Abstract
α-Defensins are mammalian antimicrobial peptides expressed mainly by cells of myeloid lineage or small intestinal Paneth cells. The peptides are converted from inactive 8.5-kDa precursors to membrane-disruptive forms by post-translational proteolytic events. Because rhesus myeloid pro-α-defensin-4 (proRMAD-4(20–94)) lacks bactericidal peptide activity in vitro, we tested whether neutrophil azurophil granule serine proteinases, human neutrophil elastase (NE), cathepsin G (CG), and proteinase-3 (P3) have in vitro convertase activity. Only NE cleaved proRMAD-4(20–94) at the native RMAD-4 N terminus to produce fully processed, bactericidal RMAD-4(62–94). The final CG cleavage product was RMAD-4(55–94), and P3 produced both RMAD-4(55–94) and RMAD-4(57–94). Nevertheless, NE, CG, and P3 digests of proRMAD4 and purified RMAD-4(62–94), RMAD-4(55–94), and RMAD-4(57–94) peptides had equivalent in vitro bactericidal activities. Bactericidal peptide activity assays of proRMAD-4(20–94) variants containing complete charge-neutralizing D/E to N/Q or D/E to A substitutions showed that (DE/NQ)-proRMAD-4(20–94) and (DE/A)-proRMAD-4(20–94) were as active as mature RMAD-4(62–94). Therefore, proregion Asp and Glu side chains inhibit the RMAD-4 component of full-length proRMAD-4(20–94), perhaps by a combination of charge-neutralizing and hydrogen-bonding interactions. Although native RMAD-4(62–94) resists NE, CG, and P3 proteolysis completely, RMAD-4(62–94) variants with disulfide pairing disruptions or lacking disulfide bonds were degraded extensively, evidence that the disulfide array protects the α-defensin moiety from degradation by the myeloid converting enzymes. These in vitro analyses support the conclusion that rhesus macaque myeloid pro-α-defensins are converted to active forms by serine proteinases that co-localize in azurophil granules.
α-Defensins are effectors of mammalian innate immunity in phagocytic leukocytes of myeloid origin and in small intestine following secretion by epithelial Paneth cells (1). Myeloid and Paneth cell α-defensins are synthesized as ∼10-kDa pre-propeptides that have canonical signal sequences, acidic proregions, and a 3.5–4.5-kDa mature α-defensin peptide component in the C-terminal moiety of the precursor molecule (1). Biosynthesis of functional α-defensins requires proteinase-mediated conversion of inactive precursors to membrane-disruptive, bactericidal peptides (2, 3). In human small bowel, evidence shows that the Paneth cell pro-α-defensin pro-HD5(20–94) is activated after secretion by anionic and meso trypsin isoforms that cleave the precursor at Arg-62 ↓ Ala-63 to produce HD5(62–94), the predominant form of HD5 in the ileum (4). By contrast, mouse enteric pro-α-defensins (pro-Crps) are converted to active forms by matrix metalloproteinase-7 (MMP-7), which co-localizes with pro-α-defensins in Paneth cell secretory granules (2, 5). Pro-Crps lack in vitro bactericidal activity until the proregion is cleaved by MMP-7 at three sites, including the Crp N terminus (2, 3), and the cleavage event at Ser-43 ↓ Ile-44 enables full bactericidal activity and membrane disruptive behavior (2, 3, 6). Post-translational processing of inactive human neutrophil pro-α-defensin 1 (pro-HNP-1) produces major intermediate forms of 75 and 56 amino acids as well as mature HNP-1 (7), but the convertases that mediate pro-α-defensin activation in primate pro-myelocytes remain unknown.
Rhesus macaque myeloid α-defensins (RMADs)3 are broad spectrum bactericidal peptides (8–10). RMADs 1–3 and 8 have similar primary structures that resemble human neutrophil HNPs 1–3, but RMADs 4/5 and 6/7 are very different from HNP-1, yet differ from each other only by a S28F polymorphism (10). proRMAD primary structures deduced from cDNA sequences predict that RMADs 4 and 5 and RMADs 6 and 7 arise by alternative post-translational processing, consistent with the N-terminal Arg in RMADs 4 and 6. Although RMAD-3 and RMAD-4 are both highly bactericidal in vitro, RMAD-4 has anti-HIV activity that RMAD-3 lacks (8).
The activating proteinases of human and mouse Paneth cell pro-α-defensins co-localize with their substrates in secretory granules (1, 11), suggesting that rhesus myeloid pro-α-defensins may be activated by proteinases that also occur in neutrophil azurophil granules. Human neutrophil granules form sequentially during myeloid cell differentiation with azurophilic, myeloperoxidase-positive granules being the first to appear at the myeloblast and promyelocyte stage of neutrophil development (12). Myeloid pro-α-defensins are synthesized and accumulate coincidentally with neutrophil elastase (NE), cathepsin G (CG), or proteinase-3 (P3) serine proteases in pro-myelocytes during myelopoiesis and are localized to the azurophil granules (13) as shown by gene expression profiling and proteomics analyses of neutrophil subcellular fractions (14, 15). Accordingly, we reasoned that the co-localizing serine proteinases could function as convertases for myeloid pro-α-defensins. That hypothesis was tested by exposing recombinant myeloid proRMAD-4(20–94) to human NE, CG, or P3 to test for evidence of proteolysis and in vitro activation of RMAD-4(62–94) bactericidal peptide activity. Each proteinase converted inactive proRMAD-4(20–94) to bactericidal peptides, but only NE cleaved the proRMAD-4(20–94) molecule at the known RMAD-4 N terminus, RMAD-4(62–94). The structural features of proRMAD-4(20–94) that maintain it in an inactive state have been investigated.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant α-Defensin Peptides and Variants—Recombinant RMAD-4(62–94) and precursors were expressed in Escherichia coli as N-terminal 6×-histidine-tagged fusion proteins from the EcoRI and SalI sites of the pET28a expression vector (Novagen, Inc. Madison, WI) as described (2, 9, 16, 17). The following native and variant rhesus macaque myeloid α-defensins RMAD-4(62–94) (10), (C4/11/31/32A)-RMAD-4(62–94), and (6C/A)-RMAD-4(62–94) were prepared. The natural pET-28a cloning primers for RMAD-4(62–94) are pET-RMAD-4-F (5′-ACA CAC GAA TTC ATG AGA CGC ACC TGC CGT) with pET-RMAD-4-R (5′-ACA CAC GTC GAC TCA TCA GCG ACA GCA GAG ACT) (9, 16, 18). Rhesus pro-α-defensins were prepared as follows: The proRMAD-4(20–94) coding sequence was amplified using forward primer 5′-ATA TAG AAT TCA TGA AGT CAC TCC AGG AAA CAG C (EcoRI-M proRMAD-4(20–94)) paired with reverse primer 3′-AAG TCA GAG ACG ACA GCG ACT CAG CTG ATA TA (3′-SalIproRMAD-4(20–94)).
