Abstract
The huntingtin-interacting protein family members (Hip1 and Hip1R in mammals and Sla2p in yeast) link clathrin-mediated membrane traffic to actin cytoskeleton dynamics. Genetic data in yeast have implicated the light chain subunit of clathrin in regulating this link. To test this hypothesis, the biophysical properties of mammalian Hip1 and Hip1R and their interaction with clathrin light chain and actin were analyzed. The coiled-coil domains (clathrin light chain-binding) of Hip1 and Hip1R were found to be stable homodimers with no propensity to heterodimerize in vitro. Homodimers were also predominant in vivo, accounting for cellular segregation of Hip1 and Hip1R functions. Coiled-coil domains of Hip1 and Hip1R differed in their stability and flexibility, correlating with slightly different affinities for clathrin light chain and more markedly with effects of clathrin light chain binding on Hip protein-actin interactions. Clathrin light chain binding induced a compact conformation of both Hip1 and Hip1R and significantly reduced actin binding by their THATCH domains. Thus, clathrin is a negative regulator of Hip-actin interactions. These observations necessarily change models proposed for Hip protein function.
Regulation of membrane traffic depends on molecules that link membrane vesicle formation to components of the actin cytoskeleton. Hip1 (huntingtin-interacting protein 1) and its homolog Hip1R (Hip1-related), which both bind clathrin and actin, are implicated in a functional connection between the two. Hip1 and Hip1R have the same domain structure, consisting of an N-terminal phospholipid binding domain (ANTH), a central coiled-coil, and a C-terminal actin-binding domain (THATCH) (1). The coiled-coil domains of both proteins comprise the binding site for clathrin light chains (2-4). The coiled-coil domain of Hip1 contains binding sites, which are not present in Hip1R, with functions linked to caspase activation, androgen receptor activation, and the pathogenesis of Huntington disease (5-9). Interactions between Hip1 and Hip1R have been shown (2), although the molecular details remain unclear. Extensive heterodimerization seems unlikely, since it would mix the functions of Hip1 and Hip1R, which are distinct in cells, and genetic deletion of each protein produces a markedly different phenotype (10, 11). To understand the molecular basis for the functions of Hip1 and Hip1R, the biophysical properties of the coiled-coil domains were analyzed with respect to their self-association and their intramolecular influence on actin binding.
Clathrin-coated vesicles mediate major membrane traffic pathways, including receptor-mediated endocytosis and protein sorting, during lysosome biogenesis (12). Clathrin is a triskelion-shaped molecule made up of three heavy chains and three light chains, which polymerizes to form a lattice-like vesicle coat that organizes cargo and proteins that control vesicle budding (12-15). In both mammalian and yeast cells, clathrin-coated vesicle budding also involves actin polymerization (1, 16). In mammalian cells, overexpression of the Hip protein-binding region of clathrin light chain causes an alteration in actin cytoskeleton structure, generating short actin protrusions tipped with cortactin (4). The complex of Hip1R and cortactin has been shown to slow or block actin assembly and is proposed to mediate the positioning of actin assembly on a budding vesicle (17). In yeast cells that are deficient in clathrin heavy chain, clathrin light chain can rescue the mobility of membrane patches containing Sla2p, the yeast homolog of Hip1 and Hip1R (18). Regulation of Sla2p-actin binding by clathrin light chain was proposed to mediate this rescue (18). Intramolecular regulation of actin binding by the Hip1 and Hip1R THATCH domains has also been shown (19, 20). Here we address how intramolecular mechanisms regulating actin binding by Hip proteins are related to the influence of clathrin light chains on Hip-actin interactions.
Many cellular functions of Hip1 and Hip1R are linked to their coiled-coil domains. Dimerization occurs through the coiled-coil, as does binding to clathrin light chain (4, 3, 21). The Hip1 coiled-coil domain also binds huntingtin and has been implicated in binding to androgen receptor (5, 6, 9). Coiled-coils have a canonical heptad repeat that allows the coils to wrap around each other and maintain a hydrophobic core (22). Changes within the heptad repeat of coiled-coils often signify a region of flexibility where protein partners bind and can potentiate conformational change (22-25). Hip1 and Hip1R coiled-coils have high similarity to each other at the primary sequence level and have virtually identical regions important for clathrin light chain binding (3, 21). The structures of fragments of the Hip1 coiled-coil showed irregularity in the heptad repeat, including the region where a hydrophobic patch predicted to bind clathrin light chain was located (21, 26). Also, these structures showed a split in the coiled-coil, suggesting increased flexibility in the clathrin light chain-binding region (21, 26), with potential for structural changes upon ligand binding.
To understand the mechanism of their functional importance, the biophysical properties of the coiled-coil domains of Hip1 and Hip1R were quantitatively analyzed. These domains showed strong homodimerization, regions of flexibility, and the ability to induce conformational changes upon clathrin light chain binding, which dramatically reduced actin affinity for the THATCH domains. These biophysical properties of Hip proteins provide a basis for understanding Hip1 and Hip1R functions in cells and for understanding the regulatory effect of clathrin light chain on Hip protein-actin interaction in endocytosis. Our findings further suggest a model for the sequential function of clathrin and actin in coated vesicle budding.
EXPERIMENTAL PROCEDURES
Expression Constructs and Peptides—Separate domains of either human Hip1 or mouse Hip1R were cloned. The coiled-coil of Hip1 (Hip1cc), corresponding to amino acids 361-637; Hip1R coiled-coil (Hip1Rcc), corresponding to amino acids 346-655; Hip1 coiled-coil and THATCH (Hip1ccth), corresponding to amino acids 400-1038; Hip1R coiled-coil and THATCH (Hip1Rccth), corresponding to amino acids 379-1068; Hip1 THATCH (Hip1th), corresponding to amino acids 600-1038; and Hip1R THATCH (Hip1Rth), corresponding to amino acids 600-1068, were cloned via PCR amplification of gene fragments with flanking Gateway (Invitrogen) recombination sequences and inserted into pRSF-duet (Novagen) or pET-duet plasmid (Novagen) modified with Gateway sequences. Full-length Hip1 or Hip1R were amplified by PCR with primers containing either the polyhistidine (His)2 tag or hemagglutinin (HA) tag sequence and cloned into the pCDNA3.1 plasmid (Invitrogen). Clathrin light chain LCb was expressed from a previously described construct (27). Cortactin Src homology 3 domain fused with glutathione S-transferase (GST) was expressed and purified as previously described (17).
A peptide representing the consensus sequence of clathrin light chains (EEDPAAAFLAQQESEIAGIEND) was synthesized commercially. Control peptides of the same length were either the clathrin light chain peptide with three previously described point mutations (underlined) that abrogate Hip1R binding (4) (EEVPAAAFLAQQESEAAGIAND) or unrelated peptide (FINKPETGAVELESPFILLADKKI) derived from the bacterial protein GroEL.
Protein Expression and Purification—For all proteins, BL21 (DE3) Escherichia coli were transformed with the appropriate plasmid, and expression was induced via standard methods. Purification was via standard methods using Talon affinity resin (all Hip protein constructs) or glutathione-Sepharose (GST-cortactin). All Hip protein constructs were further purified over a Superdex 200 column. See supplemental materials for details.
