the reactions in Fig. 5.
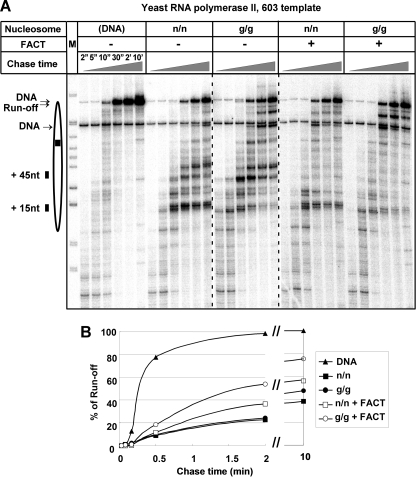
FIGURE 5.
FACT increases the rate of traversal of the 603 nucleosomal template by yeast pol II. A, 603 nucleosomes containing (n/n) or lacking (g/g) all histone tails were transcribed at 150 mm KCl in the presence or absence of FACT for the indicated time intervals. Labeled non-ligated (lower band) and ligated (upper band) DNA loading controls are also indicated. The other designations are the same as described in the legend to Fig. 2A. nt, nucleotides. B, the levels of run-off transcript, normalized to the DNA-only reaction, are given for the time courses shown in A. Note that the rate of transcript elongation on nucleosomal templates in the presence of FACT is lower than the rate on free DNA, even in the absence of the histone tails.
For studies with human pol II, 250-bp template fragments containing the adenovirus major late promoter upstream and either the 603 or 603R assembly element downstream were used (24). These constructs had been cloned into pUC18, and the working templates were amplified by PCR with the upstream primer biotinylated. The 603 and 603R templates were assembled into nucleosomes by salt dialysis, bound to streptavidin beads, and then assembled into pol II preinitiation complexes with purified pol II and TFIIH and recombinant TATA box-binding protein, TFIIB, TFIIE, and TFIIF as described (24). Human preinitiation complexes were advanced to position +20 using labeled NTPs, washed, and then chased for 5 min with excess NTPs as described (24). Some reactions were supplemented with human TFIIS at 24 μg/ml during the chase as indicated in the figures. For the kinetic experiments in Fig. 4 and supplemental Fig. S3, the published protocol was slightly modified. Preinitiation complexes were incubated with 0.25 mm CpA, 100 μm UTP, 0.7 μm [α-32P]CTP, and 50 μm dATP at 30 °C for only 1 min (with no addition of unlabeled CTP) and washed only once. This procedure yields complexes with 21-nucleotide RNAs as well as complexes with shorter transcripts. These complexes were chased for the times indicated in Fig. 4 and supplemental Fig. S3. For both yeast and human transcription reactions, labeled transcripts were resolved by electrophoresis on denaturing gels and quantified using a Storm Imager and ImageQuant software.
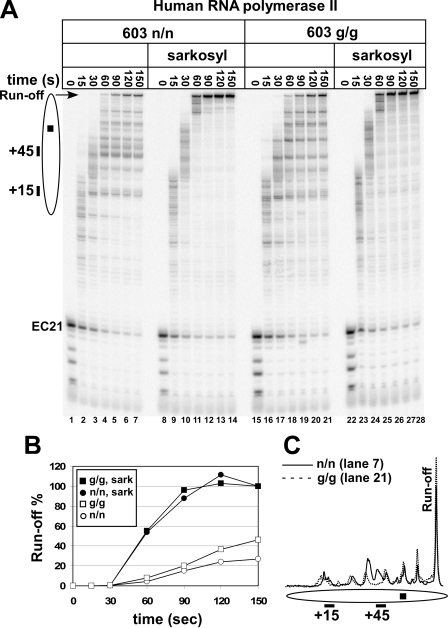
FIGURE 4.