Ala for Cys substitutions were introduced using the following primers in PCR from corresponding positions to those described above to mutagenize an existing pET28a-RMAD-4(62–94) cDNA clone (16). Primers used to mutagenize RMAD-4(62–94) included pET-RMAD-4-C4AC6A-F (5′-ACA CAC GAA TTC ATG AGA CGC ACC GCA CGT GCT), pET-RMAD-4-C31AC32A-R (5′-ACA CAC GTC GAC TCA TCA GCG AGC TGC GAG ACT), RMAD-4-C11A-F (5′-TTT GGC CGT GCC TTC AGG CGT), RMAD-4-C11A-R (5′-ACG CCT GAA GGC ACG GCC AAA), RMAD-4-C21A-F (5′-TCT GGG AGT GCT AAC ATC AAT), RMAD-4-C21-R (5′-ATT GAT GTT AGC ACT CCC AGA), and pET-RMAD-4-C4A-F (5′-ACA CAC GAA TTC ATG AGA CGC ACC GCA CGT TGC).
To test for the functional role of acidic amino acid positions in the proRMAD-4(20–94) proregion, all Asp codons in proRMAD-4(20–61), wherein residues 20–61 form the proregion of proRMAD-4(20–94) were substituted with Asn codons, and all Glu codons were substituted with Gln. Numbering from the initiating methionine and as reported previously (3, 17, 19), a series of mutagenizing PCR reactions were to prepare (E24Q/D27N/D28N/E33Q/E37Q/D38/N/D39N/D41N/E47Q/E48Q)-proRMAD-4(20–94), termed (DE/NQ)-proRMAD-4(20–94). Native proRMAD-4(20–94) cDNA served as template in the first amplification reaction using RMAD-4(62–94) (5′-CTT GCT GTC TCC TTT CAA CAA AAT GGA CTC TCT-3′) and RMAD-4-Stop-SalI-R (5′-CGC GTC GAC TCA TCA GCG ACA GCA GAG ACT GA-3′) as forward and reverse primers, respectively. Successive PCR reactions were used to extend mutagenesis toward the 5′-end of proRMAD-4(20–94). The RMAD-4-Stop-SalI reverse primer was used as the reverse primer in all PCR reactions. The forward primers for the subsequent PCR reactions were RMAD-4(37–94) (5′-CAA AAT AAT CAG AAT CTT GCT GTC TCC TTT CAA, RMAD-4(32–94) (5′-CAG CAA CAG CCT GGG CAA AAT AAT CAG AAT CTT), RMAD-4(27–94) (5′-AAT AAT GCT GCA AC CCA GCA ACA GCC TGG GCA G), RMAD-4(22–94) (5′-CTC CAG CAA ACA GCT AAT AA TGC TGC AAC CCA G-3′), RMAD-4(20–94) (5′-AAG TCA CTC CAG CAA ACA GCT AAT AAT GCT GCA-3′). The forward primer, Start D,E-N,Q proRMAD-4 (5′-CGC GAA TTC ATG AAG TCA CTC CAG CAA ACA GCT-3′) was paired with the reverse primer RMAD-4-Stop-SalI in the final PCR reaction.
To prepare (E24A/D27A/D28A/E33A/E37A/D38A/D39A/D41A/E47A/E49A)-proRMAD-4(20–94), termed (DE/A)-proRMAD-4(20–94)), Asp and Glu codons in proRMAD-4(20–61) were changed to Ala codons as described above using the following mutagenizing primers. The forward primers used to prepare (DE/A)-proRMAD-4(20–94) were proRMAD4 D,E-A(42–94) (5′-CTT GCT GTC TCC TTT GCA GCT AAT GGA CTC TCT ACT), proRMAD-4(20–94) D,E-A(35–94) (5′-CAG CCT GGG GCA GCT GCA CAG GCA CTT GCT GTC), proRMAD-4(20–94) D,E-A(29–94) (5′-GCT GCA ACC CAG GCA CAG CCT GGG), proRMAD-4(20–94) D,E-A(20–94) (5′-AAG TCA CTC CAG GAA ACA GCT GAT GAC GCT GCA ACC). Primer proRMAD-4(20–94) D,E-A Start F (5′-CGC GAA TTC ATG AAG TCA CTC CAG GCA ACA-3′) was paired with reverse primer RMAD-4-Stop-SalI in the final PCR reaction.
The order of PCR-based mutagenesis reactions and their design followed a scheme used previously to mutagenize Cys residue positions in mouse Crp4 (17). Briefly, in PCR reaction 1, a mutant forward primer containing the mutated codon flanked by three natural codons was paired with a reverse cloning primer, and PCR reaction 2 paired a primer that was the reverse-complement of the mutant forward primer with a forward cloning primer. Samples of purified products from reactions 1 and 2 were combined as templates in PCR reaction 3, using the forward and reverse cloning primers as amplimers, and these putative mutant proRMAD-4 products were cloned in pCR-2.1 TOPO and verified by DNA sequencing. Products were subcloned into SalI and EcoRI sites in pET28a plasmid DNA (Novagen, Inc., Madison, WI), and transformed into E. coli BL21 (DE3)-CodonPlus-RIL cells (Stratagene) for recombinant expression (3, 16, 17).
