Abstract
The redox state of tissues tends to become progressively more prooxidizing during the aging process. The hypothesis tested in this study was that enhancement of reductive capacity by overexpression of glucose-6-phosphate dehydrogenase (G6PD), a key enzyme for NADPH biosynthesis, could protect against oxidative stress and extend the life span of transgenic Drosophila melanogaster. Overexpression of G6PD was achieved by combining a UAS-G6PD responder transgene at one of four independent loci with either a broad expression (armadillo-GAL4, Tubulin-GAL4, C23-GAL4, and da-GAL4) or a neuronal driver (D42-GAL4 and Appl-GAL4). The mean life spans of G6PD overexpressor flies were extended, in comparison with driver and responder controls, as follows: armadillo-GAL4 (up to 38%), Tubulin-GAL4 (up to 29%), C23-GAL4 (up to 27%), da-GAL4 (up to 24%), D42-GAL4 (up to 18%), and Appl-GAL4 (up to 16%). The G6PD enzymatic activity was increased, as were the levels of NADPH, NADH, and the GSH/GSSG ratio. Resistance to experimental oxidative stress was enhanced. Furthermore, metabolic rates and fertility were essentially the same in G6PD overexpressors and control flies. Collectively, the results demonstrate that enhancement of the NADPH biosynthetic capability can extend the life span of a relatively long-lived strain of flies, which supports the oxidative stress hypothesis of aging.
Glucose-6-phosphate dehydrogenase catalyzes the oxidation of glucose-6-phosphate to 6-phosphogluconate and the reduction of NADP+ to NADPH, which is the rate-limiting step in the pentose phosphate pathway. This metabolic pathway serves multiple cellular functions, such as the supply of pentose intermediates for nucleotide synthesis, interconversion of 3–7 carbon sugars for various metabolic purposes, and the generation of reducing power in the form of NADPH. The latter metabolite plays an integral role in a broad range of cellular functions, such as (i) acting as a cofactor in reductive biosynthesis, (ii) detoxification of xenobiotics, and (iii) maintenance of the cellular redox state by providing reducing equivalents to antioxidative systems, among others (1).
The concept that maintenance of an optimal redox state is essential to cellular survival is now firmly established. Significant amounts of superoxide anion radical and its stoichiometric product, hydrogen peroxide, are generated in mitochondria and peroxisomes under normal physiological conditions (2). Hydrogen peroxide is detoxified by catalase and peroxidases. The activity of the peroxidases is dependent upon the availability of reduced forms of glutathione (GSH) or thioredoxin, and it results in the oxidation of GSH to GSSG and of reduced thioredoxinred (Trx-(SH)2)2 to oxidized thioredoxinox (Trx-(S)2). The reducing equivalents for the reductions of GSSG by glutathione reductase and thioredoxinox by thioredoxin reductase are supplied by NADPH (3). Thus, the regeneration of two major cellular antioxidants, GSH and thioredoxin, is dependent upon the supply of NADPH. In insects, glutathione reductase and glutathione peroxidase are absent, and their functions are assumed by the thioredoxin system, which consists of (i) thioredoxin, (ii) thioredoxin reductase (TR in Reaction 3), and (iii) thioredoxin peroxidase (TPx in reaction 2) (4). The mechanisms for the reduction of GSSG and removal of peroxides by the thioredoxin system are as follows (Reactions 1-3).
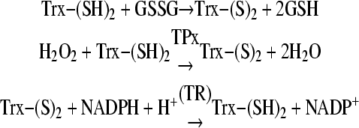 |
REACTIONS 1-3 |
There are strong indications that the level of oxidative stress is elevated during the aging process (5–8). For instance, rates of mitochondrial superoxide anion radical and hydrogen peroxide generation, as well as the steady-state amounts of the products of free radical attacks on macromolecules, such as nucleic acids, proteins, and lipids, increase in the aged organisms (5). Older flies become more prooxidizing with age, as measured by the ratio between GSH and GSSG (7). Furthermore, the old animals are relatively more susceptible to induced oxidative stress. In previous studies directed at testing the role of oxidative stress in the aging process, the most frequently used approach was to enhance the activities of antioxidant enzymes, such as superoxide dismutases, catalase, and thioredoxin reductase, in transgenic animals (9–13). In general, either there was little effect on longevity, or the life span extensions occurred in genetic backgrounds where life spans of the recipient controls were relatively short. Such effects were not replicated in long-lived backgrounds (14).
In this context, we hypothesized that transgenic overexpression of glucose-6-phosphate dehydrogenase (G6PD) in Drosophila melanogaster and the consequent increase in the ability to synthesize NADPH would enhance resistance to oxidative stress and extend the life span of flies. The underlying assumption was that compared with the elevations of activities of particular antioxidative enzymes, augmentation of reductive capacity by the efficient supply of NADPH would support a relatively broader range of mechanisms of antioxidative defenses. Results indicate that the life span of Drosophila can indeed be significantly extended by overexpression of G6PD and thereby support the concept that oxidative stress plays a causal role in the aging process.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—HPLC calibration standards were obtained from Sigma and prepared exactly as described previously (15). Milli-Q grade water was prepared by reverse osmosis on a Millipore® water purification system. All chemicals were either analytical grade or of the highest purity commercially available.