Removal of the histone tails increases the rate of traversal of the 603 nucleosomal template by human pol II. A, human pol II transcription complexes with 21-nucleotide (and shorter) transcripts on 603 n/n or 603 g/g nucleosomal templates were chased in 150 mm KCl for the indicated times in the presence or absence of Sarkosyl. The other designations are the same as described in the legend to Fig. 1A. B, the levels of run-off transcript, normalized to the plus-Sarkosyl (sark) reactions, are given for the time courses shown in A. Note that the rate of transcript elongation on the tailless templates is lower than the rate on the histone-free DNA templates. C, the relative distribution of transcripts paused within the nucleosome, or run-off, is shown for the 150-s reactions on the n/n (A, lane 7) and g/g (A, lane 21) templates.
RESULTS
We measured the extent to which either yeast or human pol II can cross one downstream nucleosome using an approach based on our earlier study (24). Nucleosomes were reconstituted over sequence elements that direct nucleosome assembly to single, precise locations. Yeast pol II transcript elongation complexes were assembled using DNA/RNA hybrid scaffolds and then ligated to short DNA fragments bearing preassembled nucleosomes. Human pol II preinitiation complexes were obtained by incubating the general pol II transcript initiation factors and templates bearing a promoter and a single downstream nucleosome. In both cases, transcription was initiated with limiting, labeled NTPs, and pol II was stalled before reaching the nucleosome. The complexes were then chased with excess unlabeled NTPs. We used the 603 nucleosome assembly sequence (28, 29) for the studies reported here. We showed previously that the same nucleosome can present a very different barrier height depending on the transcriptional orientation (24). Based on our earlier observations (24), one transcriptional orientation (designated 603) was expected to be much more permissive for transcript elongation than the other (designated 603R).
We began our study with the simple question: will the complete absence of some or all of the N-terminal tails have a significant effect on nucleosome traversal by pol II? We assembled nucleosomes with four different combinations of histones: all histones full-length (native H2A and H2B/native H3 and H4, abbreviated “n/n”), no H2A/H2B tails (globular H2A and H2B/native H3 and H4, abbreviated “g/n”), no H3/H4 tails (native H2A and H2B/globular H3 and H4, abbreviated “n/g”), and no tails on any of the histones (globular H2A and H2B/globular H3 and H4, abbreviated “g/g”). For experiments with human pol II, recombinant Xenopus histones were used. Histones for yeast pol II experiments were purified from H1-stripped chicken erythrocyte chromatin after treatment with trypsin, followed by purification of H2A/H2B dimers and H3/H4 tetramers. Preliminary tests showed that histones from either source were interchangeable. Human pol II gave identical results when transcribing nucleosomal templates assembled with recombinant tailless Xenopus histones or with trypsin-truncated chicken histones. Similarly, transcription results with yeast pol II were the same regardless of the source of tailless histones (data not shown). Transcription reactions were performed at low (40 mm), roughly physiological (150 mm), or high (300 mm) KCl concentrations; maximal traversal was obtained by removing the nucleosomes with 1% Sarkosyl (human pol II) or 1 m KCl (yeast pol II). In all cases, we normalized traversal in any given condition to traversal after nucleosome removal.
As we observed in our initial study (24), the majority of human pol II complexes failed to completely traverse the 603 nucleosome in 5-min reactions at low or moderate salt. Most of the paused complexes resided in the proximal half of the nucleosome (Fig. 1A, lanes 1 and 2). This pausing was only partially relieved by raising the KCl concentration to 300 mm (Fig. 1A, lane 3; quantitative results shown in Fig. 1B). Full traversal increased, but remained incomplete, when the pol II transcript elongation factor TFIIS was included in the reactions (Fig. 1A, lanes 6 and 7). Removal of some or all of the tails led to increased traversal of the 603 nucleosome at all salt concentrations (Fig. 1, A, lanes 8–10; and B). Most of the effect of tail removal on traversal of the 603 nucleosome by human pol II, particularly at higher ionic strengths, was obtained by removing only the H2A/H2B tails (Fig. 1B). Removal of either the H2A/H2B or H3/H4 tails did not cause specific changes in the pausing pattern but apparently slightly reduced the tendency of pol II to pause at most positions along nucleosomal DNA, leading to an increase in complete traversal. The effect of tail removal was similar to the effect of increasing the concentration of monovalent ions. TFIIS stimulated traversal of 603 templates assembled with partially or completely tailless histones at 40 mm KCl. TFIIS stimulation was not synergistic with tail removal, i.e. the -fold increase provided by TFIIS was not significantly different for nucleosomes with all tails present and for those that lacked some or all tails.