Analytical Ultracentrifugation—Hip1cc or Hip1Rcc was placed in a six-channel cell at 1 mg/ml. Concentration filtrate was used for reference cells. Protein was centrifuged at four speeds, 8,000, 10,000, 15,000, and 20,000 rpm, in an AN Ti60 rotor in a Beckman Optima XL-A analytical ultracentrifuge. Data were collected after 8 h of spinning to equilibrium via absorption optics and analyzed using Ultrascan 9.0 (28). Equilibrium data were globally fitted to a monomer-dimer equilibrium or single species of either the calculated monomer or dimer size.
Circular Dichroism—CD spectra were collected using a Jasco J-710 spectrapolarimeter. Hip1cc and Hip1Rcc were diluted in 20 mm phosphate, pH 7.0, 150 mm NaCl to a concentration of 0.035 mg/ml and 0.036 mg/ml, respectively. Individual melting analysis for Hip1cc and Hip1Rcc was performed starting at 22 °C and increasing to 80 °C while monitoring ellipticity at 222 nm. The rate of heating was 0.5 °C/min.
For melting of the mixture of Hip1cc and Hip1Rcc the rate of heating was 1 °C/min. Annealing time for the mixture of Hip1cc and Hip1Rcc was between 45 min and 1 h, until the ellipticity at 222 nm was back to the starting base line. Data were collected and analyzed to 65 °C, well past the major melting transitions of both Hip1cc and Hip1Rcc, which should include dimer unfolding. The data were plotted as the mean residue molar ellipticity.
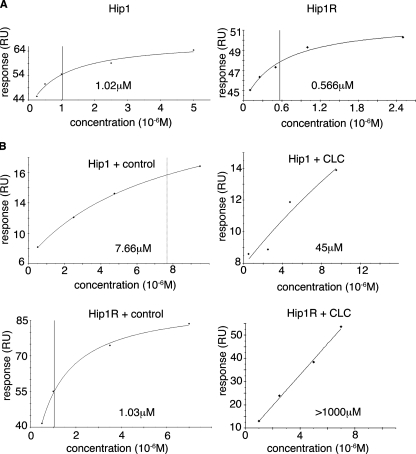
Surface Plasmon Resonance—Clathrin light chain surface was prepared by immobilizing ∼400 response units of protein via amine coupling to a CM5 chip (Biacore). For binding experiments, either Hip1cc or Hip1Rcc was flowed at the concentrations indicated in Fig. 5 with MES, pH 6.7, running buffer. The actin surface was prepared by immobilizing ∼1000 response units of rabbit muscle F-actin to the chip surface via amine coupling. Hip1ccth or Hip1Rccth at the indicated concentrations (Fig. 5) and with or without 20 μm clathrin light chain peptide or control peptide were flowed over the surface in 20 mm Tris pH 8.0, 1 mm MgCl2, 150 mm KCl running buffer.
FIGURE 5.
SPR binding of Hip1 and Hip1R to clathrin light chain and actin. Representative SPR plots for steady state binding between Hip proteins and clathrin light chains or actin are shown. Fitted binding affinities are noted on each plot, with a vertical line also indicating fitted binding affinity. A, purified clathrin light b (LCb neuronal isoform) was immobilized on a CM5 chip by amine coupling. Purified coiled-coil domains of Hip1 or Hip1R at the concentrations indicated were flowed over the clathrin light chain surface until a steady state was reached, and the steady state response units (RU) were plotted. B, actin purified from rabbit muscle was assembled and immobilized on a CM5 chip by amine coupling. Purified Hip1 or Hip1R fragments containing the coiled-coil and THATCH domains (Hip1ccth or Hip1Rccth) were flowed over the F-actin surface at the concentrations indicated in the presence of saturating amounts (20 μm) of clathrin light chain peptide (CLC) or GroEL control peptide. All data were collected on a Biacore T100. BiaEvaluation software (Biacore) was used for steady state analysis of binding data. Curves with clathrin light chain peptide have no line indicating binding affinity, because fitted binding affinity was off the scale.
GST-cortactin was immobilized with an anti-GST antibody (Biacore), and Hip1Rccth at the indicated concentrations was flowed over (Fig. S5). Running buffer was HBS-EP (Biacore).
All protein concentrations were determined in at least duplicate to ensure accurate concentrations. All data were collected on a Biacore T100 biosensor at 25 °C. Steady state binding analysis was performed using BiaEvaluation software (Biacore).
Partial Proteolysis—12.5 μg of HIP1 or HIP1R in gel filtration buffer was mixed with 0.01 μg/ml pure subtilisin (Roche Applied Science) in 20 mm HEPES, pH 7.2, 200 mm NaCl. Incubation times are as shown in the figures. At each time point, 20 μl of protein was taken, and the proteolysis reaction was stopped by mixing with SDS gel loading buffer and boiling. Proteolysis products were resolved on a 12% SDS NuPAGE gel in MES running buffer (Invitrogen).
Mass Spectrometry—Spectra of tryptic fragments excised from gels were taken on a Voyager-DE STR spectrometer (Applied Biosystems) in reflector mode. See supplemental materials for details. Spectra were processed and analyzed using the Data Explorer program (Applied Biosystems). Peptide peaks were identified using MS-Fit and MS-Digest within the Protein Prospector suite.
Immunoprecipitation and Immunoblotting—HeLa cells were co-transfected with HA- or His-tagged full-length Hip1 and Hip1R using Lipofectamine 2000 (Invitrogen). DNA concentrations used for transfection were adjusted to yield equal levels of tagged protein expression (Fig. 2) or a 4-fold excess of one tagged protein over the other (Fig. S1). Immunoprecipitation (5 μg of each antibody) and immunoblotting were with either His6 monoclonal antibody (BD Biosciences) or HA.11 monoclonal antibody (Covance) using standard methods. Immunoblotting for clathrin light chain was with a previously described anti-serum against a conserved region of LCa and LCb (29). For details, see the supplemental materials.
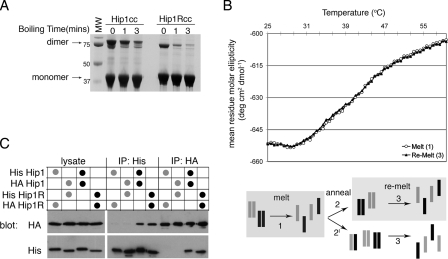
FIGURE 2.