Construction of pP[UAST]-G6PD Responder Lines—A 2074-bp EcoRI-XhoI G6PD cDNA fragment was subcloned into pP[UAST]. Twelve pP[UAST]-G6PD transgenic responder lines were established by co-injecting pP[UAST]-G6PD and pTurbo DNA (16) into Drosophila, y w early stage embryos using standard P element-mediated methodology. G1 progeny of injected flies were scored for the presence of the w+ marker gene. The presence of single pP[UAST]-G6PD insertions was confirmed by chromosome mapping and Southern blot analysis for six distinct transgenic responder lines, which were then backcrossed with y w for 6 generations to obtain isogenic stocks. The driver lines used for subsequent crosses were similarly backcrossed into the same background.
G6PD Overexpression—Male flies, homozygous with respect to the pP[UAST]-G6PD transgene, were crossed with virgin females containing one of six distinct GAL4 drivers: da-GAL4, Tubulin-GAL4, armadillo-GAL4, or C23-GAL4 for broad overexpression in multiple tissues and D42-GAL4 or Appl-GAL4 for neuronal overexpression. The driver lines were provided by Dr. B. Rogina (University of Connecticut Health Science Center). Control flies were obtained from crosses in which the yw parental strain was substituted for either the driver or the pP[UAST]-G6PD responder.
G6PD enzymatic activity was quantified in flies aged 7–70 days. Flies (four) were homogenized in 100 μl of ice-cold 0.01 m KH2PO4, K2HPO4, 1 mm EDTA, pH 7.4. After centrifugation for 10 min at 12,500 rpm, 4 °C, the protein concentration was measured using the Bradford assay. Rates of NADPH production were determined by measuring the change in absorbance at A340 for 10 min (modified from Refs. 17 and 18).
Evaluation of Reporter Gene Expression—A transgenic line carrying a UAS-AUG-DsRed insertion (Bloomington stock number 6282) was crossed to six different driver lines: da-GAL4, Tubulin-GAL4, armadillo-GAL4, C23-GAL4, D42-GAL4, and Appl-GAL4. The expression of the red fluorescent protein (RFP) reporter gene was evaluated in whole-mount organ preparations or cryosections made from 10–14-day-old adult progeny using fluorescence microscopy (Nikon) and MetaMorph software.
Life Span—Life span studies were conducted as described previously (16). Flies tested in mortality experiments were reared at a constant density and collected within 24 h posteclosion. At least two replicate experiments were performed at 25 °C and 50% relative humidity for four responder lines combined with each of the six drivers, using ∼200 flies/line. Flies (25/vial) were transferred every 1–2 days into 8-dram shell vials containing a sugar-yeast-corn meal-agar medium.
Experimental Oxidative Stress—Resistance to oxidative stress was measured by exposing 10-day-old male flies (n = 100; 25 flies/vial) continuously to reactive oxygen species generators, including 100% oxygen and paraquat, and scoring for mortality once or twice daily. For measurements of resistance to hyperoxia, flies were placed in vials containing standard medium, covered with nylon to facilitate the passage of oxygen, and transferred to a plastic container, through which 100% oxygen was passed at a constant rate. Flies were exposed to paraquat in vials containing six layers of Kimwipe soaked in 1% sucrose, 5 mm paraquat; they were scored once or twice daily and transferred every 1–2 days into vials containing a freshly prepared sucrose/paraquat solution. Stress experiments were conducted twice.
Oxygen Consumption—The rate of respiration was determined using an Oxzilla Dual Absolute and Differential Oxygen Analyzer (Sable Systems International, Las Vegas, NV), as described previously (19), except (i) groups of 50 male flies were used, with 25 each in two parallel chambers, (ii) the air stream, which was pumped from outdoors and scrubbed to remove water and carbon dioxide, was rehydrated immediately prior to entry into the respiration chambers and dehydrated after exiting the chambers but before entering the respirometer, and (iii) the calculated rate of oxygen consumption for each group was the mean of four consecutive chamber minus base-line estimations, with chamber sampling for 12 min and base lining for 8 min, and estimation for at least 4 min after stabilization of the oxygen concentration in the chamber versus reference channel in each interval. For da-GAL4 and C23-GAL4 genotypes, carbon dioxide scrubbing was repeated during the dehydration of air leaving the respiration chambers.
Fertility—The lifetime reproductive output was determined using the armadillo-GAL4 driver. Reciprocal outcrosses of G6PD overexpressors and respective control flies to yw virgins were performed within 24–72 h posteclosion. Individual female flies were crossed with three yw males in vials containing standard medium supplemented with dry yeast. Fresh vials were provided daily for the first 2 weeks and every other day subsequently, and adult progeny were counted.