FIGURE 1.
Removal of core histone tails results in increased traversal of both the 603 and 603R templates by human pol II. A, 603 (permissive) and 603R (nonpermissive) nucleosomal templates containing all histone tails (n/n) or missing H2A/H2B (g/n) or all (g/g) tails were transcribed at the indicated concentrations of KCl in the presence or absence of TFIIS. Sarkosyl (Sark.) was added to the indicated lanes after the initial pause at U21 (1st) or after an initial 5-min chase (2nd); in the latter case, chase was continued for 5 more min. The position of the nucleosome on the template is indicated by the oval, and the nucleosomal dyad is shown as a black square. The locations of the run-off RNA and the major pause sites at 15 and 45 bp within the nucleosome are indicated. B, the graphs show the levels of nucleosome traversal by human pol II on native, partially tailless, and fully tailless 603 and 603R templates under the indicated conditions in the presence or absence of TFIIS. All results were normalized to the level of run-off at 40 mm KCl when Sarkosyl was added to the U21 complexes prior to chase. Each bar indicates the average of two to five separate experiments; error bars indicate the mean ± S.D.
The 603R nucleosomal template provided a much stronger and more discrete barrier to transcription by human pol II in comparison with the 603 template, as expected from our earlier work (24). Most polymerases paused at a group of sites 45–55 bp within the 603R nucleosome, at the entry point into the H3/H4 tetramer, whereas the small fraction that successfully crossed this barrier proceeded to the end of the nucleosome (Fig. 1A, lanes 15–17 and 22–24). Tail removal generally increased traversal on 603R by human pol II (Fig. 1B). At low salt, pol II could advance somewhat farther into the major 603R transcriptional barrier in the absence of tails (Fig. 1A, compare the +45–55 region in lanes 15 and 22). As with the 603 template, this resembles the effect of increasing the ionic strength of the reaction.
We performed comparable experiments using yeast pol II and partially or completely tailless nucleosomes. As expected (24), yeast pol II traversed each nucleosome less efficiently than the human enzyme. Results for the 603 (permissive) template are shown in Fig. 2 (A and B). Apart from a small stimulation of the very low traversal level at 40 mm salt, tail removal did not significantly increase complete traversal of the 603 nucleosome. It is important to note, however, that pausing consistently occurred farther into 603 nucleosomes lacking some or all tails; this effect was most pronounced at 40 mm KCl (Fig. 2A, compare, for example, lanes 1 and 7). The effect of tail removal on progression of yeast pol II through the nucleosome was roughly additive and was not dominated by the H2A/H2B tails. The primary effect of tail removal was relief of pausing at the major site 15 bases within the nucleosome. As a secondary effect of tail removal, pausing shifted to more downstream locations, but novel pause sites were not generally observed. The effect of tail removal roughly mirrored the effect of adding KCl to the reactions and resulted in qualitatively similar changes in the pausing pattern upon removal of different combinations of tails; compare, for example, the 40 and 150 mm KCl lanes for the control (Fig. 2A, lanes 1 and 2) and fully tailless (lanes 7 and 8) nucleosomes. TFIIS increased traversal of partially or completely tailless 603 nucleosomes, particularly at low salt, but as with the human polymerase, the -fold increase was not significantly different in the presence or absence of histone tails.
FIGURE 2.
Removal of core histone tails relieves pausing at position +15 and allows further progression by yeast pol II on the 603 nucleosomal template. A, 603 nucleosomes containing all histone tails (n/n) or missing various combinations of the tails (g/g, g/n, and n/g) were transcribed at the indicated concentrations of KCl in the presence or absence of TFIIS. Lane M contained end-labeled, MspI-digested pBR322 as size markers. The other designations are the same as described in the legend to Fig. 1A. nt, nucleotides. B, the graphs show the levels of nucleosome traversal by yeast pol II on native, partially tailless, or fully tailless 603 and 603R templates under the indicated conditions in the presence or absence of TFIIS or FACT. All results were normalized to the level of run-off at 1 m KCl. Each bar indicates the average of two to five separate experiments; error bars indicate the mean ± S.D.