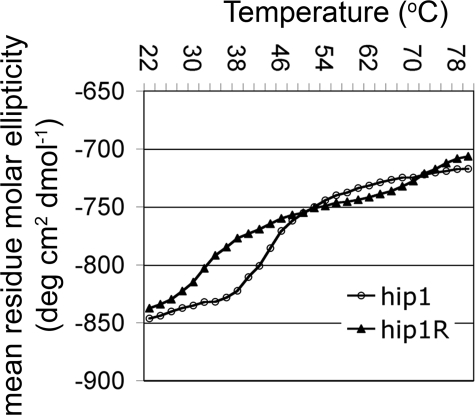
Hip1 and Hip1R are stable homodimers. A, Hip1cc or Hip1Rcc, amino acids 361-637 and 346-655, respectively, were expressed and purified. Purified Hip proteins were diluted into reducing SDS-PAGE loading buffer and run on an SDS-polyacrylamide gel after boiling for the indicated times. The migration positions of monomers and dimers and molecular weight (MW) markers (in kDa) are indicated. B, circular dichroism melting analysis of a 1:1 mixture of Hip1cc and Hip1Rcc. Shown is the molar ellipticity versus temperature as determined by monitoring the change in helical content of proteins at 222 nm. Homodimers of Hip1 and Hip1R were mixed at a 1:1 ratio, and then a thermal denaturation profile was obtained (corresponding to melting step 1 in the diagram; open circles). The mixture of denatured proteins was allowed to anneal slowly back to 20 °C (step 2 or 2′ in the diagram). The thermal denaturation profile was collected for the newly annealed population (step 3 in the diagram; filled triangles). Both melting curves precisely overlap, showing that path 2 (not 2′) was the primary route followed. C, co-immunoprecipitation of His- or HA-tagged full-length Hip1 or Hip1R. HeLa cells were co-transfected with the two constructs indicated in the table above the blot to achieve equivalent levels of expression of each construct. HeLa cells were lysed, and tagged Hip proteins were isolated by immunoprecipitation (IP) with either anti-His antibody or anti-HA antibody as shown. Immunoprecipitates and transfected cell lysate were separated by SDS-PAGE and transferred to nitrocellulose for immunoblotting. Nitrocellulose membranes were probed with anti-His, stripped, and then probed with anti-HA. Black dots indicate homotypic transfection combinations. Gray dots indicate heterotypic transfection combinations.
Immunofluorescence and siRNA Depletion—HeLa cells cultured in Dulbecco's modified Eagle's medium containing 10% bovine growth serum (Hyclone) were transfected with a final concentration of 10 nm siRNA targeted to clathrin light chain or control siRNA using the HiPerfect reagent (Qiagen). Target sequences for LCa and LCb knockdown were as described (30). Both control and the clathrin light chain siRNAs were synthesized by Qiagen. Cells were fixed and labeled for immunofluorescence 72 h post-siRNA treatment using rabbit anti-Hip1R antiserum (Millipore), monoclonal antibody to clathrin heavy chain (X22, (31)), and Alexa Fluor-conjugated phalloidin (Invitrogen) to visualize F-actin. The samples were viewed by confocal laser-scanning microscopy using a Leica TCS-SP5 operating system.
Electron Microscopy—5 μl of 0.2 μm purified Hip1ccth or Hip1Rccth incubated with 20 μm control peptide, clathrin light chain peptide, or no peptide was dropped on a carbon-coated grid and stained with uranyl formate. Samples were imaged at ×67,000 using a Philips Tecnai T20 electron microscope operated at an acceleration voltage of 120 kV with a defocus range of 1.2-1.5 μm. Images were taken using a 4K × 4K CCD (GATAN) with a pixel size of 2.25 Å/pixel. Images were selected and measured using EMAN.
8-Anilino-1-napthalenesulfonic Acid (ANS) Fluorescence—0.7 μm purified Hip coiled-coils or coiled-coil THATCH domains were incubated with 20 μm clathrin light chain peptide or control peptide and 100 μm ANS. Excitation of ANS was at 360 nm, and emission was scanned from 400 to 600 nm. Raw data were averaged over three independent experiments, and S.E. values were calculated for each condition. Difference spectra of Hip coiled-coil THATCH and Hip coiled-coils were plotted with propagated S.E.
RESULTS
Hip1 and Hip1R Are Stable Homodimers—Homodimers of Hip1R have an elongated dumbbell shape, with the N- and C-terminal globular domains in variable positions (32). This implies the primary dimerization determinants are within the coiled-coil domain. Interactions between Hip1 and Hip1R have been shown by GST pull-down and co-immunoprecipitation (2). Whether this interaction arises from direct heterodimerization or another mode of interaction has yet to be explicitly determined and would have different functional implications. Coiled-coil domains (Hip1 residues 361-637 and Hip1R residues 346-655; Fig. 1) from Hip proteins were expressed and purified, and their dimerization properties were analyzed by equilibrium sedimentation analytical ultracentrifugation (Table 1). Assuming a monomer-dimer equilibrium model, the data analysis suggested that there was no equilibrium under standard conditions and that all of the coiled-coils for both Hip1 and Hip1R are dimeric in solution. The fitted molecular masses were 67.4 and 67.7 kDa for the Hip1 and Hip1R coiled-coils, respectively, corresponding closely to calculated homodimeric sizes of 66.79 and 73.45 kDa (Table 1). Fitting the sedimentation data to a single species with the calculated molecular weight of the dimers (data not shown) also fit well with the experimental data. Attempts to fit the sedimentation data to the true calculated size of the monomer, as either a single species or as a monomer-dimer equilibrium, resulted in poor fits (data not shown). These data indicate that homodimers of Hip1 and Hip1R are stable and that there is virtually no dissociation to the monomer form of these proteins in vitro.
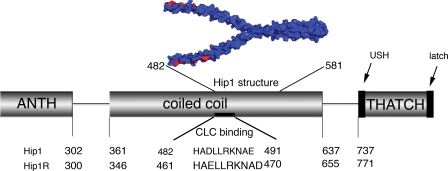
FIGURE 1.
Schematic diagram of the functional domains of Hip proteins showing amino acid numbers that mark domain boundaries. The sequences of residues influencing clathrin light chain (CLC) binding regions as determined by mutagenesis are shown in alignment (3). The structure (Protein Data Bank code 2NO2) of a fragment of the Hip1 coiled-coil is shown in blue (21). Red amino acids make up the clathrin light chain binding site predicted by Ybe et al. (21). The ANTH domain binds phospholipids, and the THATCH domain binds actin, subject to intramolecular regulation by a USH (19) and a C-terminal segment (latch) (20).
TABLE 1.
Hip coiled-coil molecular mass determined by analytical ultracentrifugation fitted to monomer-dimer equilibrium model
| Protein | Calculated dimer molecular mass | Calculated monomer molecular mass | Experimental molecular mass | Fitting variance |
|---|---|---|---|---|
| kDa | kDa | kDa | ||
| Hip1cc | 66.79 | 33.39 | 67.4 | 2.51 × 10−5 |
| Hip1Rcc | 73.45 | 36.73 | 67.7 | 2.57 × 10−5 |
This stability was also evident from biochemical treatments. Dimers of purified Hip1 and Hip1R coiled-coils survived standard denaturing, reducing SDS-polyacrylamide gels (Fig. 2A). Coiled-coil domains of Hip1 and Hip1R were mixed with SDS-PAGE sample buffer and heated to 95 °C for increasing time intervals (Fig. 2A). After only short heating times, large amounts of Hip1 or Hip1R coiled-coils remained complexed. This SDS-resistant behavior is similar to that of the four-helix bundles of SNARE proteins that require an ATP-hydrolyzing chaperone, N-ethylmaleimide sensitive factor (NSF) to dissociate the complex (33). Although both Hip1 and Hip1R coiled-coils were very stable, Hip1 remained a dimer for longer heating times (Fig. 2A).