Analysis of Oxidized and Reduced Pyridine Dinucleotides—Amounts of NAD+, NADH, NADP+, and NADPH were measured in whole fruit fly homogenates by HPLC with fluorescent detection, by a modification of the method described previously (15). Briefly, to prevent degradation and oxidation of pyridine dinucleotides, flies were frozen in liquid nitrogen and stored at -80 °C until analysis. Ten frozen flies were homogenized in 200 μl of freshly prepared ice-cold solution, containing 0.2 m KCN, 0.06 m KOH, and 1 mm bathophenanthrolinedisulfonic acid, using 1.5-ml plastic tubes and pestles obtained from RPI (Mt. Prospect, IL). The homogenates were incubated for 10 min on ice and centrifuged at 18,000 × g for 10 min at 4 °C. Supernatants were then extracted twice with 0.8 ml of ice-cold chloroform, and the samples were filtered through a 0.45-μm PTFE Acrodisk CR 4-mm syringe filters obtained from Gelman Laboratory (Ann Arbor, MI). Filtrates were transferred to sampling vials and either analyzed immediately or stored at -80 °C for up to 1 week. NADH, NADPH, and cyanide adducts with NAD+ and NADP+ (CN1 and CN2 derivatives) were separated by HPLC, fitted with a Shimadzu Call VP solvent delivery system, using a reverse phase C18 Luna (II) column (4.6 × 150 mm, 3-μm particle size) obtained from Phenomenex (Torrance, CA). A guard cartridge, C18 Luna (II) (3 × 4 mm), was used for column protection and replaced after 100–200 injections. The mobile phases for gradient elution were 0.2 m ammonium acetate, pH 5.9, containing 3.5% (v/v) methanol (solvent A) and 0.2 m ammonium acetate, pH 5.9, containing 6.0% (v/v) methanol (solvent B). A linear gradient from 100% solvent A to 100% solvent B over a 30-min separation time was used with a flow rate of 0.6 ml/min. Pyridine dinucleotides were detected with a RF10-AXL fluorescent detector from Shimadzu (excitation wavelength 330 nm and emission wavelength 460 nm). Calibration standards of pyridine dinucleotides (1 mg/ml stock solutions stored at -80 °C) were prepared by appropriate dilutions of stock solutions in 0.2 m KCN, 0.06 m KOH, and 1 mm bathophenanthrolinedisulfonic acid. Linear relationships between the peak area and pyridine nucleotide amounts were established between 1 and 1000 ng for each standard. Each sample was injected twice, and the average of the peak areas was used for quantification.
HPLC Analysis of GSH, GSSG, and Methionine—25–50 flies were immobilized on ice for 1–2 min, weighed, and homogenized in 10 volumes of freshly prepared ice-cold 5% (w/v) meta-phosphoric acid (MPA), using 1.5-ml plastic tubes and pestles obtained from RPI (Mt. Prospect, IL). The homogenates were incubated for 30 min on ice and centrifuged at 18,000 × g for 20 min at 4 °C. Supernatants were filtered using 0.45-μm PTFE Acrodisc® CR 4-mm syringe filters, obtained from Gelman Laboratory (Ann Arbor, MI); filtrates were transferred to sampling vials and either analyzed immediately or stored at -80 °C for up to 1 month.
The procedure for detection and quantification of aminothiols (GSH, GSSG, and methionine) used here and the precautionary measures taken to minimize spontaneous GSH oxidation have been described by us previously (7, 8), with the following modifications. Briefly, aminothiols were separated by HPLC, fitted with a Shimadzu Class VP solvent delivery system, using a reverse phase C18 Luna (II) column (3μ; 4.6 × 150 mm), obtained from Phenomenex (Torrance, CA). The mobile phase for isocratic elution consisted of 25 mm monobasic sodium phosphate, a 0.120 mm concentration of the ion-pairing agent 1-octane sulfonic acid, 1% (v/v) acetonitrile, pH 2.75, adjusted with 85% phosphoric acid. The flow rate was 0.7 ml/min. Under these conditions, the separation was completed in 30 min; GSSG was the last eluting peak, with a retention time of ∼22 min. Calibration standards were prepared in 5% (w/v) MPA. Aminothiols were detected with a model 5600 CoulArray® electrochemical detector (ESA Inc., Chelmsford, MA), equipped with an eight-channel analytical cell, using potentials from +500 mV in 100-mV increments. GSH was monitored at +800 mV, whereas GSSG and methionine were monitored at +900 mV. Each sample was injected twice, and the average of the peak areas was used for quantification.
Statistical Analysis—All analyses were performed using SYSTAT 10 or Microsoft EXCEL software. Differences in G6PD enzymatic activities, rates of oxygen consumption, and NADP+ and NADPH content were compared between groups by analysis of variance, with post hoc pairwise comparisons where appropriate. In studies of life span and resistance to oxidative stress, differences between survivorship curves were assessed using semiparametric log rank tests. Analysis of variance was used to assess differences in mean life spans, using vial mean life spans as input data, in order to minimize the effects of pseudoreplication, as discussed previously (20, 21).
RESULTS
Overexpression of G6PD—A total of six transgenic lines (responders) containing single pP[UAST]-G6PD insertions were subjected to preliminary studies of survivorship, prior to backcrossing. In the presence of the Tubulin-GAL4 driver, each responder line exhibited extension of life span; however, one of the responder lines was recessive lethal, raising the likelihood of an adverse, insertional position effect, and two of the remaining lines were derived from a single transformant. Consequently, four completely independent responder lines were selected for biochemical and physiological characterization.
Enhancement of G6PD enzymatic activity was observed in all of the transgenic lines containing GAL4 driver. As expected, the magnitude of G6PD overexpression varied considerably among the drivers. In whole body homogenates, activity was increased 27–65-fold (Tubulin-GAL4), 33–97-fold (da-GAL4), 1.9–3.8-fold (arm-GAL4), 11–20-fold (C23-GAL4), 9–20-fold (D42-GAL4), and 1.8–4-fold (Appl-GAL4) (Table 1). In isolated heads and bodies (thoraces plus abdomens), the activity was increased 14–55-fold and 0.75–7.5-fold, respectively, in the presence of the Appl-GAL4 driver (results not shown).
TABLE 1.