The nucleosomal 603R template provided a very strong barrier for yeast pol II at low or moderate salt (Fig. 2B and supplemental Fig. S2). Most pausing on 603R occurred 45–55 bases into the nucleosome (supplemental Fig. S2), as seen with human pol II. Traversal of the 603R nucleosome by yeast pol II was very slightly increased by removing the H2A/H2B tails, whereas removal of the H3/H4 tails had no apparent effect on traversal (Fig. 2B). However, the nucleosomal pauses upstream of the +45 nucleosomal region were relieved to different degrees by removal of different combinations of tails, as seen with the 603 template. The reduction in upstream pausing upon tail removal contributed to increased pausing at position +45. (In supplemental Fig. S2, compare the relative level of +15 to +45 pausing on the n/n template with the same ratio on the three partially or completely tailless templates.) Tail removal did not reveal novel pause sites downstream of the major +45 stop on the 603R template. The effect of the loss of H2A/H2B tails for yeast pol II was made more apparent in the context of traversal stimulation by TFIIS, particularly at 150 mm KCl (Fig. 2B).
The weak effect of tail removal on traversal of nonpermissive 603R nucleosomes by yeast pol II is at least partially explained by the exceptional tendency of the yeast polymerase to arrest at position +45 even on histone-free DNA templates (supplemental Fig. S2, 1 m KCl lanes). Assembly of the 603R DNA sequence into a nucleosome greatly exaggerates the arrest at position +45. The strong barrier to elongation at this location is apparently based on the unusually rapid rate of backtracking upon arrest at this site. Kireeva et al. (30) found that backtracking by yeast pol II after pausing at most locations within a nucleosome has a t½ of almost 1 h. In contrast, we have observed that backtracking at position +45 on 603R nucleosome is completed in <1 min (data not shown). Given this very high likelihood of backtracking, many rounds of transcript cleavage and resynthesis mediated by TFIIS would still be insufficient to allow a majority of polymerases to cross the +45 site, as we observed (supplemental Fig. S2). Strong DNA-specific arrest of human pol II was not detected at the +45 region of the 603R template (Fig. 1A, lanes 18 and 25). This suggests a slower rate of backtracking by human pol II at the +45 site in the 603R nucleosome, but it is important to note that we have not measured backtracking rates for the human enzyme.
Complete traversal of the nucleosome by pol II is generally accompanied by the loss of one of the H2A/H2B dimers (2). This suggests that the increase in traversal seen with N-terminal tail removal could be based, at least in part, on an increased tendency of H2A/H2B dimers to be lost when tails are absent. The FACT complex facilitates nucleosome traversal at normal salt concentrations and is thought to act by functioning as a chaperone for the H2A/H2B dimer (31). We therefore tested the possibility that tail removal might increase the effectiveness of FACT in mediating nucleosome traversal by yeast pol II. As shown in Fig. 3 (quantitative results in Fig. 2B), FACT strongly stimulated traversal of both the 603 and 603R nucleosomes by yeast pol II, but this effect did not depend on the histone tails. In most cases, there was no further increase in FACT-mediated traversal as a result of tail removal; the apparent increase at 40 mm KCl on the 603 template resulted from the increase in traversal of that nucleosome due to tail removal alone, in the absence of FACT. These results do not support the idea that the stimulation of traversal by tail removal results from an increased tendency of the tailless nucleosomes to lose an H2A/H2B dimer.
FIGURE 3.
Stimulation of yeast pol II traversal of the 603 nucleosome by TFIIS or FACT is largely independent of the histone tails. Nucleosomal templates containing (n/n) or missing (g/g) all histone tails were transcribed at the indicated concentrations of KCl in the presence or absence of TFIIS or FACT. The other designations are the same as described in the legend to Fig. 2A. Quantitation of the effect of the elongation factors is shown in Fig. 2B. nt, nucleotides.