Given the high stability of the coiled-coil domains of Hip1 and Hip1R, it is likely that homodimers are the preferred mode of interaction. To test the possibility of heterodimerization in a stringent way, a 1:1 mixture of Hip1 and Hip1R coiled-coils was thermally denatured through their primary melting transition to 65 °C while monitoring the melting transition by circular dichroism (Fig. 2B). The denatured mixture was then allowed to slowly reanneal. Melting and annealing should overcome any thermodynamic or kinetic barrier to monomer formation and allow heterodimers to form during reannealing if feasible. Following reannealing, thermal denaturation of the resulting molecules was measured, and the melting curve was identical to that of the mixture of the Hip1 and Hip1R coiled-coil homodimers (Fig. 2B). Thus, no new stable heterodimers had formed upon reannealing. These would have generated a melting curve that deviated from that of the original mixture because the heterodimers would contribute differently to the shape of the melting curve.
These biophysical results indicate there is no heterodimerization of Hip1 and Hip1R through their coiled-coil domains in vitro. To follow up the biophysical analysis in a cellular context, four constructs encoding full-length HA- or His-tagged Hip1 and Hip1R were produced and expressed in HeLa cells (Fig. 2C). After co-transfection of Hip1 and Hip1R, each with a different tag (in either combination), immunoprecipitation of either Hip1 or Hip1R gave negligible amounts of the other protein. However, when Hip1 constructs with two different tags or Hip1R constructs with two different tags were transfected into cells, homotypic interactions were readily detected by immunoprecipitation. Varying the expression level of each construct relative to the other did not change this result (Fig. S1). Our biochemical analysis and its correlation with cellular data thus suggest that the preferred mode of Hip protein interaction in vivo and in vitro is homodimeric. These results are compatible with reported functional segregation of Hip1 and Hip1R in cells (10, 11).
Flexibility Is a Mechanism for Conformational Regulation in Hip1 and Hip1R—Given the preference for homodimerization and the segregation of biological functions of Hip1 and Hip1R, further biophysical properties of their coiled-coil domains were analyzed to determine whether these might account for functional differences. Separate crystallographic analyses of the N- and C-terminal halves of the coiled-coil domain of Hip1 showed classical coiled-coil segments from residues 371-430 and from residues 540-581. These structures also suggested a segment, around residues 445-539, of decreased stability and increased flexibility that included residues implicated in binding clathrin light chain as well as in binding of HIPPI (Hip1 protein interactor) (21) (Fig. 1). To investigate whether this region might function in regulatory aspects of the Hip proteins, biophysical properties related to stability of the Hip protein coiled-coil domains were analyzed. First, using the program COILS (34), subdomains of high and low coiled-coil propensity were mapped within the coiled-coil domains of both Hip1 and Hip1R (Fig. 3, A and B). COILS suggested that both domains have N-terminal regions with high propensity to form coils. At the (D/E)LLRKN sequences implicated in binding clathrin light chains (3), the predicted propensity dropped for both Hip1 and Hip1R. However, for Hip1, this decrease was slight, whereas for Hip1R, there was a significant decrease in predicted coil propensity. C-terminal to the clathrin light chain binding sequence, both Hip1 and Hip1R had predicted regions of significantly lower coiled-coil propensity interspersed with regions of high coiled-coil propensity, again with Hip1R having less coil propensity.
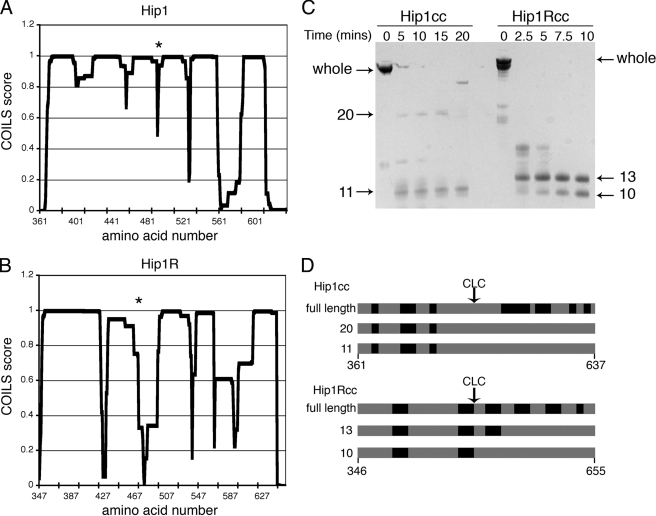
FIGURE 3.
Flexibility within the Hip1 and Hip1R coiled-coils. A and B, coiled-coil propensity of Hip1 and Hip1R coiled-coil domains, respectively, as predicted by the program COILS (34). An asterisk indicates the approximate position of residues known to be important for light chain binding, as determined by mutagenesis (3). C, partial proteolysis of Hip1 and Hip1R coiled-coil domains. 12.5 μg of Hip1 or Hip1R was digested with 0.01 μg/ml of subtilisin protease for the indicated times. Reactions were stopped by boiling in SDS-PAGE loading buffer. Proteolysis products were separated by SDS-PAGE. Fragment sizes in kDa are indicated. D, whole Hip1 or Hip1R coiled-coils or subtilisin proteolysis products indicated by arrows in C were excised from the SDS-polyacrylamide gel and further digested with trypsin. Peptides from the tryptic digests were identified by MALDI mass spectrometry. The black regions of the diagram indicate tryptic peptides identified by mass spectrometry in each digestion reaction, labeled according to the molecular weight of the band digested. Tryptic peaks missing (gray) in the subtilisin fragments indicate which regions were digested during subtilisin proteolysis. CLC marks the approximate position of clathrin light chain binding.
Partial proteolysis was used to establish the regions of stability and flexibility in Hip1 and Hip1R. The coiled-coil domains of Hip1 and Hip1R were partially digested with subtilisin over time and analyzed by SDS-PAGE. Hip1 was resistant to proteolysis for ∼10-15 min, whereas Hip1R was almost completely digested in the same time period (Fig. 3C). The coiled-coil domains of both proteins had subfragments that were resistant to proteolysis. Peptide mass fingerprints from the protease-resistant fragments of Hip1 (11 and 20 kDa) and Hip1R (10 and 13 kDa) showed that peptides C-terminal to the clathrin light chain binding region were missing from these fragments (Fig. 3D). The increased susceptibility to degradation of the C-terminal subdomains of the Hip1 and Hip1R coiled-coils agrees with the COILS analysis predicting decreased coiled-coil propensity in the same region. Thus, although the static crystal structure indicated that coiled-coil interactions can occur in this region, it is functionally less stable (more flexible) than the coiled-coil region that is N-terminal to the clathrin light chain binding site. The differences in the relative rates of partial proteolysis indicate that Hip1R has increased flexibility in the C-terminal subdomain compared with Hip1.