Purification of Recombinant Proteins—Recombinant proteins were expressed at 37 °C in Terrific Broth medium by induction with 0.1 mm isopropyl-β-d-1-thiogalactopyranoside for 6 h at 37 °C, lysed by sonication in 6 m guanidine-HCl in 100 mm Tris-Cl, pH 8.1, clarified by centrifugation (2, 6, 20), and His-tagged fusion peptides were purified as described (8, 16, 17). After CNBr cleavage of fusions, the recombinant peptides were purified by C18 reverse-phase high performance liquid chromatography (RP-HPLC), and quantitated by absorbance at 230 nm. Molecular masses of purified peptides were determined using matrix-assisted laser desorption ionization mode mass spectrometry (Voyager-DE MALDI-TOF, PE-Biosystems, Foster City, CA) in the UCI Physical Sciences Mass Spectroscopy Facility. Peptide homogeneity was confirmed by acidurea polyacrylamide gel electrophoresis (AU-PAGE), (21) and analytical C18 RP-HPLC at 230 nm.
Bactericidal Peptide Assays—Recombinant peptides were tested for microbicidal activity against E. coli ML35, Salmonella enterica serovar Typhimurium (S. typhimurium) ΔphoP, wild-type S. typhimurium strains CS022, JSG210, and 14082 (from Dr. Samuel I. Miller, University of Washington), Vibrio cholerae, Staphylococcus aureus 710a, and Listeria monocytogenes 104035 (22). Bacteria growing exponentially in trypticase soy broth at 37 °C, were deposited by centrifugation at 1700 × g for 10 min, washed in 10 mm PIPES (pH 7.4), and resuspended in 10 mm PIPES (pH 7.4) supplemented with 0.01 volumes of trypticase soy broth (2, 6). Bacteria (1–5 × 106 colony-forming units per ml (CFU/ml)) were incubated with test peptides in 50 μl for 1 h in a shaking incubator at 37 °C; then 20-μl samples of incubation mixtures were diluted 1:100 with 10 mm PIPES (pH 7.4), and 50 μl of the diluted samples were plated on trypticase soy agar plates using an Autoplate 4000 (Spiral Biotech Inc., Bethesda, MD). Surviving bacteria were counted as CFU/ml after incubation at 37 °C for 12–18 h. Bactericidal assays performed in Fig. 1B are against a defensin-sensitive strain S. typhimurium ΔphoP whereas the activities shown in Fig. 2B are against three different species of bacteria. Although the peptide concentrations were indicated in μm concentrations in Fig. 1B and as μg/ml in 2B, the data in Figs. 1B and 2B are not discordant.
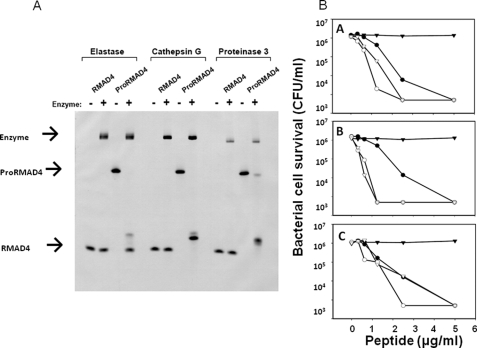
FIGURE 1.
In vitro cleavage and activation of proRMAD-4(20–94) by NE, CG, and P3. In A, samples of proRMAD-4(20–94) (10 μg) were incubated overnight in vitro with (+) or without (–) NE, CG, and P3, and digests were resolved by AU-PAGE and stained with Coomassie Blue (see “Experimental Procedures”). proRMAD-4(20–94) is digested by NE, and the digested product runs alongside native RMAD-4(62–94). CG and P3 digest proRMAD-4(20–94), and the products have lower mobilities in AU-PAGE analysis. In B, bactericidal activities against S. typhimurium ΔphoP of recombinant proRMAD-4(20–94), RMAD-4(62–94), and the complete digests of proRMAD-4(20–94) incubated with each of the azurophil granule serine proteases. Panels A, peptides exposed to NE; B, peptides exposed to CG; C, peptides exposed to P3. Symbols:(-▾-), proRMAD-4(20–94) with no serine protease; (-○-), proRMAD-4(20–94) with serine protease; (-▿-), RMAD-4(62–94) with no serine protease; (-•-), RMAD-4(62–94) exposed to serine protease. proRMAD-4(20–94) lacks bactericidal activity, native RMAD-4(62–94) is bactericidal. The three serine proteases convert inactive proRMAD-4(20–94) to a bactericidal molecule.
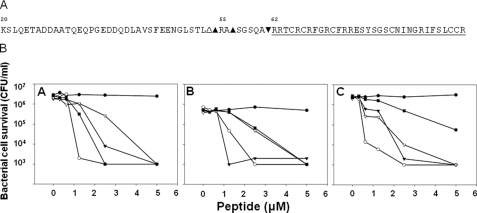
FIGURE 2.
The major serine protease cleavage products of proRMAD-4(20–94) have equivalent activity. In A, the final azurophil granule serine protease cleavage sites of proRMAD-4(20–94) deduced by MALDI-TOF MS. The native RMAD4(62–94) primary structure is underlined. The major products of proRMAD4(20–94) proteolysis are: NE, RMAD4(62–94) (▾); CG, RMAD4(55–94) (▵); P 3-RMAD4(55–94) and RMAD4(57–94) (▴). B, bactericidal activity of recombinant proRMAD-4(20–94), RMAD-4(62–94), and the final serine proteinase cleavage products against E. coli ML35. Exponentially growing E. coli ML35 (A), S. aureus (B) and V. cholerae (C) were exposed to the peptides at 37 °C in 50 μl of PIPES-TSB buffer for 1 h (see “Experimental Procedures”). Consistent with Fig. 1B, proRMAD-4(20–94) (-•-) lacks bactericidal activity, and native RMAD-4(62–94) (-○-), RMAD4(62–94) (-▾-)RMAD4(55–94), (-▿-), and RMAD4(55–94) +RMAD-4(57–94) (-▪-) are equally bactericidal.