As noted above, under some conditions, human pol II can achieve near-complete nucleosome traversal in our standard 5-min transcription reaction, particularly at 150 mm KCl in the absence of histone tails (Fig. 1B). This suggested that the rates of transcript elongation on DNA and the tailless templates could be comparable. To test this idea, transcript elongation rates were analyzed at 150 mm salt for the human enzyme on 603 DNA and on native and tailless nucleosomal 603 templates. When the 603 nucleosome was removed by Sarkosyl, some human pol II complexes generated 200-nucleotide run-off RNA in 1 min. Essentially all polymerases had run-off in 90 s (Fig. 4A). Elongation was much slower on the nucleosomal template with all tails present (Fig. 4, A and B). Significantly, in the complete absence of tails, the rate of accumulation of run-off was increased by ∼2-fold in comparison with the fully tailed templates; however, the rate on the tailless templates was still considerably slower than that on free DNA (Fig. 4B). Comparison of lanes 4–7 and 18–21 in Fig. 4A suggests that the more rapid rate of run-off accumulation was accompanied by a downstream shift in the population of paused polymerases, such that on the tailless templates, the longer paused products predominated. This is shown quantitatively through a comparison of scans of lanes 7 and 21 (Fig. 4C). Addition of TFIIS also caused an increase in the rate of full traversal and a downstream shift in the population of paused complexes (supplemental Fig. S3). The results in Fig. 4 and supplemental Fig. S3 suggest that tail removal and TFIIS addition increase nucleosome traversal by reducing the tendency of pol II to pause at multiple, nucleosome-induced sites. TFIIS helps to maintain pol II in a catalytically active state (32, 33), whereas tail removal presumably reduces the lifetime of nucleosome-induced pauses. However, even under conditions that effectively minimized pausing, the rate of transcript elongation by human pol II on a nucleosomal template remained 2–3-fold lower than the rate that could be achieved on the same DNA sequence in the absence of a nucleosome (Fig. 4B).
We obtained further insight into the mechanistic basis of FACT stimulation of nucleosome traversal by examining the kinetics of transcript elongation through the 603 nucleosome by yeast pol II as a function of FACT and the nucleosome tails (Fig. 5). FACT increased the rate of traversal of both tail-containing and tailless nucleosomes. This increase was ∼1.5-fold for the n/n template and 2-fold for the g/g template as judged by the 2-min time points in Fig. 5. FACT strongly reduced pausing throughout the body of the nucleosome, in contrast to tail removal, which primarily shifted pausing to downstream locations. The sum of the two effects can be seen by comparing the n/n and g/g lanes in the presence of FACT: on both tailed and tailless templates, there was relatively little pausing observed over most of the central segment of the nucleosome; but in the absence of tails, more polymerases were paused in the distal segment of the nucleosome, and fewer were paused near the entry into the nucleosome (Fig. 5A). These results suggest that FACT alters the entire nucleosomal barrier to transcript elongation, as opposed to simply reducing pausing at internal locations. This is consistent with the proposal that FACT functions to mediate removal of one of the H2A/H2B dimers as transcription proceeds (31). FACT allowed high levels of traversal of the 603 nucleosome in reactions of 2 min or longer, particularly in the absence of tails (Fig. 5B; see also Fig. 2B). However, as we observed with TFIIS and human pol II, the rate of transcription on the nucleosomal template for yeast pol II remained much lower than the rate on free DNA.
DISCUSSION
Removal of the N-terminal histone tails resulted in facilitated transcription through a nucleosome by both yeast and human pol II, an effect most noticeable at lower ionic strength (40 mm KCl). For human pol II, tail removal reduced nucleosome-specific pausing over the entire length of the 603 nucleosome, leading to both increased traversal and the accumulation of complexes paused at more distal locations (see Fig. 4C). Removal of the H2A/H2B tails had the strongest impact for the human polymerase. In the case of yeast pol II, tail removal did not lead to a significant increase in full traversal of either nucleosomal template. However, removal of the H2A/H2B or H3/H4 tails significantly reduced the strong barrier that the yeast polymerase encounters ∼15 bases within the 603 nucleosome. The effects of removing either pair of tails on yeast pol II pausing at position +15 on the 603 template were roughly additive. Increased traversal of downstream pause sites by the yeast enzyme was similarly facilitated by removal of some or all of the histone tails.