The relative stability of Hip1 and Hip1R coiled-coils was determined by reversible thermal denaturation while measuring the ellipticity at 222 nm by circular dichroism (Fig. 4). Thermal melting curves for Hip1 and Hip1R showed significantly different profiles for unfolding. Hip1 had a single smooth melting transition (Tm) at 43 °C, whereas Hip1R had a multistep unfolding profile. The first melting transition for Hip1R occurred at 32 °C, much lower than that for Hip1. The second Hip1R Tm was much higher, near 72 °C. The first melting transition for Hip1R probably represents most of the protein unfolding, given its significant change. The second transition occurs at high temperature and may reflect residual bits of helical structure unfolding or temperature dependent changes in absorption, not related to Hip1R denaturation. These melting data correlate with the partial proteolysis, gel denaturation, and COILS data, suggesting that the dimeric coiled-coil region of Hip1 is more stable than that of Hip1R.
FIGURE 4.
Thermal stability of Hip1 and Hip1R. Purified coiled-coil domains of Hip1 and Hip1R were subjected to increasing temperature while the change in ellipticity at 222 nm was monitored by circular dichroism. The heating rate was 0.5 °C/min. The mean residue molar ellipticity versus temperature was plotted. The transition for the thermal denaturation of Hip1 coiled-coil domain had a Tm = 43 °C (○). The thermal denaturation profile of Hip1R had a Tm = 32 °C (▴) for the first and primary transition.
Since the flexible region started at the clathrin light chain binding site, we measured whether the differences in flexibility of Hip1 and Hip1R correlated with their binding affinity for clathrin light chain. Using surface plasmon resonance (SPR), the binding affinity between Hip1 or Hip1R and full-length clathrin light chain b (LCb neuronal isoform) was determined (Fig. 5A). Hip1 bound LCb with an affinity of 1.02 μm. Hip1R bound LCb with an affinity of 0.566 μm, about 2-fold better than Hip1, correlating with the greater flexibility of Hip1R in the LCb binding region.
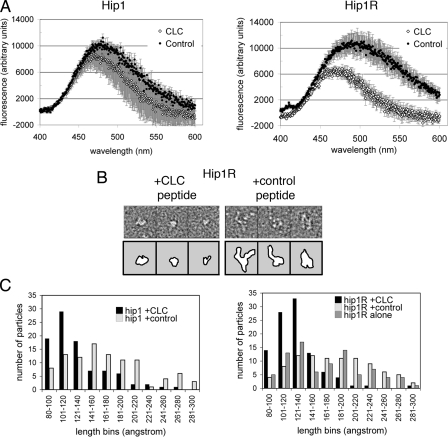
Hip1 and Hip1R Adopt Compact Conformations in Response to Clathrin Binding—Since flexibility of the coiled-coil domains correlated with clathrin light chain interaction, we addressed whether their flexibility might influence additional functions of Hip1 and Hip1R. For the yeast homolog Sla2p, sequences in the vicinity of the coiled-coil domain were shown by two-hybrid and in vitro binding assays to have the capacity for intramolecular interaction with the actin-binding THATCH domain (35). The flexibility of the coiled-coils starting at the clathrin light chain binding region could provide a means to induce this intramolecular interaction, which would affect the surface properties of the THATCH domain. To investigate this possibility, we assessed changes in the surface properties of the THATCH domains by monitoring fluorescence of the hydrophobic dye ANS (Fig. 6A). The C-terminal fragments of Hip1 and Hip1R, including the coiled-coil and THATCH domain (Hip1 residues 400-1038, Hip1ccth; or Hip1R residues 379-1068, Hip1Rccth), were expressed, and ANS fluorescence spectra were collected in the presence of clathrin light chain peptide or control peptide. The clathrin light chain peptide is a 22-residue peptide representing the Hip-binding region of clathrin light chains (common to LCa and LCb) (see “Experimental Procedures” for the sequence) (4). ANS spectra in the presence of the two peptides were also collected for the coiled-coil domains alone. Coiled-coil spectra were subtracted from the coiled-coil THATCH spectra to get the net change in ANS fluorescence, reflecting changes due to conformational rearrangement of the THATCH domain alone. Hip1R showed a significant change in the ANS fluorescence difference spectrum for the THATCH domain in the presence of the clathrin light chain peptide compared with the control peptide. The Hip1 THATCH domain difference spectrum had a similar trend but was not above the error (Fig. 6A). These assays reveal a change in the accessible hydrophobic surfaces in the THATCH domain when clathrin light chains are bound to coiled-coil domains, suggesting an intramolecular conformational rearrangement in mammalian Hip proteins similar to that predicted for Sla2p (35). The limited change detected in Hip1 is probably due to ANS being insensitive to the conformational changes induced by the clathrin light chain peptide, since electron microscopy data (see below) clearly show altered conformations for both Hip1 and Hip1R.
FIGURE 6.
Hip1 and Hip1R change conformation upon clathrin light chain binding. A, purified Hip coiled-coils (Hip1cc and Hip1Rcc) or Hip coiled-coil plus THATCH fragments (Hip1ccth and Hip1Rccth) were incubated with ANS in the presence of clathrin light chain (CLC) or control peptide. ANS fluorescence spectra of the coiled-coils were subtracted from ANS spectra of the coiled-coil THATCH fragments. The difference between the spectra of the two fragments is plotted, indicating ANS fluorescence due to the THATCH domains alone in the presence of either peptide. B, representative electron microscopy images of Hip1Rccth in the presence of control or clathrin light chain peptide, stained with uranyl formate. Box edge, 270 Å. Top panels show micrographs, and bottom panels show outlines of protein images. See Fig. S2 for representative images of Hip1ccth after peptide binding. C, length distributions of particles of Hip1ccth and Hip1Rccth in the presence of clathrin light chain peptide, control peptide, or no peptide from images of uranyl formate-stained samples. Distribution was determined by measuring ∼100 particles for each condition. The control peptide is the clathrin light chain peptide with three mutations that abrogate Hip protein binding (4).
To visualize the conformational changes in Hip1 and Hip1R that were implied by ANS fluorescence, negative stain electron microscopy was used. Hip1 and Hip1R coiled-coil THATCH domains (Hip1ccth and Hip1Rccth) were combined with clathrin light chain peptide or control peptide and stained with uranyl formate, and electron microscopy images were collected. The control peptide was the clathrin light chain peptide containing three mutations that abrogate Hip protein binding (4). To avoid bias, random single particles were preselected from fields without knowing which peptide had been added, and the longest dimension of these particles was measured. Measurements were binned in groups of 20 Å and plotted as a histogram (Fig. 6, B and C). The length of single particles of both Hip1 and Hip1R with clathrin light chain peptide bound had a narrower distribution than seen with control peptide and peaked near 120 Å. In the presence of control peptide (and no peptide for Hip1R), both proteins showed a much broader length distribution and many more particles with dimensions greater than 120 Å. This indicates that clathrin light chain binding to their coiled-coil domains can induce compaction of Hip1 and Hip1R (Figs. 6B and S2).