G6PD enzyme activity/mg of protein
| Genotype | Rate | S.D. | S.E. |
|---|---|---|---|
| μmol/min | |||
| +/G6PD4c | 0.16 | 0.06 | 0.04 |
| +/G6PD5f | 0.18 | 0.05 | 0.03 |
| +/G6PD7b | 0.15 | 0.04 | 0.02 |
| +/G6PD9g | 0.11 | 0.02 | 0.01 |
| arm/+ | 0.12 | 0.02 | 0.02 |
| arm/4c | 0.30 | 0.03 | 0.02 |
| arm/5f | 0.45 | 0.06 | 0.04 |
| arm/7b | 0.32 | 0.08 | 0.06 |
| arm/9g | 0.44 | 0.06 | 0.04 |
| Tubulin/+ | 0.16 | 0.05 | 0.03 |
| Tubulin/4c | 6.71 | 0.26 | 0.02 |
| Tubulin/5f | 8.43 | 0.45 | 0.26 |
| Tubulin/7b | 4.45 | 1.02 | 0.59 |
| Tubulin/9g | 7.37 | 0.49 | 0.28 |
| da/+ | 0.15 | 0.06 | 0.04 |
| da/4c | 8.58 | 2.96 | 1.71 |
| da/5f | 6.13 | 1.33 | 0.76 |
| da/7b | 5.55 | 1.06 | 0.61 |
| da/9g | 10.97 | 2.79 | 1.61 |
| Appl/+ | 0.20 | 0.02 | 0.02 |
| Appl/4c | 0.38 | 0.04 | 0.02 |
| Appl/5f | 0.72 | 0.12 | 0.08 |
| Appl/7b | 0.38 | 0.04 | 0.02 |
| Appl/9g | 0.35 | 0.01 | 0.01 |
| D42/+ | 0.13 | 0.07 | 0.04 |
| D42/4c | 2.10 | 0 | 0 |
| D42/5f | 1.93 | 0.46 | 0.26 |
| D42/7b | 1.40 | 0.33 | 0.19 |
| D42/9g | 2.25 | 0.92 | 0.65 |
| 2703/+ | 0.14 | 0.03 | 0.02 |
| C23/4c | 2.22 | 0.68 | 0.48 |
| C23/5f | 2.26 | 0.47 | 0.34 |
| C23/7b | 1.65 | 0.35 | 0.25 |
| C23/9g | 2.28 | 0.88 | 0.63 |
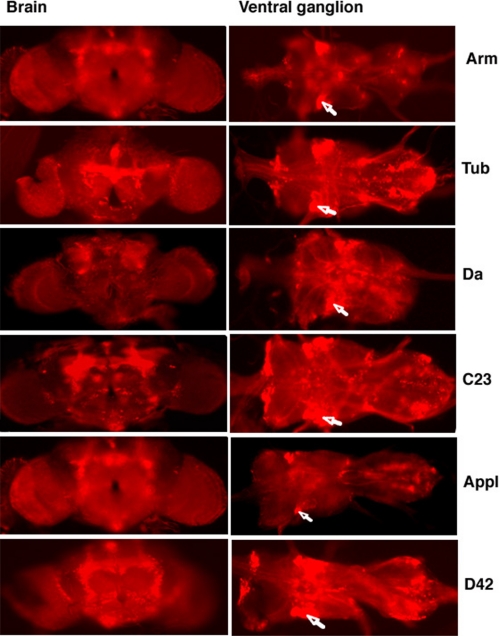
Evaluation of Driver Tissue Specificity—RFP was used as a reporter to evaluate patterns of tissue-specific expression elicited by the GAL4 driver lines used in the present study. A transgenic line containing the UAS-AUG-DsRed insertion was crossed to driver lines, and red fluorescence was evaluated in whole mount organ preparations and/or cryosections made from adult flies. The UAS-AUG-DsRed transgenic line crossed to the reference y w strain was used as a control. The data are summarized in Table 2. Three of the drivers have been reported to exhibit a relatively broad expression pattern in adults (armadillo, tubulin, and da), and this was confirmed, although expression was by no means uniform. For instance, as observed in Fig. 1, there were distinct brain and ventral ganglion expression patterns for each of the drivers. Expression driven by the C23 driver was also relatively broad in adult flies, whereas previously it had only been characterized in embryonic tissue, where it was localized to transverse muscle. Regarding the neuronal D42 and Appl drivers, our results are consistent with earlier studies (11, 22), which reported expression in both the brain and the ventral ganglion. As expected, RFP expression driven by D42 was quite prominent in motor neurons but was also observed in adult cardia and salivary gland. Distinct neuronal expression patterns were noted for all six drivers with overlap in the motor neurons and certain areas of the midbrain. Overall, the global drivers selected for this study exhibited a more patterned than uniform expression of the reporter gene, whereas candidate tissue-specific drivers permitted broader expression than expected.
TABLE 2.
Tissue-specific expression of RFP in adult flies carrying a reporter gene construct in combination with different GAL4 driver lines
|
Tissue
|
Driver
|
|||||
|---|---|---|---|---|---|---|
| Arm | Tub | Da | C23 | Appl | D42 | |
| Brain | ++ | ++ | + | + | +++ | +++ |
| Ventral gangliona | + | + | + | + | + | + |
| Cardia | ± | ++ | ++ | ++ | + | |
| Gut | + | + | ||||
| Malpighian tubules | + | |||||
| Salivary gland | ± | + | +++ | +++ | +++ | |
| Muscles | + | +++ | ++ | + | ||
| Fat bodyb | ± | ± | ± | ± | ± | ± |
| Oenocytes | + | + | ||||
A pronounced expression of RFP was observed in motor neurons
A slight nonspecific fluorescence was observed in fat bodies isolated from both control flies and flies expressing RFP
FIGURE 1.