How could deletion of only one pair of tails affect pausing throughout the nucleosome? Reduction in pausing due to tail removal was most pronounced at lower ionic strength, mimicking the results seen when additional KCl was added to the reactions. This suggests that electrostatic tail-DNA interactions contribute to the inhibiting effect of tails on transcript elongation. Given the lengths of the H3 and H2B N-terminal tails in particular, removal of either the H2A/H2B or H3/H4 tails could conceivably affect progress by the polymerase through the entire nucleosome.
In addition to the global effect, the tails had rather specific effects that differed for the two polymerases. The very strong tendency of yeast pol II to pause ∼15 bases into the nucleosome was responsive to removal of either pair of tails. In this context, it is useful to note two recent studies on the effect of tail removal on nucleosome structure and stability. Experiments with single nucleosomes demonstrated that removal of either set of tails facilitated the unwrapping of ∼30 bp of DNA from the ends of the nucleosome (34). This effect was greater for H3/H4 tail removal, perhaps because the H3 tails emerge from the nucleosome near the entry and exit points for the wrapped DNA (3). In a different study, Bertin et al. (35) concluded that unwrapping DNA from the region bound by the H2A/H2B dimers is facilitated by removal of the H2A/H2B tails. These results support the idea that, in addition to any simple “coating” of the DNA, both sets of tails can control the stability of the H2A/H2B dimer-DNA interaction. Thus, the absence of either pair of tails would be expected to reduce pausing early in nucleosome traversal, e.g. at position +15.
Reducing the major +15 pause by tail removal did not cause a significant increase in complete nucleosome traversal by yeast pol II, presumably because strong pausing downstream (particularly at the +45 region) continued to capture nearly all of the yeast polymerases before traversal was complete. In contrast, TFIIS addition not only increased the ability of yeast pol II to pass the +15 barrier but also facilitated complete traversal of the 603 nucleosome. FACT also stimulated full traversal at low salt, but it did so by a different mechanism. Addition of FACT almost eliminated pausing by yeast pol II at most locations over the central segment of the 603 nucleosome (Fig. 5). This result is consistent with the observations of Rhoades et al. (36), who showed that FACT-nucleosome interaction increases the accessibility of the nucleosomal DNA primarily at internal locations. It is also consistent with previous observations on the effect of FACT on transcription in vitro (31).
Human pol II also recognizes the +15 pause, particularly on the 603 template, but the major pause site for the human enzyme occurs at 45–55 bases into the nucleosome on both 603 and 603R. Although this location corresponds to the point at which the polymerase enters the segment of the nucleosome organized by the H3/H4 tetramer, under most conditions, the +45 pause was more sensitive to the loss of H2A/H2B tails than to the loss of the H3/H4 tails. Nucleosome-unwinding experiments (34) have identified two distinct transitions as DNA is removed from the surface of the histone octamer: an initial unwrapping of the DNA segments primarily associated with the H2A/H2B dimers and a second transition in which the DNA 45–55 bp into the nucleosome disassociates from the underlying histones. Like the corresponding reduction in the transcriptional barrier, this latter transition was most strongly influenced by removal of the H2A/H2B dimer tails (34). Destabilization of the H3/H4-DNA interactions resulting from removal of the H2A/H2B tails could have resulted in facilitated passage through the +45 barrier by human pol II.