Analyses of proRMAD-4(20–94) and RMAD-4(62–94) in in Vitro Proteolysis—Recombinant RMAD-4(62–94), proRMAD-4(20–94), and disulfide variants digested with NE, P3, or CG (Elastin Products Company, Inc) as described below were analyzed for susceptibility to proteolysis by AU-PAGE, and samples of the proteolytic digests were tested in bactericidal peptide assays and analyzed by MALDI-TOF MS (17). Samples (11 μg) of proRMAD-4(20–94) and disulfide variants and 5-μg samples of RMAD-4(62–94) were incubated with proteinases at 37 °C for 2 h at a substrate to enzyme molar ratio of 4:1 as follows: NE and P3 in 50 mm Tris, 150 mm NaCl (pH 7.5), and CG in the same buffer at pH 8.3. Equimolar quantities of all digests were analyzed by AU-PAGE. The biological effects of enzyme-specific proteolysis of proRMAD-4(20–94) molecules were determined using bactericidal peptide assays as described (9, 16, 17). To test for proteinase-mediated activation of bactericidal peptide activity, equimolar quantities (0 to 20 μg/ml) of RMAD-4(62–94), proRMAD-4(20–94), or their variants were incubated for 2 h or 18 h at 37 °C with or without proteinases as noted in individual legends. Subsequently, digests were incubated with exponentially growing bacterial cells (∼1–5 × 106 CFU/ml) for 60 min at 37 °C (2). Samples (20 μl) of each incubation mixture were diluted 1:100 with 10 mm PIPES (pH 7.4) and 50 μl of the diluted, peptide- or digest-exposed bacteria were plated on trypticase soy agar using a Spiral Biotech Autoplate 4000 device (Spiral Biotech Inc., Bethesda, MD). Surviving bacteria were quantitated as CFU/ml on plates after incubation at 37 °C for 12–18 h.
Purification of Digestion Products of proRMAD-4(20–94)—40 μg of proRMAD-4(20–94) digested with 1 μg of NE, CG, or P3 and the products of proteolysis were separated and purified using C18-RP HPLC, quantitated by absorbance 230 nm, and molecular masses of digested peptides were determined using MALDI-TOF MS. Peptide homogeneity was assessed by AU-PAGE (21).
Permeabilization of Live E. coli ML35 Cells—Exponentially growing E. coli ML35 cells were washed and resuspended in 10 mm PIPES (pH 7.4) supplemented with 0.01 volume of trypticase soy broth. Bacteria were exposed in triplicate to RMAD-4(62–94), proRMAD-4(20–94), and (DE/NQ)-proRMAD-4(20–94) in the presence of 2.5 mm ONPG for 2 h at 37 °C. E. coli ML35 express β-galactosidase constitutively but are permease-negative and do not take up ONPG unless permeabilized by external factors, such as defensins. β-Galactosidase hydrolysis was measured at 405 nm on a 96-well Spectra-Max plate spectrophotometer (Molecular Devices, Sunnyvale, CA).
RESULTS
In Vitro Activation of the Rhesus Myeloid proRMAD-4-(20–94)— The precursors of mouse Paneth cell α-defensin Crp4 and human neutrophil α-defensins 1–3 (HNP-1–3) lack bactericidal activity until enzymatic conversion to their active forms (2, 3, 23–25). To test whether rhesus macaque myeloid pro-α-defensins also require proteolytic conversion from inactive precursors to active forms, the bactericidal peptide activities of recombinant RMAD-4(62–94) and proRMAD-4(20–94) molecules were compared. In contrast to the robust in vitro bactericidal activity of mature RMAD-4(62–94) and consistent with findings for proCrp4 and proHNP-1, equimolar quantities of proRMAD-4(20–94) had no bactericidal effects against varied bacterial cell targets (Fig. 1B). Thus, RMAD-4(62–94) bactericidal activity also is dependent on protease-mediated precursor activation.
The mouse and human Paneth cell α-defensin-activating convertases are abundant secretory granule constituents (4, 5, 20). Accordingly, we tested whether neutrophil serine proteinases, which co-localize with α-defensins in azurophil granules, can activate proRMAD-4(20–94) in vitro. Digests of proRMAD-4(20–94) with NE, CG, or P3 were equally bactericidal and as active as native, fully processed RMAD-4(62–94), evidence that each enzyme activated proRMAD-4(20–94) to the same extent under these in vitro conditions (Fig. 1B). None of the proteases cleaved native RMAD-4(62–94) or the α-defensin component of proRMAD-4(20–94) as is evident from the approximate co-migration of the major cleavage products with native RMAD-4(62–94) in AU-PAGE (Fig. 1A). The major product of proRMAD-4(20–94) cleavage with NE co-migrated with RMAD-4(62–94), showing that it cleaved the proform at the RMAD-4 N terminus (Fig. 1A). On the other hand, the major products of CG and P3 digestion did not, even though NE, CG, and P3 digests of proRMAD-4(20–94) were equally bactericidal (Fig. 1B).
The Major Products of proRMAD-4(20–94) Serine Proteolysis Have Equivalent Activity—Because samples of complete in vitro digests of proRMAD-4(20–94) with NE, CG, or P3 were bactericidal, the active bactericidal molecules in those digests were characterized by purification of the major serine proteinase cleavage products using C18 RP-HPLC (see “Experimental Procedures”). Based on the known C18 elution profiles of proRMAD-4(20–94) and RMAD-4(62–94), the major RMAD-containing fractions were identified using MALDI-TOF MS. To summarize, the major product purified from NE digests had a mass of 3964.6 AMU, which corresponds precisely to native RMAD-4(62–94). Similarly, the major CG digestion product was 4627 AMU, and the two main products of P3 cleavage had masses of 4627 and 4396 AMU. Based on these analyses, the final cleavage events mediated by these proteinases occur at Ala-61 ↓ Arg-62 for NE, the N terminus of native RMAD-4, at Leu-54 ↓ Arg-55 for CG, corresponding to RMAD-4(55–94), and at Leu-54 ↓ Arg-55 and Ala-56 ↓ Ser-57 for P3, corresponding to RMAD-4(55–94) and RMAD-4(57–94), respectively (Fig. 2A). Although the NE and CG enzymes have endogenous antimicrobial activity, NE and P3 lack activity under the conditions of these bactericidal peptide assays (supplemental Fig. S1). Similarly, CG has modest bactericidal activity at 10 μg/ml, but that concentration is ten times greater than those used in these experiments (supplemental Fig. S1).