Fluorescence microscopy analysis of the tissues of flies expressing Red Fluorescent Protein. Whole mount preparations of the brain (left) and ventral ganglion (right) were dissected from flies carrying a UAS-RFP transgene and the different GAL4 enhancers indicated on the right. Motor neurons are indicated by arrows. No fluorescence patterns were observed in controls carrying transgene or driver only (not shown).
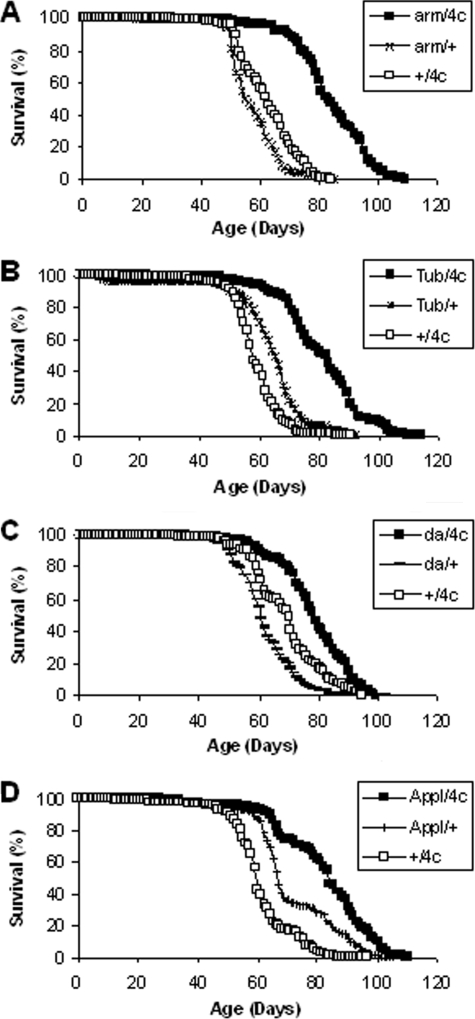
Overexpression of G6PD Extends Life Span—Life span experiments were performed using all four responders in combination with all of the six drivers, typically utilizing 200–225 flies in each of two independent experiments for a total of more than 20,000 flies. The mean life span was greater in each experiment for every G6PD overexpressor genotype than for the corresponding driver/+ and +/responder controls (Fig. 2 and Table S1). These results represent a total of 102 comparisons, among which 78 individual comparisons revealed statistically significant differences (p < 0.05). The magnitude of the increase, in comparison with the stronger driver control in each case, was up to 38% for arm/G6PD, 29% for Tubulin/G6PD, 27% for C23/G6PD, 24% for da/G6PD, 18% for D42/G6PD, and 16% for Appl/G6PD. The maximum life span (time to 90% mortality) was also extended (Table S1). Moreover, the longevity effects did not appear to be sex-specific. We conducted survivorship studies on females using two broad expression drivers (arm and Tubulin) and observed life span extensions that were comparable with those observed in the males (data not shown). Collectively, the results lend strong support to the hypothesis that increasing G6PD activity, either broadly or in neuronal tissue, is associated with extension of life span in Drosophila.
FIGURE 2.
Life spans of flies overexpressing G6PD. Overxpression of G6PD in arm/G6PD (A), Tubulin/G6PD (B), da/G6PD (C), and Appl/G6PD male flies (D) resulted in extension of mean and maximum life spans. Results of multiple replicate experiments are provided in Table 2. n = 189–220 flies for each panel. p < 0.0005 for comparisons of mean life spans.
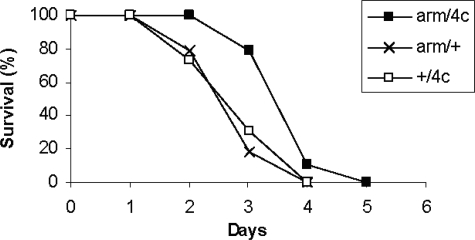
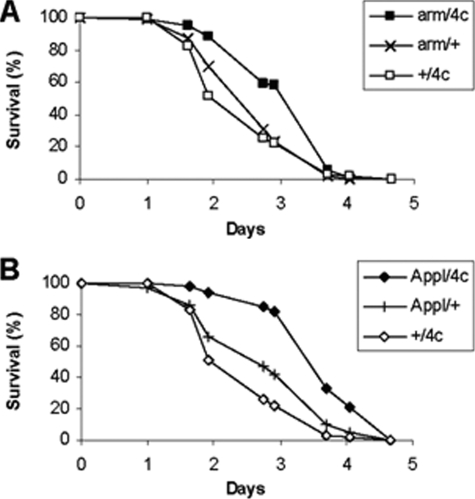
Resistance to Experimental Oxidative Stress—In order to determine the effects of G6PD overexpression on resistance to oxidative stress, flies were exposed continuously to 100% oxygen. The survival times of three arm/G6PD genotypes were increased by 19–38%, p < 0.0005 (Fig. 3). Resistance to paraquat was also enhanced by 25–47% in two Appl/G6PD genotypes (p < 0.0005) and by 9–26% in two arm/G6PD genotypes (p ≤ 0.005 for G6PD 4c and p = 0.12 for G6PD 5f) (Fig. 4).
FIGURE 3.
Resistance to hyperoxia. Male flies overexpressing G6PD driven by arm-GAL4 were exposed continuously to 100% oxygen. n = 98–100 flies per group. p < 0.0005 for comparisons of mean survival times.
FIGURE 4.