Actin Binding by Hip1 and Hip1R Is Negatively Regulated by Clathrin Binding—Our studies here have shown how clathrin light chain can induce a compact conformation of Hip proteins through binding the flexible regions of their coiled-coil domains and also influence the surface properties of their actin-binding THATCH domains. These observations suggest that clathrin light chain binding to the coiled-coil domain of Hip proteins could affect the binding of actin to their THATCH domains. To assess this possibility, SPR was used to determine the binding affinities of Hip1 and Hip1R for actin in the presence of clathrin light chain peptide or control peptide. For these experiments, Hip1ccth or Hip1Rccth were flowed over a chip with covalently bound F-actin. The steady state affinity of Hip1ccth binding to F-actin was calculated to be 7.66 μm, and for Hip1R, it was 1.03 μm in the presence of control peptide (Fig. 5B). These affinities were very similar to the affinity of Hip proteins for actin with no peptide present (Fig. S3). In agreement with our results, it has been previously reported that Hip1 binds actin with a relatively weaker affinity than Hip1R, but no specific binding constants were calculated in that study (2). Also both affinities calculated from our data are in the micromolar range, as previously reported for binding between actin and the isolated THATCH domains of Hip1 and Hip1R (lacking their regulatory upstream helix) (19).
Clathrin light chain peptide or control peptide was then introduced into the SPR assay for measuring Hip-actin interaction. In the presence of saturating amounts of clathrin light chain peptide, the determined KD(actin) for actin binding was 45 μm for the Hip1ccth-peptide complex (Fig. 5B) and >1000 μm for the Hip1Rccth-peptide complex (Fig. 5B). Its greater response to clathrin light chain peptide correlates with Hip1R being the more flexible of the two Hip proteins. Notably, there was an ∼6 to >1000-fold decrease in affinity of both Hip1 and Hip1R for actin when the clathrin light chain peptide was present. The reciprocal effect was also observed, although to a lesser extent (Fig. S6). Hip1 (Hip1ccth) had a 3-4-fold decreased affinity for clathrin light chain if it was prebound to actin. Purified THATCH domains missing the clathrin-binding coiled-coil domains (residues 600-1038 for Hip1 and residues 600-1068 for Hip1R) had little change in actin-binding affinity in the presence of clathrin light chain peptide or control peptide (Fig. S4). Thus, the clathrin light chain effects on actin binding by Hip proteins are the result of long range conformational interactions between the coiled-coil and THATCH domains.
Studies in yeast and mammalian cells indicated that clathrin light chain fragments comprising only the Hip-binding or Sla2p-binding regions affected actin-dependent membrane dynamics (4, 18). Our results explain this effect by demonstrating the profound influence of the clathrin light chain peptide on the affinity of the Hip proteins for actin. Consistent with the cellular assays showing functional efficacy of Hip-binding or Sla2p-binding domains, the clathrin light chain peptide used in our studies binds to Hip proteins with similar efficiency as full-length purified clathrin light chain (data not shown) (4, 18).
The proline-rich domain of Hip1R, found C-terminal to the THATCH domain, has been previously shown to bind cortactin (17). SPR experiments showed that the presence of clathrin light chain did not alter the affinity Hip1Rccth for cortactin (Fig. S5), demonstrating specific regulation of Hip1R actin affinity by clathrin light chain. As expected, the equivalent domain of Hip1, which does not contain a proline-rich domain, did not bind to cortactin (data not shown).
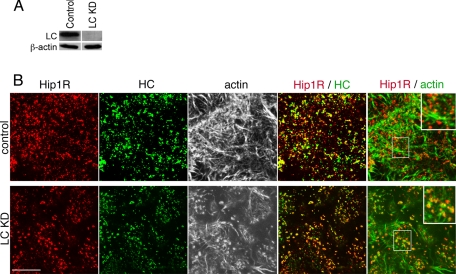
The demonstration that clathrin light chain specifically reduces Hip protein affinity for actin suggests that cellular depletion of clathrin light chains should increase Hip protein-actin interactions at sites of clathrin heavy chain-containing structures. Normally, in mammalian cells, Hip1R is seen colocalized with punctate clathrin structures that align with but do not overlap with actin (Fig. 7). As predicted, depletion of cellular clathrin light chains showed an increase in overlap between Hip1R and actin (Fig. 7). These observations indicate that the regulation of actin-Hip protein interactions by clathrin light chain is relevant to cellular control of actin-clathrin interactions.
FIGURE 7.
Immunofluorescence of Hip1R, actin, and clathrin following cellular depletion of clathrin light chains. HeLa cells were transfected with siRNAs targeting both clathrin light chain a (LCa) and clathrin light chain b (LCb) to knock down all clathrin light chains (LC KD) or with control (scrambled) siRNA at the same concentration and cultured for 72 h. A, HeLa cells were lysed after siRNA treatment, and protein levels were determined by immunoblotting. The indicated proteins were detected by a rabbit polyclonal antiserum against a conserved sequence shared by LCa and LCb (LC) (29) and β-actin (Sigma). Actin is shown as a loading control. B, cells treated with siRNA against clathrin light chain (LC KD) or with control siRNA were labeled for immunofluorescence with antibodies against clathrin heavy chain (HC) (green) and Hip1R (red). Actin was detected with fluorescent phalloidin and is shown in black and white images in the center panels or in green in the far right panels. Merged images of the indicated proteins are shown in the two right-hand panels of each row, with yellow indicating overlap of red and green labeling. In the far right panels, the central boxed area is magnified in the upper righthand corner. Bar, 10 μm.
DISCUSSION
Hip1 and Hip1R promote clathrin assembly through binding to clathrin light chains (3, 4), and Hip1R controls assembly of actin through its interaction with cortactin and actin (17, 20). Hip1 is also an actin-binding protein (19). In addition to their common properties, Hip1 and Hip1R seem to have distinct roles within cells, because separate phenotypes result from deletion of the genes encoding either Hip1 or Hip1R (10). Here we demonstrate that homodimerization is the predominant state for the Hip proteins, which allows segregation of their cellular functions. We characterize further biophysical differences between the coiled-coil domains of Hip1 and Hip1R but demonstrate that both contain flexible regions that contribute to their common functions. In particular, clathrin light chain binding to their coiled-coil regions is shown to change the conformation of both Hip proteins and to reduce their affinity for actin. These observations suggest that Hip proteins have separate interactions with clathrin and actin during membrane traffic rather than the direct bridging function between clathrin and the cytoskeleton proposed previously (32, 36). They also suggest that clathrin light chain partially rescues endocytosis in yeast, in the absence of clathrin heavy chain, by releasing actin from Sla2p to promote asymmetric attachment of actin to the budding vesicle (18).
Differential localization and binding partners suggest that Hip1 and Hip1R have varied and separate functions. Here we show strong tendencies for homodimerization of the Hip1 and Hip1R coiled-coil domains in vitro that would prevent the mixing of their separate functions. By co-immunoprecipitation experiments, we confirmed preference for homotypic interaction of the full-length proteins in cells. It is possible that earlier reports of Hip1-Hip1R heterodimerization and the very minor (unidirectional) heterotypic interaction in cells that we detect reflect an interaction between Hip1 and Hip1R that occurs under special conditions of cellular regulation. However, our data establish homodimers as the base-line state for Hip1 and Hip1R in cells. Consistent with their homodimerization and distinct binding properties, Hip1 and Hip1R can only partially compensate for each other in genetic knock-out experiments (10).