Bactericidal Activities of Major proRMAD-4 Serine Protease Cleavage Products—Bactericidal peptide activities of these C18-purified proRMAD-4 cleavage products were assayed against E. coli ML 35, S. aureus and V. cholerae (Fig. 2B). The major proRMAD-4 products of in vitro serine protease processing were equivalent in bactericidal activity, comparable to that of RMAD-4(62–94) (Fig. 2B). At low peptide concentrations, e.g. <3 μm, modest differences in the activities of native RMAD-4(62–94), RMAD-4(55–94), RMAD-4(55–94), and RMAD-4(57–94) are observed, but at 5 μm all the proRMAD-4(20–94) digest products, RMAD-4(62–94), RMAD-4(55–94), and RMAD-4(57–94) had equivalent activity. In Fig. 2B, panel C, the major P3 cleavage products of proRMAD-4 are attenuated relative to RMAD-4(62–94) at 5 μm peptide; but in replicate assays, 5 μm P3 products and RMAD-4(62–94) often have equivalent activities. Because α-defensin concentrations in human neutrophil phagolysosomes has been estimated to be in the millimolar range (26), the modest differences between these variants at low micromolar concentrations in Fig. 2B seem unlikely to be of consequence in vivo. Therefore, we conclude that the RMAD-4 variants generated by CG and P3 processing are equal to native RMAD-4(62–94) in activity.
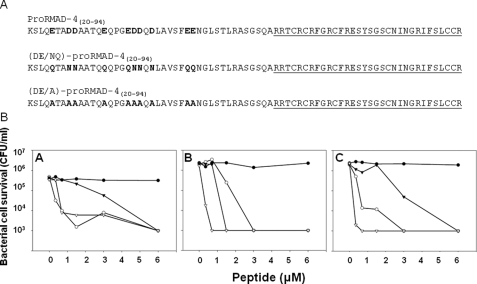
Functional Effects of Mutagenesis in proRMAD-4(20–49) at Acidic Residue Positions—In mouse proCrp4(20–92), acidic residue positions in proCrp4(20–43) fully inhibit precursor bactericidal activity (3). To test whether a similar mechanism regulates myeloid pro-α-defensin bactericidal action, Asp and Glu carboxyl groups were eliminated from the proRMAD-4(20–94) proregion by replacements with Asn and Gln amines, respectively or by Ala substitutions at all Asp and Glu positions (Fig. 3A). Equimolar quantities of RMAD-4(62–94), proRMAD-4(20–94), and (DE/NQ)-proRMAD-4(20–94) were assays (Fig. 3B). At low peptide levels, (DE/NQ)-proRMAD-4(20–94) showed greater bactericidal activity, than native proRMAD-4(20–94), which lacks activity, but showed attenuated bactericidal activity relative to native RMAD-4(62–94) (Fig. 3B). On the other hand, at concentrations ≥ 6 μm, (DE/NQ)-proRMAD-4(20–94) and RMAD-4(62–94) were equally active; under the conditions of these assays, proRMAD-4(20–94) lacks activity (Fig. 3B). Thus, removing the anionic charge of the Asp and Glu carboxyl groups was insufficient to override the inhibitory effect of the combined Asp and Glu positions completely. Accordingly, (DE/A)-proRMAD-4(20–94) (Fig. 3A) was compared with RMAD-4(62–94) and proRMAD-4(20–94) in in vitro bactericidal peptide assays. Unexpectedly, (DE/A)-proRMAD-4(20–94) displayed greater bacterial cell killing activity than native, fully processed RMAD-4(62–94), showing that the mutagenesis abolished all inhibition of proRMAD-4(20–94) activity (Fig. 3B). This finding is consistent with the effects of Gly replacements at Asp and Glu residue positions in the mouse pro-Crp4(20–92) proregion in which the resulting (DE/G)-pro-Crp4(20–92) molecule and Crp4 had equal activities (3).
FIGURE 3.
Bactericidal activities of recombinant (DE/NQ)-proRMAD-4(20–94) and (DE/A)-proRMAD-4(20–94). In A, the primary amino acid structures of recombinant (DE/NQ)-proRMAD-4(20–94) and (DE/A)-proRMAD-4(20–94) are shown. In B, exponentially growing V. cholerae (A), E. coli ML35 (B), and L. monocytogenes (C) were exposed to the peptides, and surviving bacteria were quantified as CFU/ml (see “Experimental Procedures”). Symbols: proRMAD-4(20–94) (-•-), RMAD-4(62–94) (-○-), (DE/NQ)-proRMAD-4(20–94) (-▾-), and (D,E/A)-proRMAD-4(20–94) (-▿-). Acidic proregion residues maintain proRMAD-4(20–94) in an inactive state. In panel B, the data points for the two peptides (DE/NQ)-proRMAD-4(20–94) and (D,E/A)-proRMAD-4(20–94) are superimposed.
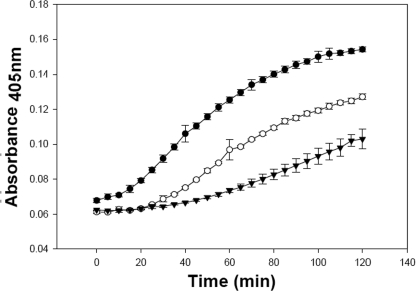
As an index of relative membrane disruptive activity, RMAD-4(62–94), proRMAD-4(20–94), and (DE/NQ)-proRMAD-4(20–94) were evaluated by performing permeabilization assays on live in E. coli ML35 cells exposed to 0.75 μm to 6.0 μm peptide levels (see “Experimental Procedures”). Consistent with the general behavior of α-defensins and their precursors, exposure to RMAD-4(62–94) induced the greatest level of ONPG conversion, and inactive proRMAD-4(20–94) did not perturb the E. coli cell membranes enough for measureable ONPG to diffuse into the cells (Fig. 4). At every concentration tested, exposure of E. coli ML35 cells to (DE/NQ)-proRMAD-4(20–94) induced greater ONPG conversion than equimolar native proRMAD-4(20–94) but less permeabilization than that caused by RMAD-4(62–94) (Fig. 4). Thus, in both bactericidal and membrane permeabilization assays, eliminating Asp and Glu carboxyl groups alone is sufficient to relieve proregion inhibition of proRMAD-4(20–94) activity at ≥ 6 μm peptide levels, but below 6 μm, the Asn and Gln replacements in (DE/NQ)-proRMAD-4(20–94) inhibit the precursor RMAD-4 moiety at an intermediate level. Possibly, although Asp and Glu mutagenesis eliminates charge neutralizing effects of carboxyl groups in the proform, hydrogen bonding interactions between Asn and Gln side chains and RMAD-4 basic residues also may contribute to blocking proRMAD-4(20–94) bactericidal activity. Alternatively, proregion amino acids other than Asp and Glu may also inhibit proRMAD-4(20–94) bactericidal action. These collective results suggest that proRMAD-4(20–94) lacks bactericidal activity because proregion Asp and Glu side chains prevent electrostatic RMAD-4 interactions with bacterial cell targets through a combination of charge neutralizing and hydrogen-bonding interactions (3, 6, 16).