The effects of tail removal on nucleosome traversal can be rationalized for both yeast and human pol II based on changes in nucleosome stability upon tail removal. However, the tails are clearly not the entire basis of the nucleosomal transcription barrier. Even in the complete absence of tails, transcript elongation rates on nucleosomal templates are much lower than those seen on free DNA. Widom and co-workers (37) have recently determined that DNA spontaneously releases from the nucleosome surface and then rapidly rebinds; rewrapping occurs in only 10–50 ms. During nucleosome traversal, pol II may spend much of its time waiting for the downstream template to be made available by this spontaneous “breathing” of DNA away from the histone octamer. If the rate at which transcription resumes after pausing is much slower than the rate at which the window of template availability closes, then overall transcript elongation rates on nucleosomal templates must be considerably slower than elongation rates on free DNA. It is worth noting that, under the conditions used in this study, elongation rates for either yeast or human pol II did not exceed 200–300 nucleotides/min, strongly suggesting that inefficiency in exploiting the temporary availability of template DNA is an important aspect of the overall slow transcript elongation rate on nucleosomal templates. This also explains why TFIIS addition and tail removal can be additive for traversal by human pol II: tail removal could slow the reassociation of DNA with the nucleosome surface, and TFIIS can increase the likelihood of bond formation; but both may still leave the effective rate of bond formation well below the average rate of reassociation of DNA with the nucleosome surface.
Supplementary Material
Acknowledgments
We thank Guohong Li and Danny Reinberg for purified human FACT; Caroline Kane for the TFIIS-expressing pET15bHMK plasmid; and Karolin Luger and colleagues for plasmids expressing the Xenopus histones, advice on histone expression and purification, and an initial gift of purified tailless histones. HeLa cells used for preparation of human RNA polymerase II and TFIIH were provided by the United States National Cell Culture Center.
This work was supported, in whole or in part, by National Institutes of Health Grant GM58650 (to V. M. S.). This work was also supported by National Science Foundation Grants 0646019 and 0743298 (to D. S. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: pol II, RNA polymerase II; TF, transcription factor.
References
- 1.Izban, M. G., and Luse, D. S. (1991) Genes Dev. 5 683–696 [DOI] [PubMed] [Google Scholar]
- 2.Kireeva, M. L., Walter, W., Tchernajenko, V., Bondarenko, V., Kashlev, M., and Studitsky, V. M. (2002) Mol. Cell 9 541–552 [DOI] [PubMed] [Google Scholar]
- 3.Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997) Nature 389 251–260 [DOI] [PubMed] [Google Scholar]
- 4.Ausio, J., Dong, F., and van Holde, K. E. (1989) J. Mol. Biol. 206 451–463 [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Ramirez, M., Dong, F., and Ausio, J. (1992) J. Biol. Chem. 267 19587–19595 [PubMed] [Google Scholar]
- 6.Morse, R. H., and Cantor, C. R. (1986) Nucleic Acids Res. 14 3293–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Widlund, H. R., Vitolo, J. M., Thiriet, C., and Hayes, J. J. (2000) Biochemistry 39 3835–3841 [DOI] [PubMed] [Google Scholar]
- 8.Hill, C. S., and Thomas, J. O. (1990) Eur. J. Biochem. 187 145–153 [DOI] [PubMed] [Google Scholar]
- 9.Lee, K. M., and Hayes, J. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8959–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng, C. Y., and Hayes, J. J. (2003) J. Biol. Chem. 278 24217–24224 [DOI] [PubMed] [Google Scholar]