Although strong dimerization is clearly a property of the coiled-coil regions of the Hip proteins, there have been suggestions of flexibility within the coiled-coil regions from electron microscopy and crystallography studies (21, 26, 32). Here we characterized the extent of flexibility within the coiled-coil regions of Hip proteins in solution and addressed its function. The binding constants reported here indicate that Hip1 and Hip1R exhibit a small difference in affinity for clathrin light chain, consistent with previous qualitative predictions that Hip1R binds clathrin light chain more avidly than Hip1 (4, 32). The lower melting temperature and higher susceptibility to proteolysis of the Hip1R coiled-coil domain compared with that of Hip1 indicates greater flexibility or conformational entropy, which could play a role in the stronger binding by Hip1R of light chain. We propose that intrinsic flexibility may modulate binding affinity by influencing the frequency that the coiled-coil is in the clathrin light chain binding-competent state, although a complete analysis of this phenomenon awaits further experimentation.
We further investigated whether coiled-coil flexibility and clathrin light chain binding play a role in actin binding by Hip proteins, because it has been previously suggested that the THATCH domain is subject to intramolecular regulation (18, 19, 20). In studies of isolated THATCH domains, Senetar et al. (19) observed that their actin binding affinity was considerably reduced in the presence of an N-terminal upstream helix (USH), and Brett et al. (20) identified a C-terminal LATCH sequence that increased Hip1R THATCH binding affinity for actin in the presence of the USH. In addition, intramolecular interactions between the coiled-coil domain of Sla2p and the THATCH domain were predicted by yeast two-hybrid studies (35). Here we measured actin binding of extended domains of Hip1 and Hip1R, fragments comprising the coiled-coil and THATCH domains with both the USH and the LATCH sequences. In the absence of the clathrin light chain peptide, Hip1 and Hip1R bound actin in the micromolar range, as if the inhibitory USH were inactive. In the presence of the light chain peptide, the KD(actin) of both proteins was significantly decreased, with a 6.4-fold change for Hip1 and a >1000-fold change for Hip1R. Thus the binding of clathrin light chain to the coiled-coil of Hip proteins seems to reduce the affinity of both proteins for actin, with a greater effect on the more flexible Hip1R. These effects of clathrin light chain peptide on actin binding by the Hip proteins correlated with conformational changes induced upon peptide binding. Both Hip proteins are able to adopt compact structures as well as extended structures. Upon clathrin light chain binding, both proteins assumed a more compact state. That these conformational changes involved the THATCH domain was confirmed by ANS spectra, which showed a change in THATCH surface contacts upon binding of clathrin light chain peptide to Hip1R, with the same tendency for Hip1. These data suggest that in the compact state, the Hip protein actin binding sites are masked by an intramolecular interaction. The compact conformation may also involve repositioning of the USH (Fig. 8A). In cells depleted of clathrin light chain, Hip1R labeling was directly co-localized with actin, consistent with Hip1R being in the extended state with higher affinity for actin. In cells treated with control siRNA, Hip1R in the compact state with low affinity for actin was colocalized with clathrin-containing structures and was “offset” (not overlapping with) adjacent labeled actin. These cellular and biophysical observations together indicate that Hip1R interactions with clathrin and actin are mutually exclusive.
FIGURE 8.
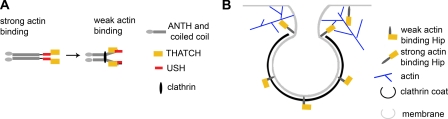
Model of Hip protein conformations and effect on vesicle budding. A, model of the mechanism for regulation of actin binding to Hip proteins by clathrin light chain. Flexibility in the coiled-coil allows large scale bending of the coiled-coil upon clathrin light chain binding. This could reduce actin binding by the THATCH domain by direct blocking resulting from an intramolecular interaction between THATCH and a site that is exposed following clathrin light chain binding or indirectly by light chain inducing a conformational change that repositions the USH to an inhibitory conformation or a combination of both mechanisms. B, the decreased affinity of Hip1 and Hip1R for actin while bound to clathrin light chain suggests that Hip proteins do not interact with actin while incorporated into the clathrin coat. Instead, Hip proteins may interact with actin at the neck of the budding vesicle or edge of the clathrin coat, promoting development of a budding vesicle.
It remains to be seen whether binding of other partner proteins to the Hip1 coiled-coil affects its coiled-coil properties and whether this in turn influences other binding partner interactions. In contrast to the multiple partners of Hip1, clathrin light chain is the only binding partner identified so far for the Hip1R coiled-coil region, suggesting that Hip1R primarily functions in clathrin-mediated endocytosis. Notably, clathrin light chain binding did not affect cortactin binding, which interacts with Hip1R C-terminal to the THATCH domain. In addition to demonstrating the specificity of clathrin light chain binding in regulating actin binding, this result indicates that cortactin should remain bound to Hip1R whether or not clathrin is bound.
The work reported here has novel implications for the influence of clathrin and Hip proteins on actin organization during coated vesicle formation. During clathrin coat formation, Hip interactions with clathrin and actin are likely to be sequential, since clathrin-bound Hip proteins have considerably reduced affinity for actin (Fig. 8B). The comparable affinities of Hip proteins for clathrin light chains or for actin and the reciprocal influence of each ligand on the other's binding are compatible with the scenario that Hip proteins, when captured by a clathrin coat, will not be encumbered by Hip-actin interactions. Consistent with this prediction, electron micrographs localizing Hip proteins relative to clathrin and actin show that clathrin-associated Hip does not co-localize with actin, except at the neck of the budding vesicle, near the clathrin lattice edge (32). At the neck, Hip proteins can interact directly with membrane via their ANTH domains and dissociate from clathrin light chains, enhancing their affinity for actin and thereby regulating actin filament growth at this site. As suggested by Le Clainche et al. (17), the binding of cortactin by Hip1R could control the localization of actin polymerization at the budding vesicle neck and contribute to vesicle scission. Since cortactin binding is not affected by clathrin light chain, it could be constitutively bound to lattice-associated Hip1R.
In summary, our findings show that although overall stability of the coiled-coil domains of Hip proteins favors homodimerization and segregation of Hip1 and Hip1R function, their intrinsic flexibility allows regulation of partner protein binding through conformational changes. In particular, these changes mandate sequential interaction of Hip proteins with clathrin and actin, redefining the proposed mechanism of action of Hip proteins during membrane traffic.
Supplementary Material
Acknowledgments
We thank D. Drubin and C. Le Clainche for the cortactin constructs and expression methods and Daniel Southworth for expert technical assistance with electron microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant GM038093 (to F. M. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S6.
Footnotes
The abbreviations used are: His, polyhistidine; HA, hemagglutinin; GST, glutathione S-transferase; MES, 2-(N-morpholino)ethanesulfonic acid; siRNA, small interfering RNA; ANS, 8-anilino-1-napthalenesulfonic acid; NSF, N-ethylmaleimide sensitive factor; SPR, surface plasmon resonance.