FIGURE 4.
Permeabilization of live E. coli ML35 cells by RMAD-4(62–94), proRMAD-4(20–94), and (DE/NQ)-proRMAD-4(20–94). E. coli ML35 cells growing in log-phase were exposed to 6 μm peptide concentrations in the presence of ONPG at 37 °C (see “Experimental Procedures”). (DE/NQ)-proRMAD-4(20–94) (-○-) induces greater membrane permeabilization and ONPG conversion than proRMAD-4(20–94) (-▾-) but less than RMAD-4(62–94) (-•-).
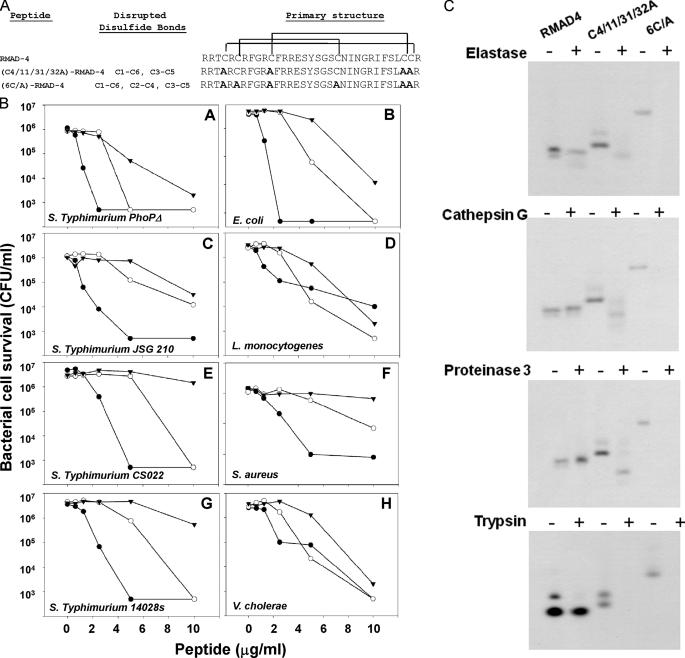
The Disulfide Array Protects RMAD-4(62–94) from NE, CG, and P3 Proteolysis—Analyses of mouse Crp4 have shown that the canonical α-defensin disulfide connectivities are not required for bactericidal activity but confer protection against proteolysis by the activating convertase MMP-7 (17). To test whether that function is a conserved feature of α-defensins, Ala for Cys substitutions were made in RMAD-4(62–94) (Fig. 5A) to retain only the CII-CIV pairing ((C4/11/31/32A)-RMAD-4(62–94)) or to disrupt all disulfide bonds ((6C/A)-RMAD-4(62–94)). The effects of mutagenesis on bactericidal peptide activities and on the NE, CG, P3, and trypsin sensitivities of recombinant (C4/11/31/32A)-RMAD-4(62–94 and (6C/A)-RMAD-4(62–94) variants were compared with native RMAD-4(62–94) (10, 16). Consistent with Crp4 disulfide mutagenesis (17), disrupting disulfide bonds in RMAD-4(62–94) did not eliminate in vitro bactericidal activities in relation to the parent molecule (Fig. 5B). Against S. typhimurium CS022, S. aureus, and S. typhimurium 14028S, eliminating RMAD-4(62–94) disulfide bonds attenuated bactericidal activity slightly, and, in contrast to findings with Crp4, none of the disulfide variants had improved activities (Fig. 5B). Ala for Cys substitutions in RMAD-4(62–94) resulted in sensitivity to in vitro proteolysis by NE, CG, and P3, the apparent convertases (Fig. 5C). Because RMAD-4(62–94) is Arg-rich and thus potentially susceptible to trypsin proteolysis, (C4/11/31/32A)-RMAD-4(62–94), and (6C/A)-RMAD-4(62–94) were exposed to trypsin. The disulfide variants were degraded so extensively that the peptides could not be visualized following proteinase exposure (Fig. 5C). In marked contrast, native RMAD-4(62–94) was completely resistant to trypsin, NE, CG, or P3, which had no effect on peptide mass or migration in AU-PAGE (Fig. 5C) as a function of its disulfide pairings. Based on these analyses of RMAD-4(62–94) and of Crp4 (17), it seems likely that α-defensin disulfide pairings evolved to protect the molecules from proteolysis during post-translational activation, regardless of the activating convertase.
FIGURE 5.
Disulfide bonds protect RMAD-4(62–94) from degradation by serine proteases. Panel A shows the primary amino acid sequences of RMAD-4(62–94) and the two disulfide variants (C4/11/31/32/A)-RMAD-4(62–94) and (6C/A)-RMAD-4(62–94) (see “Experimental Procedures”). CI-CVI and CIII-CV disulfide bonds are disrupted in (C4/11/31/32/A)-RMAD-4(62–94) and all disulfide bonds CI-CVI, CIII-CV, and CII-CIV are disrupted in (6C/A)-RMAD-4(62–94). Mutated Cys positions converted to Ala are shown in bold font (right panel). In B, the bactericidal activities of RMAD-4(62–94) disulfide variants are compared. Exponentially growing bacterial cells were exposed to (C4/11/31/32A)-RMAD-4(62–94) (-○-), (6C/A)-RMAD-4(62–94) (-▾-), and native RMAD-4(62–94) (-•-) for 1 h at 37 °C (see “Experimental Procedures”). Both disulfide variants have activities comparable with RMAD-4 against S. typhimurium ΔphoP (A), E. coli (B), S. typhimurium JSG 210 (C), L. monocytogenes (D), and V. cholerae (H). Against S. typhimurium CS022 (E), S. aureus (F), and S. typhimurium 14028s (G), (6C/A)-RMAD-4 has attenuated activity relative to RMAD-4. C, equal quantities (10 μg) of RMAD-4, (C4/11/28/29A)-RMAD-4(62–94), and (6C/A)-RMAD-4(62–94) were digested for 18 h in the presence of NE, CG, P3, or trypsin, resolved on AU-PAGE, and stained with Coomassie Blue. Disulfide bonds protect α-defensins from proteolysis during conversion to active forms.