- 11.Sivolob, A., De Lucia, F., Alilat, M., and Prunell, A. (2000) J. Mol. Biol. 295 55–69 [DOI] [PubMed] [Google Scholar]
- 12.Bancaud, A., Wagner, G., CondeeSilva, N., Lavelle, C., Wong, H., Mozziconacci, J., Barbi, M., Sivolob, A., LeCam, E., Mouawad, L., Viovy, J. L., Victor, J. M., and Prunell, A. (2007) Mol. Cell 27 135–147 [DOI] [PubMed] [Google Scholar]
- 13.Fletcher, T. M., and Hansen, J. C. (1995) J. Biol. Chem. 270 25359–25362 [DOI] [PubMed] [Google Scholar]
- 14.Tse, C., and Hansen, J. C. (1997) Biochemistry 36 11381–11388 [DOI] [PubMed] [Google Scholar]
- 15.Carruthers, L. M., and Hansen, J. C. (2000) J. Biol. Chem. 275 37285–37290 [DOI] [PubMed] [Google Scholar]
- 16.Kan, P. Y., Lu, X., Hansen, J. C., and Hayes, J. J. (2007) Mol. Cell. Biol. 27 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischle, W., Wang, Y. M., and Allis, C. D. (2003) Curr. Opin. Cell Biol. 15 172–183 [DOI] [PubMed] [Google Scholar]
- 18.Mellor, J. (2006) Trends Genet. 22 320–329 [DOI] [PubMed] [Google Scholar]
- 19.Kouzarides, T. (2007) Cell 128 693–705 [DOI] [PubMed] [Google Scholar]
- 20.Schones, D. E., Cui, K. R., Cuddapah, S., Roh, T. Y., Barski, A., Wang, Z. B., Wei, G., and Zhao, K. J. (2008) Cell 132 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Awrey, D. E., Weilbaecher, R. G., Hemming, S. A., Orlicky, S. M., Kane, C. M., and Edwards, A. M. (1997) J. Biol. Chem. 272 14747–14754 [DOI] [PubMed] [Google Scholar]
- 22.Studitsky, V. M. (1999) Methods Mol. Biol. 119 17–26 [DOI] [PubMed] [Google Scholar]
- 23.Hayes, J. J., Clark, D. J., and Wolffe, A. P. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 6829–6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bondarenko, V. A., Steele, L. M., Újvári, A., Gaykalova, D. A., Kulaeva, O. I., Polikanov, Y. S., Luse, D. S., and Studitsky, V. M. (2006) Mol. Cell 24 469–479 [DOI] [PubMed] [Google Scholar]
- 25.Luger, K., Rechsteiner, T. J., Flaus, A. J., Waye, M. M. Y., and Richmond, T. J. (1997) J. Mol. Biol. 272 301–311 [DOI] [PubMed] [Google Scholar]
- 26.Dyer, P. N., Edayathumangalam, R. S., White, C. L., Bao, Y. H., Chakravarthy, S., Muthurajan, U. M., and Luger, K. (2004) Methods Enzymol. 375 23–44 [DOI] [PubMed] [Google Scholar]
- 27.Luger, K., Rechsteiner, T. J., and Richmond, T. J. (1999) Methods Enzymol. 304 3–19 [DOI] [PubMed] [Google Scholar]
- 28.Lowary, P. T., and Widom, J. (1998) J. Mol. Biol. 276 19–42 [DOI] [PubMed] [Google Scholar]
- 29.Thåström, A., Lowary, P. T., Widlund, H. R., Cao, H., Kubista, M., and Widom, J. (1999) J. Mol. Biol. 288 213–229 [DOI] [PubMed] [Google Scholar]
- 30.Kireeva, M. L., Hancock, B., Cremona, G. H., Walter, W., Studitsky, V. M., and Kashlev, M. (2005) Mol. Cell 18 97–108 [DOI] [PubMed] [Google Scholar]
- 31.Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M., and Reinberg, D. (2003) Science 301 1090–1093 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, C. F., Yan, H. G., and Burton, Z. F. (2003) J. Biol. Chem. 278 50101–50111 [DOI] [PubMed] [Google Scholar]
- 33.Kettenberger, H., Armache, K.-J., and Cramer, P. (2004) Mol. Cell 16 955–965 [DOI] [PubMed] [Google Scholar]
- 34.Brower-Toland, B., Wacker, D. A., Fulbright, R. M., Lis, J. T., Kraus, W. L., and Wang, M. D. (2005) J. Mol. Biol. 346 135–146 [DOI] [PubMed] [Google Scholar]
- 35.Bertin, A., Durand, D., Renouard, M., Livolant, F., and Mangenot, S. (2007) Eur. Biophys. J. 36 1083–1094 [DOI] [PubMed] [Google Scholar]
- 36.Rhoades, A. R., Ruone, S., and Formosa, T. (2004) Mol. Cell. Biol. 24 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, G., Levitus, M., Bustamante, C., and Widom, J. (2005) Nat. Struct. Mol. Biol. 12 46–53 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.