References
- 1.Engqvist-Goldstein, A. E., and Drubin, D. G. (2003) Annu. Rev. Cell Dev. Biol. 19 287-332 [DOI] [PubMed] [Google Scholar]
- 2.Legendre-Guillemin, V., Metzler, M., Charbonneau, M., Gan, L., Chopra, V., Philie, J., Hayden, M. R., and McPherson, P. S. (2002) J. Biol. Chem. 277 19897-19904 [DOI] [PubMed] [Google Scholar]
- 3.Legendre-Guillemin, V., Metzler, M., Lemaire, J. F., Philie, J., Gan, L., Hayden, M. R., and McPherson, P. S. (2005) J. Biol. Chem. 280 6101-6108 [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Y., and Brodsky, F. M. (2005) J. Biol. Chem. 280 6109-6117 [DOI] [PubMed] [Google Scholar]
- 5.Wanker, E. E., Rovira, C., Scherzinger, E., Hasenbank, R., Walter, S., Tait, D., Colicelli, J., and Lehrach, H. (1997) Hum. Mol. Genet. 6 487-495 [DOI] [PubMed] [Google Scholar]
- 6.Kalchman, M. A., Koide, H. B., McCutcheon, K., Graham, R. K., Nichol, K., Nishiyama, K., Kazemi-Esfarjani, P., Lynn, F. C., Wellington, C., Metzler, M., Goldberg, Y. P., Kanazawa, I., Gietz, R. D., and Hayden, M. R. (1997) Nat. Genet. 16 44-53 [DOI] [PubMed] [Google Scholar]
- 7.Hackam, A. S., Yassa, A. S., Singaraja, R., Metzler, M., Gutekunst, C. A., Gan, L., Warby, S., Wellington, C. L., Vaillancourt, J., Chen, N., Gervais, F. G., Raymond, L., Nicholson, D. W., and Hayden, M. R. (2000) J. Biol. Chem. 275 41299-41308 [DOI] [PubMed] [Google Scholar]
- 8.Gervais, F. G., Singaraja, R., Xanthoudakis, S., Gutekunst, C. A., Leavitt, B. R., Metzler, M., Hackam, A. S., Tam, J., Vaillancourt, J. P., Houtzager, V., Rasper, D. M., Roy, S., Hayden, M. R., and Nicholson, D. W. (2002) Nat. Cell Biol. 4 95-105 [DOI] [PubMed] [Google Scholar]
- 9.Mills, I. G., Gaughan, L., Robson, C., Ross, T., McCracken, S., Kelly, J., and Neal, D. E. (2005) J. Cell Biol. 170 191-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley, S. V., Hyun, T. S., Oravecz-Wilson, K. I., Li, L., Waldorff, E. I., Ermilov, A. N., Goldstein, S. A., Zhang, C. X., Drubin, D. G., Varela, K., Parlow, A., Dlugosz, A. A., and Ross, T. S. (2007) Hum. Mol. Genet. 16 1279-1292 [DOI] [PubMed] [Google Scholar]
- 11.Metzler, M., Li, B., Gan, L., Georgiou, J., Gutekunst, C. A., Wang, Y., Torre, E., Devon, R. S., Oh, R., Legendre-Guillemin, V., Rich, M., Alvarez, C., Gertsenstein, M., McPherson, P. S., Nagy, A., Wang, Y. T., Roder, J. C., Raymond, L. A., and Hayden, M. R. (2003) EMBO J. 22 3254-3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brodsky, F. M., Chen, C. Y., Knuehl, C., Towler, M. C., and Wakeham, D. E. (2001) Annu. Rev. Cell Dev. Biol. 17 517-568 [DOI] [PubMed] [Google Scholar]
- 13.Kirchhausen, T. (2000) Annu. Rev. Biochem. 69 699-727 [DOI] [PubMed] [Google Scholar]
- 14.Traub, L. M. (2005) Biochim. Biophys. Acta 1744 415-437 [DOI] [PubMed] [Google Scholar]
- 15.Ungewickell, E. J., and Hinrichsen, L. (2007) Curr. Opin. Cell. Biol. 19 417-425 [DOI] [PubMed] [Google Scholar]
- 16.Engqvist-Goldstein, A. E., Kessels, M. M., Chopra, V. S., Hayden, M. R., and Drubin, D. G. (1999) J. Cell Biol. 147 1503-1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Clainche, C., Pauly, B. S., Zhang, C. X., Engqvist-Goldstein, A. E., Cunningham, K., and Drubin, D. G. (2007) EMBO J. 26 1199-1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newpher, T. M., Idrissi, F. Z., Geli, M. I., and Lemmon, S. K. (2006) Mol. Biol. Cell 17 4343-4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senetar, M. A., Foster, S. J., and McCann, R. O. (2004) Biochemistry 43 15418-15428 [DOI] [PubMed] [Google Scholar]
- 20.Brett, T. J., Legendre-Guillemin, V., McPherson, P. S., and Fremont, D. H. (2006) Nat. Struct. Mol. Biol. 13 121-130 [DOI] [PubMed] [Google Scholar]
- 21.Ybe, J. A., Mishra, S., Helms, S., and Nix, J. (2007) J. Mol. Biol. 367 8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkhard, P., Stetefeld, J., and Strelkov, S. V. (2001) Trends Cell Biol. 11 82-88 [DOI] [PubMed] [Google Scholar]
- 23.Brown, J. H., Cohen, C., and Parry, D. A. (1996) Proteins 26 134-145 [DOI] [PubMed] [Google Scholar]
- 24.Singh, A., and Hitchcock-DeGregori, S. E. (2003) Biochemistry 42 14114-14121 [DOI] [PubMed] [Google Scholar]
- 25.Singh, A., and Hitchcock-DeGregori, S. E. (2006) Structure 14 43-50 [DOI] [PubMed] [Google Scholar]
- 26.Niu, Q., and Ybe, J. A. (2008) J. Mol. Biol. 375 1197-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S. H., Wong, M. L., Craik, C. S., and Brodsky, F. M. (1995) Cell 83 257-267 [DOI] [PubMed] [Google Scholar]
- 28.Demeler, B. (2005) in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott, D. J., Harding, S. E., and Rowe, A. J., eds) Royal Society of Chemistry, Cambridge
- 29.Acton, S. L., Wong, D. H., Parham, P., Brodsky, F. M., and Jackson, A. P. (1993) Mol. Biol. Cell 4 647-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, F., Khvorova, A., Marshall, W., and Sorkin, A. (2004) J. Biol. Chem. 279 16657-16661 [DOI] [PubMed] [Google Scholar]
- 31.Brodsky, F. M. (1985) J. Cell Biol. 101 2047-2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engqvist-Goldstein, A. E., Warren, R. A., Kessels, M. M., Keen, J. H., Heuser, J., and Drubin, D. G. (2001) J. Cell Biol. 154 1209-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, Y. A., Scales, S. J., Patel, S. M., Doung, Y. C., and Scheller, R. H. (1999) Cell 97 165-174 [DOI] [PubMed] [Google Scholar]
- 34.Lupas, A., Van Dyke, M., and Stock, J. (1991) Science 252 1162-1164 [DOI] [PubMed] [Google Scholar]
- 35.Yang, S., Cope, M. J., and Drubin, D. G. (1999) Mol. Biol. Cell 10 2265-2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engqvist-Goldstein, A. E., Zhang, C. X., Carreno, S., Barroso, C., Heuser, J. E., and Drubin, D. G. (2004) Mol. Biol. Cell 15 1666-1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.