DISCUSSION
The three azurophil granule serine proteases, NE, CG, and P3, cleave and activate proRMAD-4(20–94) in vitro. The fact that the human and mouse Paneth cell pro-α-defensin convertases co-localize with their substrates in secretory granules (4, 5), suggested that proRMAD-4(20–94) activation also could be mediated by neutrophil proteinases associated with azurophil granules (13). To test this hypothesis, recombinant proRMAD-4(20–94) molecules were exposed to NE, CG, or P3 to test for evidence of proteolysis and bactericidal peptide activity. Each enzyme cleaved proRMAD-4(20–94) at several different sites within the proregion, at no site within the α-defensin moiety, and processing by all three enzymes resulted in activation. Although only NE cleaved proRMAD-4(20–94) at the known RMAD-4 peptide N terminus to yield RMAD4(62–94) (Fig. 2A), all final processed products of proRMAD-4(20–94) activation, RMAD-4(55–94), RMAD-4(57–94), or RMAD-4(62–94), have equivalent activities (Fig. 2B). NE and P3 cleave at the N terminus of small aliphatic residues such as Ala, and CG cleaves at the N terminus of Lys. These sites are consistent with their known recognition sites. Unexpectedly, P3 cleaves at Arg in proRMAD-4 perhaps because the tertiary structure of the folded precursor influences sterically an alternative cleavage that P3 is able to recognize (27). Thus, NE, CG, and P3 are individually capable of mediating proRMAD-4(20–94) activation to the same extent in vitro even though CG and P3 do not cleave at the RMAD-4 natural N terminus. Because they co-localize with α-defensins in neutrophil azurophilic granules, it seems likely that the three serine proteinases would participate coincidentally in pro-α-defensin activation with NE catalyzing the final Ala-61 ↓ Arg-62 cleavage event. This suggestion is supported by the fact that CG and P3 intermediates have not been isolated from macaque neutrophils (Tang et al. (10)).
The proregion of proRMAD-4(20–94) maintains the precursor molecule in an inactive state until it is cleaved by the azurophil granule serine proteases, and activation of pro-α-defensins involves the removal of the proregion acidic residues from covalent association with the RMAD-4(62–94) component of the precursor. The cationic residues in α-defensins play an essential role in α-defensin antibacterial activity as seen in mouse Crp4 (3), and the human enteric α-defensin HD5 (4). Analyses of (DE/NQ)-proRMAD-4(20–94) showed that the variant proregion inhibited bactericidal activity less than native prosegment at precursor concentrations ≤ 6 μm, but ≥ 6 μm (DE/NQ)-proRMAD-4(20–94) and processed RMAD-4(62–94) had equivalent activities (Fig. 3B). Unexpectedly, (DE/A)-proRMAD-4(20–94) showed equivalent or greater activity than native RMAD-4(62–94) (Fig. 3B), perhaps because the Ala substitutions introduced ten moderately hydrophobic side chains to the proregion that may facilitate (DE/A)-proRMAD-4(20–94) interactions with the bacterial cell envelopes. This finding is also consistent with the mouse enteric α-defensin Crp-4 and its precursor variant (DE/G)-proCrp-4, which have equivalent activity (3). These collective results suggest that proRMAD-4(20–94) lacks bactericidal activity because proregion Asp and Glu side chains prevent electrostatic RMAD-4(62–94) interactions with bacterial cell targets through a combination of charge neutralizing and hydrogen-bonding interactions (3, 16, 28).
The three intramolecular disulfide bonds that characterize α-defensins are an invariant structural feature of the peptide family (7, 29–31), but peptide bactericidal activity does not require that the array be intact (17, 32). This conclusion is supported by analyses of site-directed Ala for Cys substitutions in the mouse enteric α-defensin, where each of the Crp-4 disulfide variants were as active as native Crp-4 (17) and also seen in the human β-defensin-3 (33). Similarly, RMAD-4(62–94) variants lacking two or three disulfide bonds were as bactericidal as the native RMAD-4(62–94) against varied bacterial cell targets (Fig. 5B). Consistent with the sensitivity of Crp4 disulfide variants to MMP-7 (17), NE, CG, and P3 degraded RMAD-4(62–94) disulfide variants extensively (Fig. 5C). As myeloid α-defensins are rich in Arg, we tested proRMAD-4(20–94) sensitivity to trypsin proteolysis. Not surprisingly, trypsin cleaved within the proregion of proRMAD-4(20–94), and as seen with serine protease exposure, RMAD-4(62–94) was unaffected by incubation with trypsin, but disulfide variants were degraded (Fig. 5C). From that, we infer that the disulfide array protects RMAD-4(62–94), as well as other α-defensins, from degradation during proteolytic activation, in phagolysosomes, and perhaps following release into extracellular environment of the intestinal lumen and at the sites of inflammation.
Supplementary Material
Acknowledgments
We thank Drs. Michael E. Selsted, Wuyuan Lu, and Dat Tran for useful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants AI059346 and DK044632 and the Human Frontiers Science Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: RMAD, rhesus myeloid α-defensin; NE, human neutrophil elastase; CG, human cathepsin G; P3, human proteinase 3; MALDI-TOF-MS, matrix-assisted laser desorption ionization time-of-flight mass spectrometry; RP-HPLC, reverse-phase high performance liquid chromatography; AU-PAGE, acid-urea polyacrylamide gel electrophoresis; PIPES, 1,4-piperazinediethanesulfonic acid; AMU, atomic mass unit.