Resistance to paraquat. Male flies overexpressing G6PD driven by arm-GAL4 (A) and Appl-GAL4 (B) were exposed to 5 mm paraquat. n = 94–98 flies/group. p < 0.01 for comparisons of mean survival times.
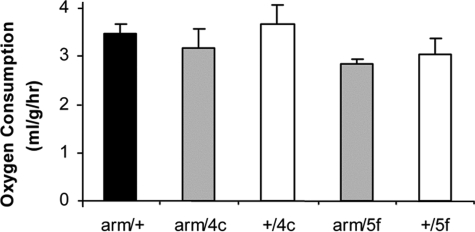
Oxygen Consumption—The rate of oxygen consumption was measured in experimental flies overexpressing G6PD driven either by Tubulin-GAL4, da-GAL4, C23-GAL4, or arm/GAL4. The Tubulin/G6PD genotypes all exhibited significantly lower rates of respiration than the corresponding +/G6PD controls (p < 0.01), by amounts ranging from 20 to 29%, but there was no significant difference in comparison with the Tubulin/+ control. Respiration rates exhibited only nonsignificant decreases in da/G6PD and C23/G6PD genotypes (results not shown). The respiration rate was slightly lower in arm/G6PD experimental versus control flies (Fig. 5), but the effect was not statistically significant with respect to both controls. These results suggest that the life span extensions displayed by flies overexpressing G6PD cannot be ascribed to reductions in metabolic rate.
FIGURE 5.
Oxygen consumption. Results for each genotype are mean ± S.D. for five groups of 50 male flies, aged 14–19 days. Black bar, arm/+ control. White bars, +/G6PD controls. Gray bars, arm/G6PD overexpressors. p = 0.003 for arm/G6PD ANOVA. p = 0.004 for arm/G6PD 5f versus arm/+, and p = 0.018 for arm/4c versus +/G6PD 4c. p > 0.05 for other arm/G6PD versus control comparisons.
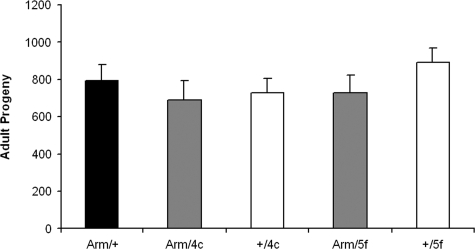
Fertility—The fertility of G6PD overexpressor and control flies was determined in reciprocal crosses with the yw parental strain by counting the number of adult progeny. There were no consistent differences in female fertility between experimental and control flies, based on two different transgenic lines (Fig. 6); nor were any differences observed in male fertility (data not shown).
FIGURE 6.
Female fertility. Lifetime reproductive outputs, measured as numbers of adult progeny, are indicated for arm/G6PD overexpressors (gray bars), the UAS-G6PD responder controls (white bars), and the armadillo-GAL4 driver control (black bar). Results are mean ± S.E. for 20 crosses involving one female fly of the stated genotype with three yw males. The outputs between experimentals and controls were not statistically distinguishable. A replicate experiment yielded similar results.
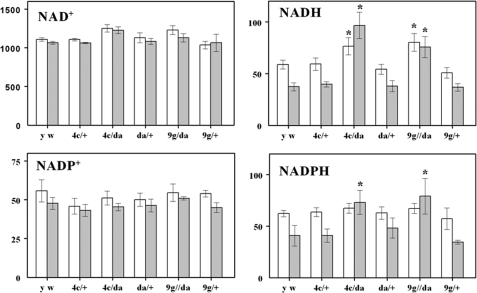
Pyridine Dinucleotide Content—Comparisons of amounts of NAD+, NADH, NADP+, and NADPH were made between da/G6PD flies and respective controls at 10 and 36 days of age. The da driver was selected for this experiment because the da/G6PD flies exhibited the highest G6PD enzyme activity among the G6PD overexpressors (Table 1). There were no significant differences in the amounts of NAD+ and NADP+ between the experimental and control groups at either age (Fig. 7). In contrast, significant increases in amounts of both NADPH and NADH were found in the older (36-day) experimental flies (da/G6PD 4c and da/G6PD 9g) versus controls. In the 10-day-old flies, small increases were also observed, but these only reached statistical significance in the case of NADH levels (Fig. 7).
FIGURE 7.
Pyridine dinucleotide content of whole body homogenates of D. melanogaster overexpressing G6PD at 10 (white bars) and 36 (gray bars) days of age. Results are mean ± S.D. of three independent preparations of 25 flies at each age and are expressed as pmol/mg of body weight. Significant differences between experimental and age-matched control flies are indicated with an asterisk (p < 0.001).
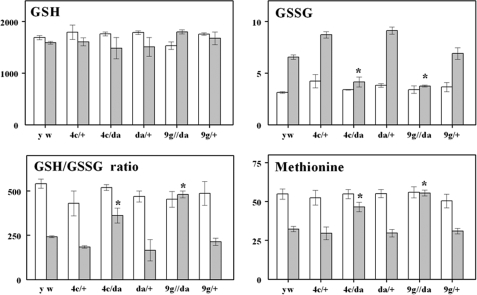
Glutathione and Methionine Content—Comparisons of amounts of GSH, GSSG, and methionine were made between da/G6PD flies and respective controls at 10 and 36 days. An increase in GSH/GSSG ratio was found in da/G6PD flies at 36 days compared with the control groups (Fig. 8). This increase was due to a significantly lower GSSG content in the da/G6PD overexpressors, whereas GSH was not significantly affected. Similarly, there were higher amounts of methionine in experimental as compared with control groups at 36 days of age (Fig. 8).
FIGURE 8.
GSH/GSSG ratio and GSH, GSSG, and methionine content of whole body homogenates of D. melanogaster overexpressing G6PD at 10 (white bars) and 36 (gray bars) days of age. Results are mean ± S.D. of three independent preparations of 25 flies at each age and are expressed as pmol/mg of body weight. Significant differences between experimental and age-matched control flies are indicated with an asterisk (p < 0.001).
DISCUSSION
The main finding of this study was that enhancement of the activity of glucose-6-phosphate dehydrogenase, either broadly or in neuronal tissues, is associated with an extension of life span in transgenic D. melanogaster, the basis for which may lie in the observed modulation of cellular reductive capacity. Additionally, the increased activity of this enzyme was associated with enhanced resistance to experimental oxidative stress and could not be explained by a decrease in the metabolic rate or fertility.
Neuronal tissue overexpression alone is sufficient to support a moderate increase in life span. Given that all six drivers confer overexpression in the motor neurons, based on RFP analysis, and in view of a previous report that D42-driven overexpression of SOD1 extends life span, it is tempting to speculate that the motor neurons are weak links wherein antioxidant overexpression has particularly beneficial effects on longevity. Notwithstanding, our analysis of driver-dependent RFP expression revealed other potential common regions in the mid-brain (Fig. 1), which could also represent candidate weak links, a possibility that is presently under investigation. Notably, the strongest longevity effects were associated with transgenic lines where G6PD is driven by armadillo and the other broad expression drivers, suggesting that one or more nonneuronal domains may also be weak links. Initial indications would point to muscle tissue as a candidate based on our RFP analysis (Table 2), which shows that muscle expression is associated with the four broad expression drivers but not the neuronal drivers. The use of drivers with greater specificity should help to test this notion.
G6PD catalyzes the committed step in the oxidative phase of the pentose phosphate pathway, which converts NADP+ to NADPH and diverts metabolites from energy production to biosynthesis. In this study, high levels of G6PD enzymatic activity were detected in an in vitro assay, but there were only modest increases in the levels of NADPH. These results are consistent with the tight regulation of both G6PD activity and NADPH levels known to occur in vivo. G6PD activity is inhibited by NADPH and is stimulated as the physiological demands for NADPH increase (23–25). Post-translational regulation of the enzyme and feedback inhibition of NADPH levels modulate the redox balance in vivo (24, 25). In this context, the observed modest increases in NADPH, especially notable in older flies, may represent the enhanced capacity of the transgenic overexpressors to resist normal, age-related oxidative stress as well as the increased oxidative stress associated with exposure to 100% oxygen or paraquat.
NADPH is an indirectly acting antioxidant, which participates as an electron donor in the regeneration of reduced glutathione and thioredoxin (26). It was therefore notable that the increase in NADPH was accompanied by an increase in the GSH/GSSG redox ratio in the G6PD overexpressors, primarily due to a decrease in the amount of GSSG. It may be parenthetically added that the latter constitutes less than 1% of the total glutathione content. Therefore, the observed decrease in GSSG amount in overexpressors would not be expected to have a readily discernible effect on glutathione content. Since flies do not have the glutathione reductase enzyme, which normally converts GSSG to GSH, the thioredoxin system serves in its stead. As depicted above, reduction of GSSG is thought to be mediated directly by reduced thioredoxin (Reaction 1), which itself can be generated from its oxidized form using NADPH (Reaction 3).
The finding that an enhanced capacity for reduction of NADPH is associated with extension of life span provides support for the oxidative stress hypothesis of aging. Such support reinforces the conclusions of a recent study demonstrating extension of life span in Drosophila by overexpression of glutamate-cysteine ligase, which catalyzes the rate-limiting step in the biosynthesis of glutathione (19). In each case, overexpression of enzymes with broad effects on the cellular redox buffer system exhibited beneficial effects. NADPH, in particular, is both a direct and indirect determinant of the redox state, via the NADPH/NADP+ couple and the GSH/GSSG couple, respectively (27). Furthermore, NADPH may also affect the levels of NADH via the activity of proton-motive force dependent transdehydrogenase (Reaction 4). The increase in NADPH in the G6PD overexpressors could dampen the forward reaction, providing a sparing effect that results in the increased NADH levels observed in this study.
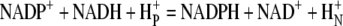 |
REACTION 4 |
Thus, redox couple fluctuations, particularly those that might be occurring under prooxidizing conditions that accompany aging, might be deleterious at the level of antioxidant defenses and/or redox-sensitive signal transduction. Bolstering the capacity to maintain critical redox couple ratios may underlie the beneficial effects on survival observed in animals overexpressing either G6PD or glutamate-cysteine ligase.
The results of both glutamate-cysteine ligase and G6PD overexpression are notable not only because the life spans were extended but also because these extensions took place in comparison with long-lived reference strains and were not accompanied by a marked depression of metabolic rate. A critical issue in the interpretation of life span studies involving transgenic Drosophila is the strength of the recipient strain. If the controls are weak, then antioxidant overexpressors could live relatively longer due to a remedial or compensatory effect. Alternatively, treatments that decrease energy utilization can extend the life span without exerting a direct effect on the underlying rate of aging (5). For flies overexpressing G6PD, mean life spans were frequently greater than 80 days and occasionally greater than 90 days, but there was not a simultaneous, statistically significant change in the rate of metabolism in comparison with both driver and responder controls. In contrast, overexpression of the antioxidative enzyme, superoxide dismutase, generally extended life spans in short- but not long-lived genetic backgrounds (28).