References
- 1.Selsted, M. E., and Ouellette, A. J. (2005) Nat. Immunol. 6 551–557 [DOI] [PubMed] [Google Scholar]
- 2.Shirafuji, Y., Tanabe, H., Satchell, D. P., Henschen-Edman, A., Wilson, C. L., and Ouellette, A. J. (2003) J. Biol. Chem. 278 7910–7919 [DOI] [PubMed] [Google Scholar]
- 3.Weeks, C. S., Tanabe, H., Cummings, J. E., Crampton, S. P., Sheynis, T., Jelinek, R., Vanderlick, T. K., Cocco, M. J., and Ouellette, A. J. (2006) J. Biol. Chem. 281 28932–28942 [DOI] [PubMed] [Google Scholar]
- 4.Ghosh, D., Porter, E., Shen, B., Lee, S. K., Wilk, D., Drazba, J., Yadav, S. P., Crabb, J. W., Ganz, T., and Bevins, C. L. (2002) Nat. Immunol. 3 583–590 [DOI] [PubMed] [Google Scholar]
- 5.Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M., and Parks, W. C. (1999) Science 286 113–117 [DOI] [PubMed] [Google Scholar]
- 6.Satchell, D. P., Sheynis, T., Shirafuji, Y., Kolusheva, S., Ouellette, A. J., and Jelinek, R. (2003) J. Biol. Chem. 278 13838–13846 [DOI] [PubMed] [Google Scholar]
- 7.Ganz, T. (2003) Nat. Rev. Immunol. 3 710–720 [DOI] [PubMed] [Google Scholar]
- 8.Tanabe, H., Ouellette, A. J., Cocco, M. J., and Robinson, W. E., Jr. (2004) J. Virol. 78 11622–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanabe, H., Yuan, J., Zaragoza, M. M., Dandekar, S., Henschen-Edman, A., Selsted, M. E., and Ouellette, A. J. (2004) Infect. Immun. 72 1470–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang, Y. Q., Yuan, J., Miller, C. J., and Selsted, M. E. (1999) Infect. Immun. 67 6139–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouellette, A. J. (2006) Curr. Top. Microbiol. Immunol. 306 1–25 [DOI] [PubMed] [Google Scholar]
- 12.Bainton, D. F., Ullyot, J. L., and Farquhar, M. G. (1971) J. Exp. Med. 134 907–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borregaard, N., Sorensen, O. E., and Theilgaard-Monch, K. (2007) Trends Immunol. 28 340–345 [DOI] [PubMed] [Google Scholar]
- 14.Cowland, J. B., and Borregaard, N. (1999) J. Leukoc Biol. 66 989–995 [DOI] [PubMed] [Google Scholar]
- 15.Arnljots, K., Sorensen, O., Lollike, K., and Borregaard, N. (1998) Leukemia 12 1789–1795 [DOI] [PubMed] [Google Scholar]
- 16.Tanabe, H., Qu, X., Weeks, C. S., Cummings, J. E., Kolusheva, S., Walsh, K. B., Jelinek, R., Vanderlick, T. K., Selsted, M. E., and Ouellette, A. J. (2004) J. Biol. Chem. 279 11976–11983 [DOI] [PubMed] [Google Scholar]
- 17.Maemoto, A., Qu, X., Rosengren, K. J., Tanabe, H., Henschen-Edman, A., Craik, D. J., and Ouellette, A. J. (2004) J. Biol. Chem. 279 44188–44196 [DOI] [PubMed] [Google Scholar]
- 18.Aarbiou, J., Ertmann, M., van Wetering, S., van Noort, P., Rook, D., Rabe, K. F., Litvinov, S. V., van Krieken, J. H., de Boer, W. I., and Hiemstra, P. S. (2002) J. Leukoc Biol. 72 167–174 [PubMed] [Google Scholar]
- 19.Rosengren, K. J., Daly, N. L., Fornander, L. M., Jonsson, L. M., Shirafuji, Y., Qu, X., Vogel, H. J., Ouellette, A. J., and Craik, D. J. (2006) J. Biol. Chem. 281 28068–28078 [DOI] [PubMed] [Google Scholar]
- 20.Ayabe, T., Satchell, D. P., Pesendorfer, P., Tanabe, H., Wilson, C. L., Hagen, S. J., and Ouellette, A. J. (2002) J. Biol. Chem. 277 5219–5228 [DOI] [PubMed] [Google Scholar]
- 21.Selsted, M. E. (1993) Genet. Eng. (N Y) 15 131–147 [DOI] [PubMed] [Google Scholar]
- 22.Lehrer, R. I., Barton, A., and Ganz, T. (1988) J. Immunol. Methods 108 153–158 [DOI] [PubMed] [Google Scholar]
- 23.Valore, E. V., and Ganz, T. (1992) Blood 79 1538–1544 [PubMed] [Google Scholar]
- 24.Valore, E. V., Martin, E., Harwig, S. S., and Ganz, T. (1996) J. Clin. Investig. 97 1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganz, T., Liu, L., Valore, E. V., and Oren, A. (1993) Blood 82 641–650 [PubMed] [Google Scholar]
- 26.Ganz, T., Selsted, M. E., Szklarek, D., Harwig, S. S., Daher, K., Bainton, D. F., and Lehrer, R. I. (1985) J. Clin. Investig. 76 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett, A. J., Rowlings, N. D., and Woessner, J. F. (2004) Handbook of Proteolytic Enzymes, 2nd Ed., Elsevier Academic Press, New York
- 28.Satchell, D. P., Sheynis, T., Kolusheva, S., Cummings, J., Vanderlick, T. K., Jelinek, R., Selsted, M. E., and Ouellette, A. J. (2003) Peptides 24 1795–1805 [DOI] [PubMed] [Google Scholar]
- 29.Lehrer, R. I., Ganz, T., and Selsted, M. E. (1991) Cell 64 229–230 [DOI] [PubMed] [Google Scholar]
- 30.Selsted, M. E., and Harwig, S. S. (1989) J. Biol. Chem. 264 4003–4007 [PubMed] [Google Scholar]
- 31.White, S. H., Wimley, W. C., and Selsted, M. E. (1995) Curr. Opin. Struct. Biol. 5 521–527 [DOI] [PubMed] [Google Scholar]
- 32.Wu, Z., Li, X., de Leeuw, E., Ericksen, B., and Lu, W. (2005) J. Biol. Chem. 280 43039–43047 [DOI] [PubMed] [Google Scholar]
- 33.Wu, Z., Hoover, D. M., Yang, D., Boulegue, C., Santamaria, F., Oppenheim, J. J., Lubkowski, J., and Lu, W. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8880–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.