Collectively, the results of this study suggest that an augmentation of G6PD activity and supply of NADPH in Drosophila improves the in vivo response to age-related oxidative stress. The fact that the life span of the flies is extended, both in the presence and absence of experimental stress and in a long-lived genetic background, provides powerful evidence supporting the oxidative stress hypothesis of aging.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health, NIA, Grant RO1 AG15122. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: Trx-(SH)2, reduced thioredoxinred; Trx-(S)2, oxidized thioredoxinox; G6PD, glucose-6-phosphate dehydrogenase; HPLC, high pressure liquid chromatography; RFP, red fluorescent protein; MPA, meta-phosphoric acid.
References
- 1.Kletzien, R. F., Harris, P. K., and Foellmi, L. A. (1994) FASEB J. 8 174-181 [DOI] [PubMed] [Google Scholar]
- 2.Fridovich, I. (1978) Science 201 875-880 [DOI] [PubMed] [Google Scholar]
- 3.Wolin, M. S., Ahmad, M., and Gupte, S. A. (2005) Am. J. Physiol. 289 L159-L173 [DOI] [PubMed] [Google Scholar]
- 4.Kanzok, S. M., Fechner, A., Bauer, H., Ulschmid, J. K., Muller, H.-M., Botella-Munoz, J., Schneuwly, S., Schirmer, R. H., and Becker, K. (2001) Science 291 643-646 [DOI] [PubMed] [Google Scholar]
- 5.Sohal, R. S., Mockett, R. J., and Orr, W. C. (2002) Free Radic. Biol. Med. 33 575-586 [DOI] [PubMed] [Google Scholar]
- 6.Stadtman, E. R. (1992) Science 257 1220-1224 [DOI] [PubMed] [Google Scholar]
- 7.Rebrin, I., Bayne, A.-C. V., Mockett, R. J., Orr, W. C., and Sohal, R. S. (2004) Biochem. J. 382 131-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebrin, I., Kamzalov, S., and Sohal, R. S. (2003) Free Radic. Biol. Med. 35 626-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seto, N. O. L., Hayashi, S., and Tener, G. M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 4270-4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orr, W. C., and Sohal, R. S. (1994) Science 263 1128-1130 [DOI] [PubMed] [Google Scholar]
- 11.Parkes, T. L., Elia, A. J., Dickinson, D., Hilliker, A. J., Phillips, J. P., and Boulianne, G. L. (1998) Nat. Genet. 19 171-174 [DOI] [PubMed] [Google Scholar]
- 12.Mockett, R. J., Orr, W. C., Rahmandar, J. F., Benes, J. J., Radyuk, S. N., Klichko, V. I., and Sohal, R. S. (1999) Arch. Biochem. Biophys. 371 260-269 [DOI] [PubMed] [Google Scholar]
- 13.Sun, J., and Tower, J. (1999) Mol. Cell. Biol. 19 216-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orr, W. C., Mockett, R. J., Benes, J. J., and Sohal, R. S. (2003) J. Biol. Chem. 278 26418-26422 [DOI] [PubMed] [Google Scholar]
- 15.Klaidman, L. K., Leung, A. C., and Adams, J. D., Jr. (1995) Anal. Biochem. 228 312-317 [DOI] [PubMed] [Google Scholar]
- 16.Mockett, R. J., Orr, W. C., and Sohal, R. S. (2002) Methods Enzymol. 349 213-220 [DOI] [PubMed] [Google Scholar]
- 17.Clark, A. G., and Keith, L. E. (1988) Genetics 119 565-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beutler, E. (1971) Red Cell Metabolism: A Manual of Biochemical Methods, pp. 62-66, Grune and Stratton, New York
- 19.Orr, W. C., Radyuk, S. N., Prabhudesai, L., Toroser, D., Benes, J. J., Luchak, J. M., Mockett, R. J., Rebrin, I., Hubbard, J. G., and Sohal, R. S. (2005) J. Biol. Chem. 280 37331-37338 [DOI] [PubMed] [Google Scholar]
- 20.Tatar, M. (1999) Am. Nat. 154 S67-S81 [DOI] [PubMed] [Google Scholar]
- 21.Bayne, A.-C. V., Mockett, R. J., Orr, W. C., and Sohal, R. S. (2005) Biochem. J. 391 277-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapahi, P., Zid, B. M., Harper, T., Koslover, D., Sapin, V., and Benzer, S. (2004) Curr. Biol. 14 885-890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luzzatto, L. (1967) Biochim. Biophys. Acta 146 18-25 [DOI] [PubMed] [Google Scholar]
- 24.Stanton, R. C., Seifter, J. L., Boxer, D. C., Zimmerman, E., and Cantley, L. C. (1991) J. Biol. Chem. 266 12442-12448 [PubMed] [Google Scholar]
- 25.Corcoran, C. M., Fraser, P., Martini, G., Luzzatto, L., and Mason, P. J. (1996) Gene (Amst.) 173 241-246 [DOI] [PubMed] [Google Scholar]
- 26.Kirsch, M., and De Groot, H. (2001) FASEB J. 15 1569-1574 [DOI] [PubMed] [Google Scholar]
- 27.Schafer, F. Q., and Buettner, G. R. (2001) Free Radic Biol. Med. 30 1191-1212 [DOI] [PubMed] [Google Scholar]
- 28.Orr, W. C., and Sohal, R. S. (2003) Exp. Gerontol. 38 227-230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.